Strengthening Surveillance Systems for Malaria Elimination by Integrating Molecular and Genomic Data
Abstract
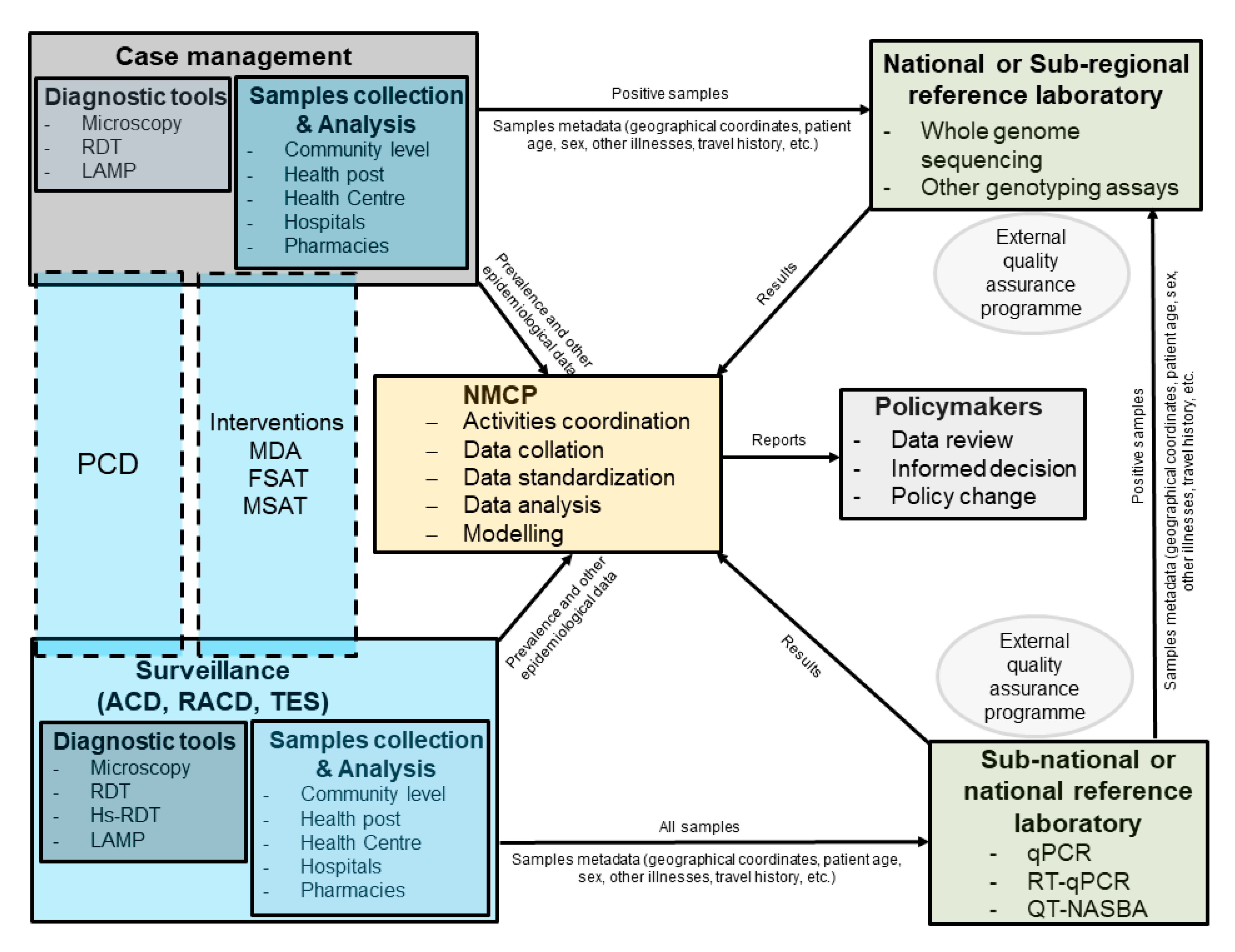
:1. Background
2. Molecular Diagnosis
3. Population Genetics
4. Discussion
5. Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. World Malaria Report 2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- WHO. World Malaria Report 2015; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- WHO. Global Technical Strategy for Malaria 2016–2030; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- malERA Refresh Consultative Panel on Characterising the Reservoir and Measuring Transmission. malERA: An updated research agenda for characterising the reservoir and measuring transmission in malaria elimination and eradication. PLoS Med. 2017, 14, e1002452. [Google Scholar]
- malERA Refresh Consultative Panel on Tools for Malaria Elimination. malERA: An updated research agenda for diagnostics, drugs, vaccines, and vector control in malaria elimination and eradication. PLoS Med. 2017, 14, e1002455. [Google Scholar]
- Das, S.; Jang, I.K.; Barney, B.; Peck, R.; Rek, J.C.; Arinaitwe, E.; Adrama, H.; Murphy, M.; Imwong, M.; Ling, C.L.; et al. Performance of a High-Sensitivity Rapid Diagnostic Test for Plasmodium falciparum Malaria in Asymptomatic Individuals from Uganda and Myanmar and Naive Human Challenge Infections. Am. J. Trop. Med. Hyg. 2017, 97, 1540–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vásquez, A.M.; Medina, A.C.; Tobón-Castaño, A.; Posada, M.; Vélez, G.J.; Campillo, A.; González, I.J.; Ding, X. Performance of a highly sensitive rapid diagnostic test (HS-RDT) for detecting malaria in peripheral and placental blood samples from pregnant women in Colombia. PLoS ONE 2018, 13, e0201769. [Google Scholar] [CrossRef] [Green Version]
- Landier, J.; Haohankhunnatham, W.; Das, S.; Konghahong, K.; Christensen, P.; Raksuansak, J.; Phattharakokoedbun, P.; Kajeechiwa, L.; Thwin, M.M.; Jang, I.K.; et al. Operational Performance of a Plasmodium falciparum Ultrasensitive Rapid Diagnostic Test for Detection of Asymptomatic Infections in Eastern Myanmar. J. Clin. Microbiol. 2018, 56, e00565-18. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, N.E.; Gruenberg, M.; Nate, E.; Ura, A.; Rodriguez-Rodriguez, D.; Salib, M.; Mueller, I.; Smith, T.A.; Laman, M.; Robinson, L.J.; et al. Assessment of ultra-sensitive malaria diagnosis versus standard molecular diagnostics for malaria elimination: An in-depth molecular community cross-sectional study. Lancet Infect. Dis. 2018, 18, 1108–1116. [Google Scholar] [CrossRef]
