An Assessment of InP/ZnS as Potential Anti-Cancer Therapy: Quantum Dot Treatment Increases Apoptosis in HeLa Cells
Abstract
:1. Introduction
2. Experimental Section
2.1. InP/ZnS Quantum Dots
2.2. Cell Culture
2.3. XTT Viability Assay
2.4. Calculation of IC50 Value
2.5. Reactive Oxygen Species Assay
2.6. Apoptosis Assay
2.7. Total RNA Extraction and cDNA Conversion
2.8. Transcriptome Analysis
2.9. RT-qPCR
2.10. Chemophysical Properties
2.11. Statistical Analysis
3. Results
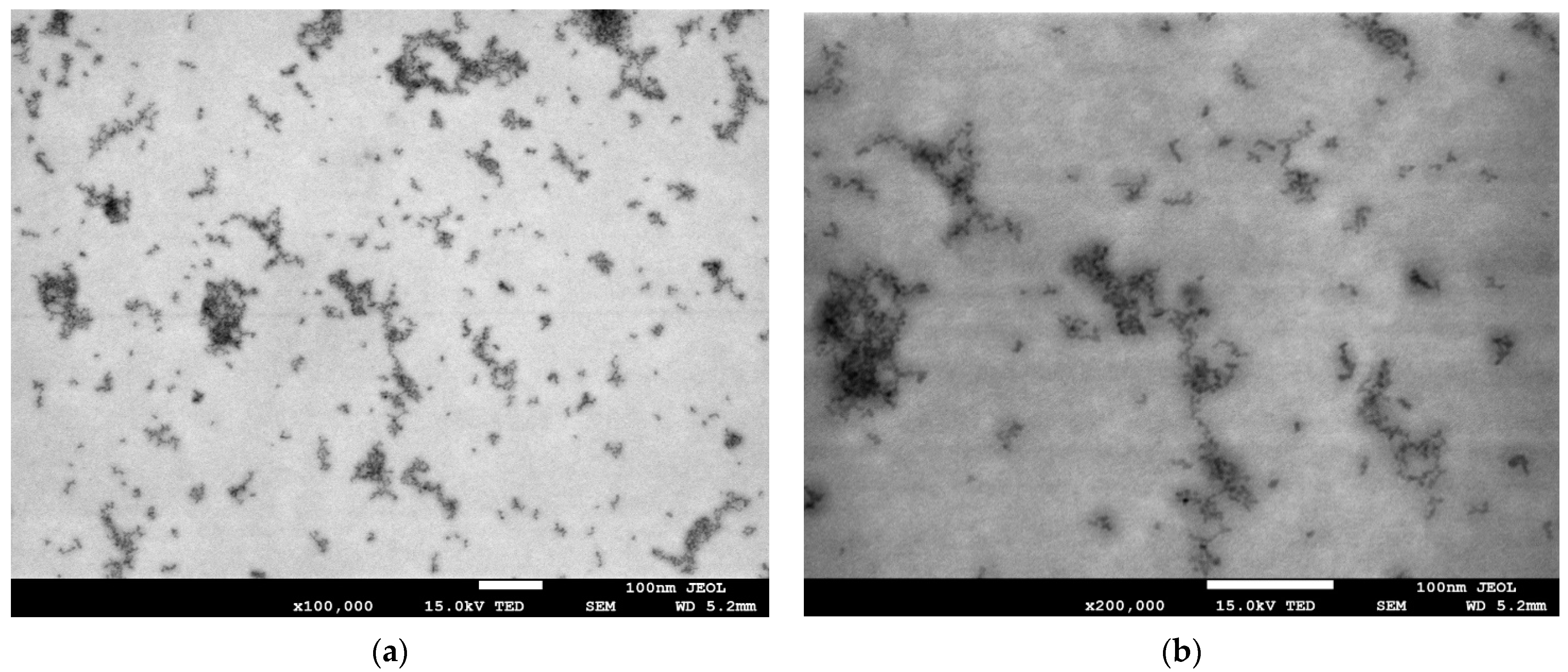
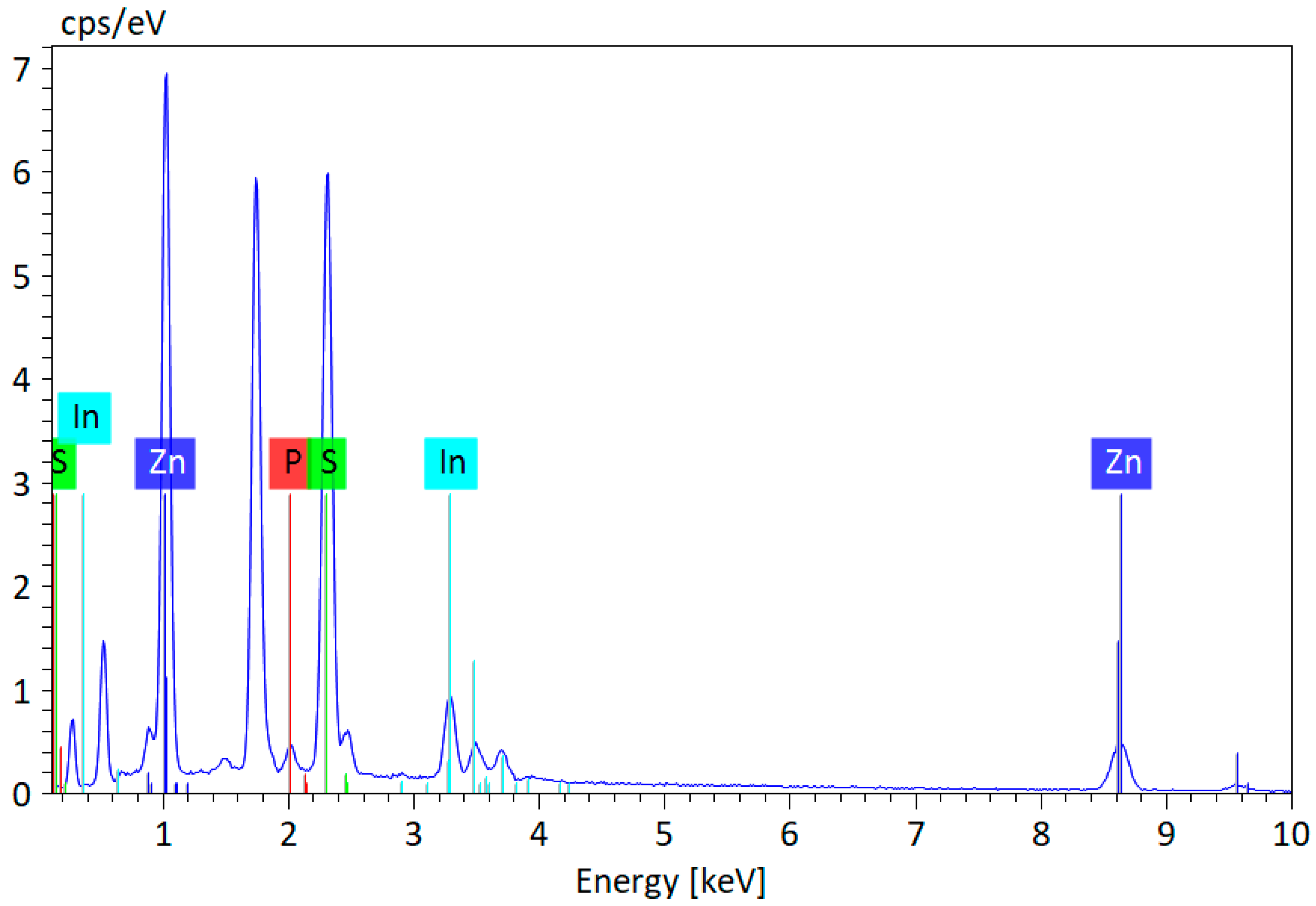
3.1. Chemophysical Properties of Green InP/ZnS
3.2. Effect on Cell Viability when Treated with InP/ZnS QDs
3.3. Increased Production of Peroxynitrite Radicals in InP/ZnS Presence
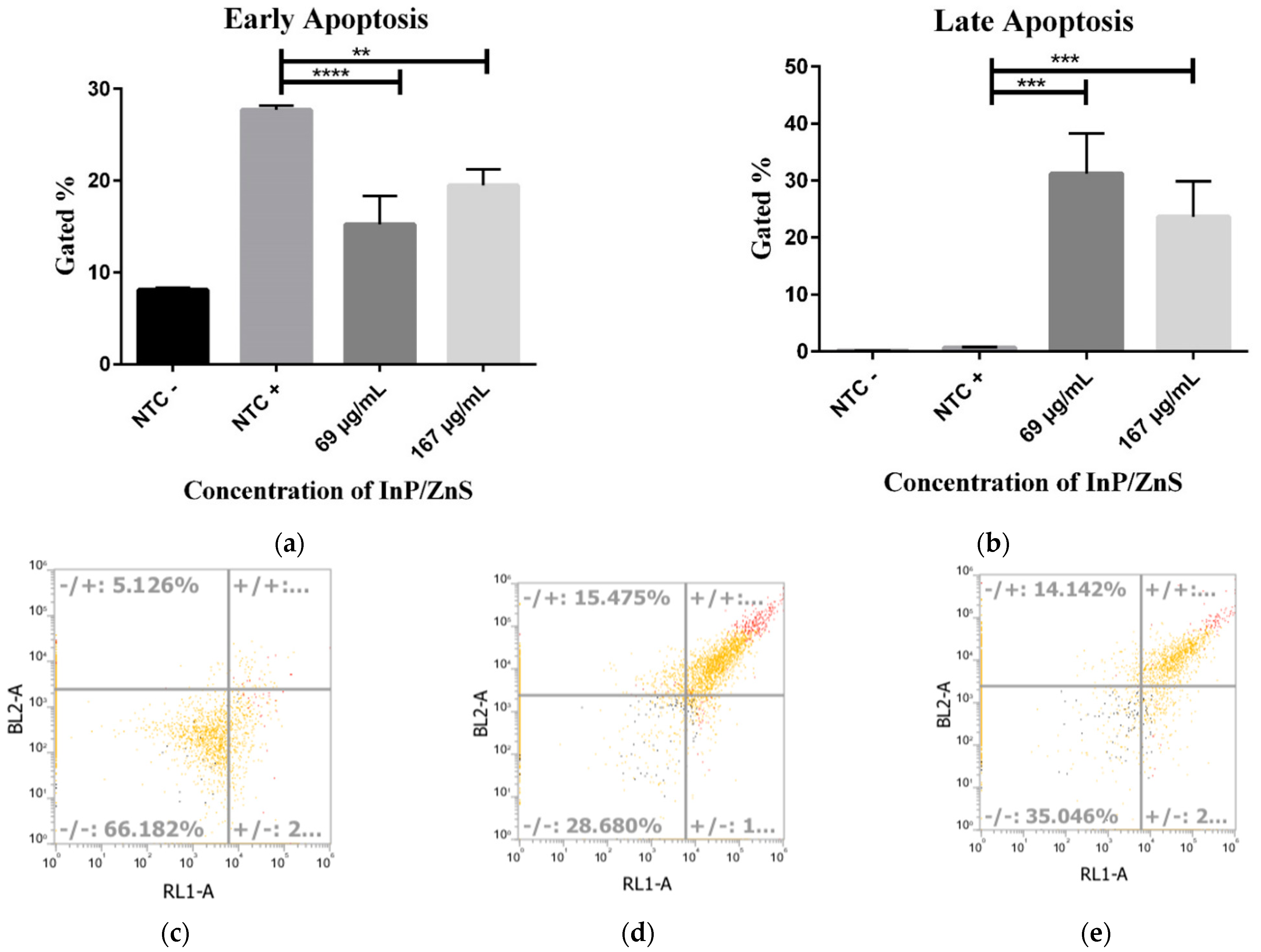
3.4. Increased Late Apoptosis with InP/ZnS Treatment
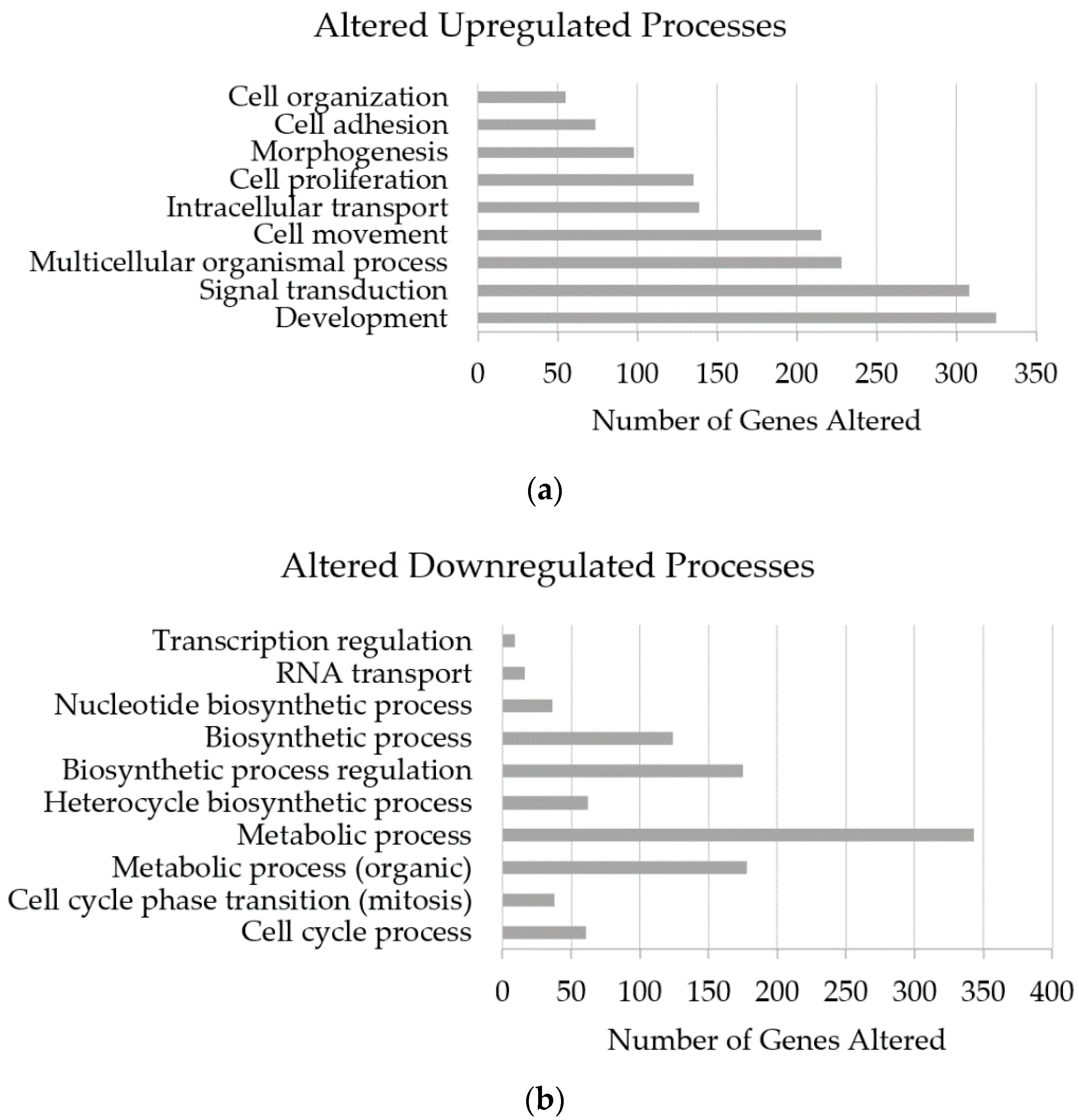
3.5. Altered Genome with InP/ZnS Treatment
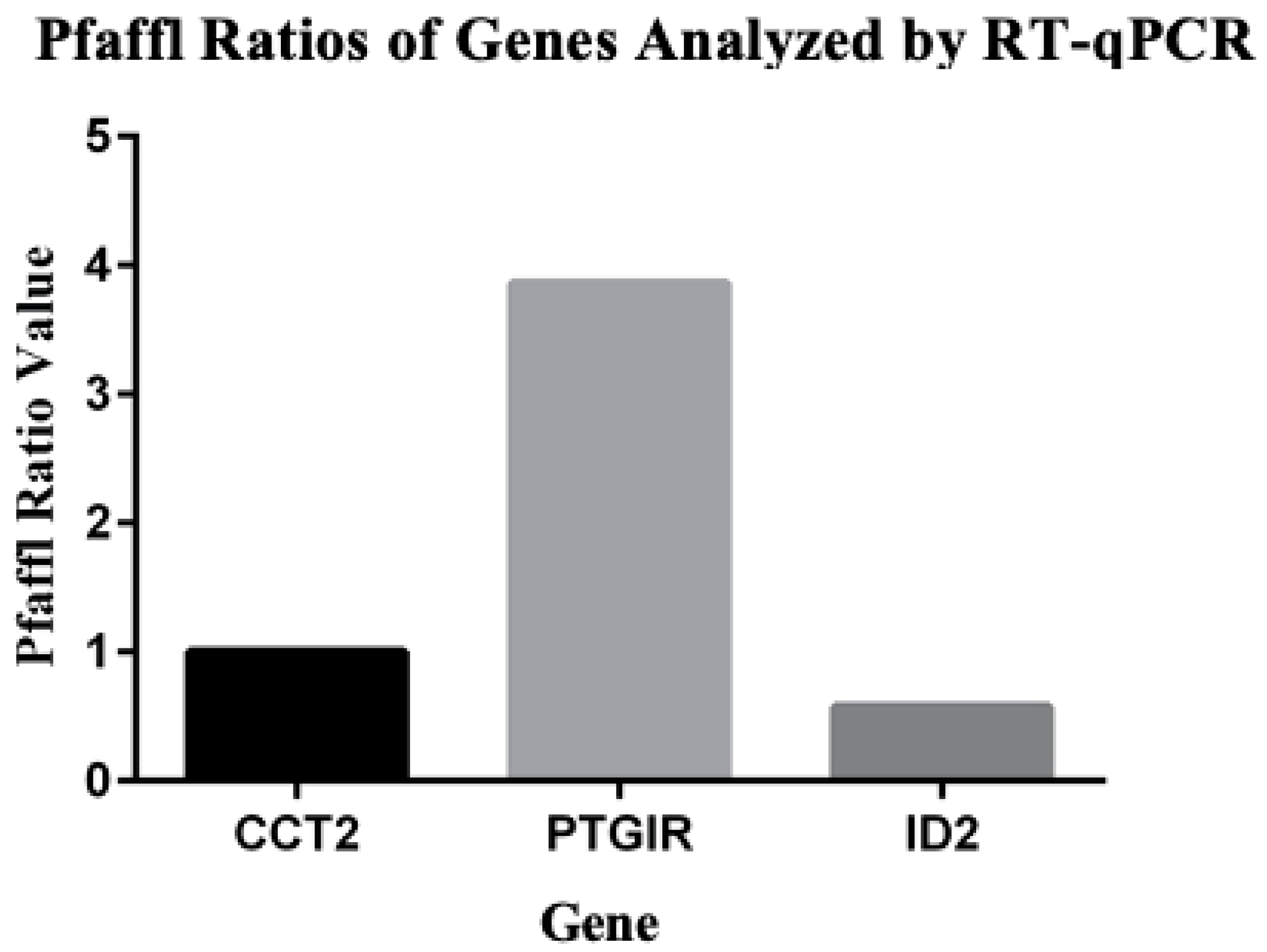
3.6. RNAseq Confirmation with RT-qPCR
4. Discussion
4.1. Changes to HeLa Cell Viability, ROS, and Apoptosis Levels with InP/ZnS Treatment
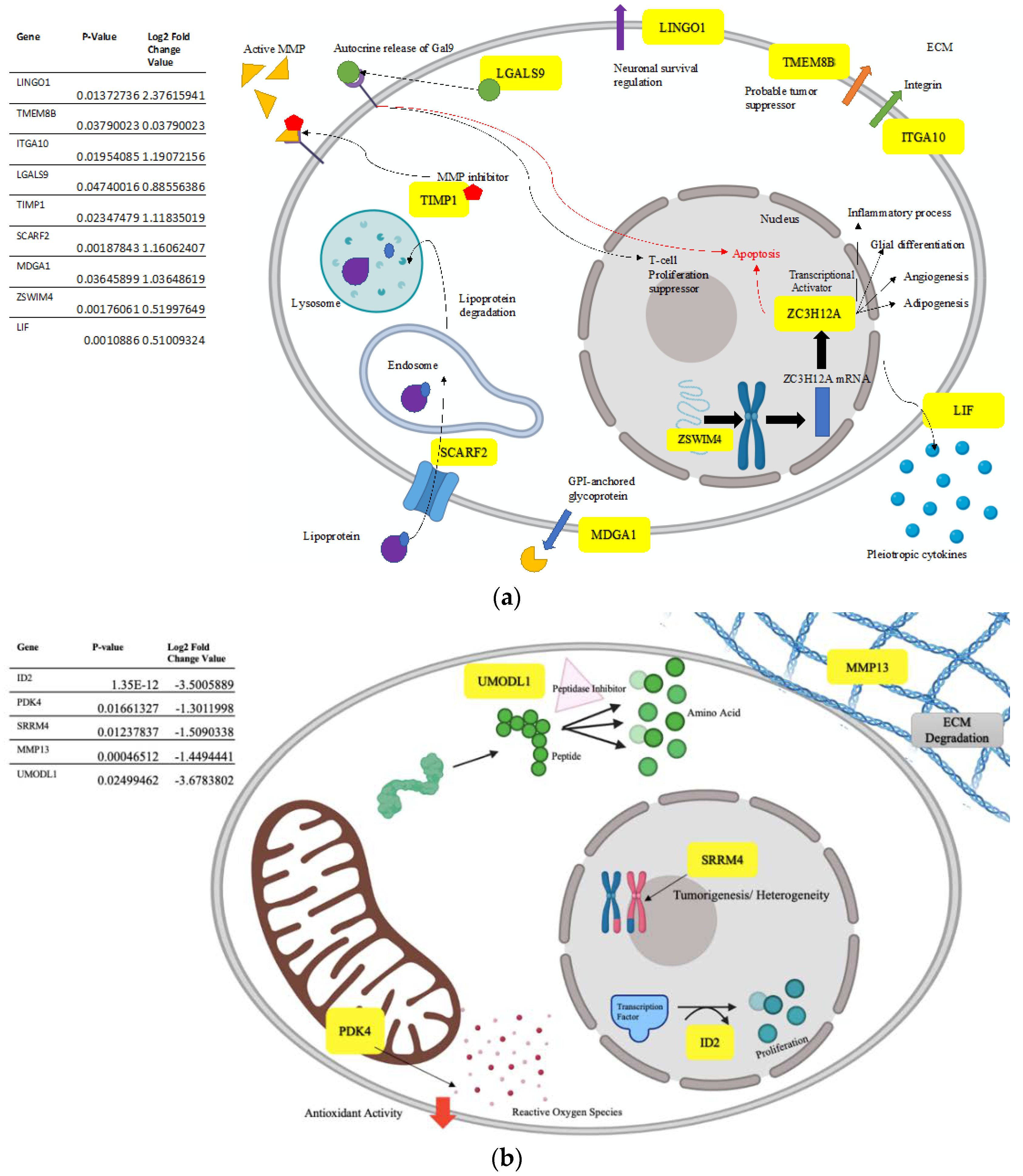
4.2. Upregulated Gene Processes Induce Apoptosis and Inhibit Metastasis
4.3. Downregulated Processes also Prevent Tumor Growth and Spread
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lucey, B.P.; Nelson-Rees, W.A.; Hutchins, G.M. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch. Pathol. Lab. Med. 2019, 133, 1463–1467. [Google Scholar]
- Skloot, R. The Immortal Life of Henrietta Lacks; Broadway Paperbacks: New York, NY, USA, 2011. [Google Scholar]
- Michalet, X.; Pinaud, F.F.; Bentolila, L.A.; Tsay, J.M.; Doose, S.; Li, J.J.; Sundaresan, G.; Wu, A.M.; Gambhir, S.S.; Weiss, S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 