- Girma, S.; Cheaveau, J.; Mohon, A.N.; Marasinghe, D.; Legese, R.; Balasingam, N.; Abera, A.; Feleke, S.M.; Golassa, L.; Pillai, D.R. Prevalence and Epidemiological Characteristics of Asymptomatic Malaria Based on Ultrasensitive Diagnostics: A Cross-sectional Study. Clin. Infect. Dis. 2019, 69, 1003–1010. [Google Scholar] [CrossRef]
- Mwesigwa, J.; Slater, H.; Bradley, J.; Saidy, B.; Ceesay, F.; Whittaker, C.; Kandeh, B.; Nkwakamna, D.; Drakeley, C.; Van Geertruyden, J.-P.; et al. Field performance of the malaria highly sensitive rapid diagnostic test in a setting of varying malaria transmission. Malar. J. 2019, 18, 288. [Google Scholar] [CrossRef]
- Watson, O.J.; Slater, H.C.; Verity, R.; Parr, J.B.; Mwandagalirwa, M.K.; Tshefu, A.; Meshnick, S.R.; Ghani, A.C. Modelling the drivers of the spread of Plasmodium falciparum hrp2 gene deletions in sub-Saharan Africa. Elife 2017, 6, e25008. [Google Scholar] [CrossRef]
- Thomson, R.; Beshir, K.B.; Cunningham, J.; Baiden, F.; Bharmal, J.; Bruxvoort, K.J.; Maiteki-Sebuguzi, C.; Owusu-Agyei, S.; Staedke, S.G.; Hopkins, H. pfhrp2 and pfhrp3 gene deletions that affect malaria rapid diagnostic tests for Plasmodium falciparum: Analysis of archived blood samples from three African countries. J. Infect. Dis. 2019, 220, 1444–1452. [Google Scholar] [CrossRef]
- Kamau, E.; Tolbert, L.S.; Kortepeter, L.; Pratt, M.; Nyakoe, N.; Muringo, L.; Ogutu, B.; Waitumbi, J.N.; Ockenhouse, C.F. Development of a Highly Sensitive Genus-Specific Quantitative Reverse Transcriptase Real-Time PCR Assay for Detection and Quantitation of Plasmodium by Amplifying RNA and DNA of the 18S rRNA Genes. J. Clin. Microbiol. 2011, 49, 2946–2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bousema, T.; Okell, L.; Felger, I.; Drakeley, C. Asymptomatic malaria infections: Detectability, transmissibility and public health relevance. Nat. Rev. Microbiol. 2014, 12, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Chaumeau, V.; Kajeechiwa, L.; Fustec, B.; Landier, J.; Naw Nyo, S.; Nay Hsel, S.; Phatharakokordbun, P.; Kittiphanakun, P.; Nosten, S.; Thwin, M.M.; et al. Contribution of Asymptomatic Plasmodium Infections to the Transmission of Malaria in Kayin State, Myanmar. J. Infect. Dis. 2019, 219, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Slater, H.C.; Ross, A.; Felger, I.; Hofmann, N.E.; Robinson, L.; Cook, J.; Gonçalves, B.P.; Björkman, A.; Ouedraogo, A.L.; Morris, U.; et al. The temporal dynamics and infectiousness of subpatent Plasmodium falciparum infections in relation to parasite density. Nat. Commun. 2019, 10, 1433. [Google Scholar] [CrossRef] [Green Version]