2005, 307, 538–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deerinck, T.J. The application of fluorescent quantum dots to confocal, multiphoton, and electron microscopic imaging. Toxicol. Pathol. 2008, 36, 112–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juzenas, P.; Chen, W.; Sun, Y.P.; Coelho, M.A.N.; Generalov, R.; Generalova, N.; Christensen, I.L. Quantum dots and nanoparticles for photodynamic and radiation therapies of cancer. Adv. Drug Deliv. Rev. 2018, 60, 1600–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Zheng, C.; Zhao, H.; Wang, K.; Tao, W. Tumor cell pH detection based on CdSe quantum dots’ fluorescence charateristics. Technol. Health Care 2019, 27, 239–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Wang, Y.; Kong, L.; Xue, Y.; Tang, M. Threshold dose of three types of quantum dots (QDs) induces oxidative stress triggers DNA damage and apoptosis in mouse fibroblast L929 cells. Int. J. Eviron. Res. Public Health 2015, 12, 13435–13454. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, L.; Xu, G.; Wang, X.; Wang, J.; Chen, Y.; Jiang, W.; Yang, Z.; Lin, G. Cytotoxicity of InP/ZnS quantum dots with different surface functional groups toward two lung-derived cell lines. Front. Pharmacol. 2018, 9, 763. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; He, Y.; Lu, H.; Sai, L.; Li, Q.; Li, W.; Wang, L.; Shen, P.; Huang, Q.; Fan, C. The cytotoxicity of cadmium based, aqueous phase—Synthesized, quantum dots and its modulation by surface coating. Biomaterials 2009, 30, 19–25. [Google Scholar] [CrossRef]
- Perini, G.; Palmieri, V.; Ciasca, G.; De Spirito, M.; Papi, M. Unravelling the potential of graphene quantum dots in biomedicine and neuroscience. Int. J. Mol. Sci. 2020, 21, 3712. [Google Scholar] [CrossRef]
- Perini, G.; Palmieri, V.; Ciasca, G.; D’Ascenzo, M.; Gervasoni, J.; Primiano, A.; Rinaldi, M.; Fioretti, D.; Prampolini, C.; Tiberio, F. Graphene quantum dot’s surface chemistry modulates the sensitivity of glioblastoma cells to chemotherapeutics. Int. J. Mol. Sci. 2020, 21, 6301. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, K.; Zhao, J.; Liu, X.; Bu, J.; Yan, X.; Huang, R. Multifunctional mesoporous silica-coated graphene nano sheet used for chemo-photothermal synergistic targeted therapy of glioma. J. Am. Chem. Soc. 2013, 135, 4799–4804. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, Y.; Chen, Y.; Yao, Z. InP/ZnS quantum dots cause inflammatory response in macrophages through endoplasmic reticulum stress and oxidative stress. Int. J. Nanomed. 2019, 14, 9577–9586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horstmann, C.; Campbell, C.; Kim, D.; Kim, K. Transcriptome profile with 20 nm silver nanoparticles in yeast. FEMS Yeast Res. 2019, 19, foz003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horstmann, C.; Kim, D.S.; Campbell, C.; Kim, K. Transcriptome profile alteration with cadmium selenide/zinc sulfide quantum dots in Saccharomyces cerevisiae. Biomolecules 2019, 9, 653. [Google Scholar] [CrossRef] [Green Version]
- Hens, B.; Smothers, J.; Rizvanovic, H.; Patel, R.; Wu, Q.; Kim, K. The future of anticancer drugs: A cytotoxicity assessment study of CdSe/ZnS quantum dots. J. Nanotheranostics 2020, 1, 19–38. [Google Scholar] [CrossRef]
- Pfaffl, M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- NNCrystal US Corporation. Technical Specifications: InP/ZnS Core-Shell Quantum Dots in Water. Available online: https://cdn.shopify.com/s/files/1/0255/7306/4776/files/INPW-Tech-Specs.pdf?162 (accessed on 12 December 2020).