- Dalmat, R.; Naughton, B.; Kwan-Gett, T.S.; Slyker, J.; Stuckey, E.M. Use cases for genetic epidemiology in malaria elimination. Malar. J. 2019, 18, 163. [Google Scholar] [CrossRef] [Green Version]
- Volkman, S.K.; Ndiaye, D.; Diakite, M.; Koita, O.A.; Nwakanma, D.; Daniels, R.F.; Park, D.J.; Neafsey, D.E.; Muskavitch, M.A.T.; Krogstad, D.J.; et al. Application of genomics to field investigations of malaria by the international centers of excellence for malaria research. Acta Trop. 2012, 121, 324–332. [Google Scholar] [CrossRef] [Green Version]
- Escalante, A.A.; Ferreira, M.U.; Vinetz, J.M.; Volkman, S.K.; Cui, L.; Gamboa, D.; Krogstad, D.J.; Barry, A.E.; Carlton, J.M.; van Eijk, A.M.; et al. Malaria Molecular Epidemiology: Lessons from the International Centers of Excellence for Malaria Research Network. Am. J. Trop. Med. Hyg. 2015, 93, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Nag, S.; Kofoed, P.-E.; Ursing, J.; Lemvigh, C.K.; Allesøe, R.L.; Rodrigues, A.; Svendsen, C.A.; Jensen, J.D.; Alifrangis, M.; Lund, O.; et al. Direct whole-genome sequencing of Plasmodium falciparum specimens from dried erythrocyte spots. Malar. J. 2018, 17, 91. [Google Scholar] [CrossRef]
- Neafsey, D.E.; Volkman, S.K. Malaria Genomics in the Era of Eradication. Cold Spring Harb. Perspect. Med. 2017, 7, a025544. [Google Scholar] [CrossRef] [Green Version]
- Auburn, S.; Barry, A.E. Dissecting malaria biology and epidemiology using population genetics and genomics. Int. J. Parasitol. 2017, 47, 77–85. [Google Scholar] [CrossRef]
- Schaffner, S.F.; Taylor, A.R.; Wong, W.; Wirth, D.F.; Neafsey, D.E. hmmIBD: Software to infer pairwise identity by descent between haploid genotypes. Malar. J. 2018, 17, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, A.R.; Schaffner, S.F.; Cerqueira, G.C.; Nkhoma, S.C.; Anderson, T.J.C.; Sriprawat, K.; Pyae Phyo, A.; Nosten, F.; Neafsey, D.E.; Buckee, C.O. Quantifying connectivity between local Plasmodium falciparum malaria parasite populations using identity by descent. PLoS Genet. 2017, 13, e1007065. [Google Scholar] [CrossRef] [PubMed]