- NNCrystal US Corporation. Optical Spectra of InP/ZnS Nanocrystals Coated with Carboxylic Acid Ligands. Available online: https://cdn.shopify.com/s/files/1/0255/7306/4776/files/INPW-sample-spectra-updated.pdf?162 (accessed on 12 December 2020).
- Radi, R. Oxygen radicals, nitric oxide, and peroxynitrite: Redox pathways in molecular medicine. Proc. Natl. Acad. Sci. USA 2018, 115, 5839–5848. [Google Scholar] [CrossRef] [Green Version]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta Mol. Cell. Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef]
- Kaczanowski, S. Apoptosis: Its origin, history, maintenance and the medical implications for cancer and aging. Phys. Biol. 2016, 13, 3. [Google Scholar] [CrossRef] [Green Version]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Moussa, H.; Merlin, C.; Dezanet, C.; Balan, L.; Medjahdi, G.; Ben-Attia, M.; Schneider, R. Trace amounts of Cu2+ ions influence ROS production and cytotoxicity of ZnO quantum dots. J. Hazard. Mater. 2016, 304, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Owusu, E.; MacRobert, A.; Naasani, I.; Parkin, I.; Allan, E.; Yaghini, E. Photoactivable polymers embedded with cadmium-free quantum dots and crystal violet: Efficient bactericidal activity against clinical strains of antibiotic-resistant bacteria. ACS Appl. Mater. Interf. 2019, 11, 12367–12378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chibli, H.; Carlini, L.; Park, S.; Dimitrijevic, N.; Nadeau, J. Cytotoxicity of InP/ZnS quantum dots related to reactive oxygen species generation. Nanoscale 2011, 3, 2552–2559. [Google Scholar] [CrossRef] [PubMed]
- Corce, V.; Chamoreau, L.; Derat, E.; Goddard, J.; Ollivier, C.; Fensterbank, L. Silicates as latent alkyl radical precursors: Visible-light photocatalytic oxidation of hypervalant bis-catecholato silicon compounds. Angew. Chem. Int. Ed. 2015, 54, 11414–11418. [Google Scholar] [CrossRef]
- Ayupova, D.; Dobhal, G.; Laufersky, G.; Nann, T.; Goreham, R.V. An in vitro investigation of cytotoxic effects of InP/ZnS quantum dots with different surface chemistries. Nanomaterials 2019, 9, 135. [Google Scholar] [CrossRef] [Green Version]
- Dumortier, M.; Ladam, F.; Damour, I.; Vacher, S.; Bièche, I.; Marchand, N.; de Launoit, Y.; Tulasne, D.; Chotteau-Lelièvre, A. ETV4 transcription factor and MMP13 metalloprotease are interplaying actors of breast tumorigenesis. Breast Cancer Res. 2018, 20, 73. [Google Scholar] [CrossRef] [Green Version]
- Saftencu, M.; Braicu, C.; Cojocneanu, R.; Buse, M.; Irimie, A.; Piciu, D.; Berindan-Neagoe, I. Gene expression patterns unveil new insights in papillary thyroid cancer. Medicina 2019, 55, 500. [Google Scholar] [CrossRef] [Green Version]
- Holderried, T.A.W.; De Vos, L.; Bawden, E.G.; Vogt, T.J.; Dietrich, J.; Zarbl, R.; Bootz, F.; Kristiansen, G.; Brossart, P.; Landsberget, J.; et al. Molecular and immune correlates of TIM-3 (HAVCR2) and galectin 9 (LGALS9) mRNA expression and DNA methylation in melanoma. Clin. Epigenetics 2019, 11, 161. [Google Scholar] [CrossRef]
- Chen, T.; Du, D.; Chen, J.; Zhou, P.; Weinstein, J.N.; Yao, L.; Liu, Y. ZC3H12A expression in different stages of colorectal cancer. Oncoscience 2018, 6, 301–311. [Google Scholar] [CrossRef] [Green Version]
- Vazquez, J.M.; Sulak, M.; Chigurupati, S.; Lynch, V.J. A zombie LIF gene in elephants is upregulated by TP53 to induce apoptosis in response to DNA damage. Cell Rep. 2018, 24, 1765–1776. [Google Scholar] [CrossRef] [Green Version]
- Roszik, J.; Ring, K.L.; Wani, K.M.; Lazar, A.J.; Yemelyanova, A.V.; Soliman, P.T.; Frumovitz, M.; Jazaeri, A.A. Gene expression analysis identifies novel targets for cervical cancer therapy. Front. Immunol. 2018, 9, 2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Pandey, P.R.; Sharma, S.; Xing, F.; Wu, K.; Chittiboyina, A.; Wu, S.Y.; Tyagi, A.; Watabe, K. ID2 and GJB2 promote early-stage breast cancer progression by regulating cancer stemness. Breast Cancer Res. Treat. 2019, 175, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Frattini, V.; Bansal, M.; Castano, A.M.; Sherman, D.; Hutchinson, K.; Bruce, J.N.; Califano, A.; Liu, G.; Cardozo, T.; et al. An ID2-dependent mechanism for VHL inactivation in cancer. Nature 2016, 529, 172–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimojo, M.; Kasahara, Y.; Inoue, M.; Tsunoda, S.; Shudo, Y.; Kurata, T.; Obika, S. A gapmer antisense oligonucleotide targeting SRRM4 is a novel therapeutic medicine for lung cancer. Sci. Rep. 2019, 9, 7618. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Shirane, M.; Nakayama, K.I. SRRM4-dependent neuron-specific alternative splicing of protrudin transcripts regulates neurite outgrowth. Sci. Rep. 2017, 7, 41130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.R.; Gan, Y.; Tang, Y.; Dong, X. A novel mechanism of SRRM4 in promoting neuroendocrine prostate cancer development via a pluripotency gene network. EBioMedicine 2018, 35, 167–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhang, S.L.; Hu, X.; Tam, K.Y. Targeting tumor metabolism for cancer treatment: Is pyruvate dehydrogenase kinases (PDKs) a viable anticancer target? Int. J. Biol. Sci. 2015, 11, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Liu, X.; Xie, J.; Meng, J.; Ni, X. PDK-1 mediated Hippo–YAP–IRS2 signaling pathway and involved in the apoptosis of non-small cell lung cancer cells. Biosci. Rep. 2019, 39, 5. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Huang, R.T.; Hamanaka, R.B.; Krause, M.; Oh, M.J.; Kuo, C.H.; Nigdelioglu, R.; Meliton, A.Y.; Witt, L.; Dai, G.; et al. HIF-1α is required for disturbed flow-induced metabolic reprogramming in human and porcine vascular endothelium. ELife 2017, 6, e25217. [Google Scholar] [CrossRef]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [Green Version]
- Tan, Q.; Cui, J.; Huang, J.; Ding, Z.; Lin, H.; Niu, X.; Li, Z.; Wang, G.; Luo, Q.; Lu, S. Genomic alteration during metastasis of lung adenocarcinoma. Cell Physiol. Biochem. 2016, 38, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tang, Y.; Ni, L.; Kim, E.; Jongwutiwes, T.; Hourvitz, A.; Zhang, R.; Xiong, H.; Liu, H.C.; Rosenwaks, Z. Overexpression of Uromodulin-like1 accelerates follicle depletion and subsequent ovarian degeneration. Cell Death Dis. 2012, 3, e433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.D.; Matrisian, L.M. The other side of MMPs: Protective roles in tumor progression. Cancer Metastasis Rev. 2017, 26, 717–724. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davenport, V.; Horstmann, C.; Patel, R.; Wu, Q.; Kim, K. An Assessment of InP/ZnS as Potential Anti-Cancer Therapy: Quantum Dot Treatment Increases Apoptosis in HeLa Cells. J. Nanotheranostics 2021, 2, 16-32. https://doi.org/10.3390/jnt2010002
Davenport V, Horstmann C, Patel R, Wu Q, Kim K. An Assessment of InP/ZnS as Potential Anti-Cancer Therapy: Quantum Dot Treatment Increases Apoptosis in HeLa Cells. Journal of Nanotheranostics. 2021; 2(1):16-32. https://doi.org/10.3390/jnt2010002
Chicago/Turabian StyleDavenport, Victoria, Cullen Horstmann, Rishi Patel, Qihua Wu, and Kyoungtae Kim. 2021. "An Assessment of InP/ZnS as Potential Anti-Cancer Therapy: Quantum Dot Treatment Increases Apoptosis in HeLa Cells" Journal of Nanotheranostics 2, no. 1: 16-32. https://doi.org/10.3390/jnt2010002