- Henden, L.; Lee, S.; Mueller, I.; Barry, A.; Bahlo, M. Identity-by-descent analyses for measuring population dynamics and selection in recombining pathogens. PLoS Genet. 2018, 14, e1007279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pringle, J.C.; Tessema, S.; Wesolowski, A.; Chen, A.; Murphy, M.; Carpi, G.; Shields, T.M.; Hamapumbu, H.; Searle, K.M.; Kobayashi, T.; et al. Genetic Evidence of Focal Plasmodium falciparum Transmission in a Pre-elimination Setting in Southern Province, Zambia. J. Infect. Dis. 2019, 219, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Tessema, S.; Wesolowski, A.; Chen, A.; Murphy, M.; Wilheim, J.; Mupiri, A.-R.; Ruktanonchai, N.W.; Alegana, V.A.; Tatem, A.J.; Tambo, M.; et al. Using parasite genetic and human mobility data to infer local and cross-border malaria connectivity in Southern Africa. Elife 2019, 8, e43510. [Google Scholar] [CrossRef] [PubMed]
- Searle, K.M.; Katowa, B.; Kobayashi, T.; Siame, M.N.S.; Mharakurwa, S.; Carpi, G.; Norris, D.E.; Stevenson, J.C.; Thuma, P.E.; Moss, W.J.; et al. Distinct parasite populations infect individuals identified through passive and active case detection in a region of declining malaria transmission in southern Zambia. Malar. J. 2017, 16, 154. [Google Scholar] [CrossRef]
- Britton, S.; Cheng, Q.; McCarthy, J.S. Novel molecular diagnostic tools for malaria elimination: A review of options from the point of view of high-throughput and applicability in resource limited settings. Malar. J. 2016, 15, 88. [Google Scholar] [CrossRef] [Green Version]
- Lucchi, N.W.; Ndiaye, D.; Britton, S.; Udhayakumar, V. Expanding the malaria molecular diagnostic options: Opportunities and challenges for loop-mediated isothermal amplification tests for malaria control and elimination. Expert Rev. Mol. Diagn. 2018, 18, 195–203. [Google Scholar] [CrossRef]
- Lucchi, N.W.; Demas, A.; Narayanan, J.; Sumari, D.; Kabanywanyi, A.; Kachur, S.P.; Barnwell, J.W.; Udhayakumar, V. Real-Time Fluorescence Loop Mediated Isothermal Amplification for the Diagnosis of Malaria. PLoS ONE 2010, 5, e13733. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, N.; Mwingira, F.; Shekalaghe, S.; Robinson, L.J.; Mueller, I.; Felger, I. Ultra-Sensitive Detection of Plasmodium falciparum by Amplification of Multi-Copy Subtelomeric Targets. PLoS Med. 2015, 12, e1001788. [Google Scholar] [CrossRef] [Green Version]
- Imwong, M.; Hanchana, S.; Malleret, B.; Rénia, L.; Day, N.P.J.; Dondorp, A.; Nosten, F.; Snounou, G.; White, N.J. High-Throughput Ultrasensitive Molecular Techniques for Quantifying Low-Density Malaria Parasitemias. J. Clin. Microbiol. 2014, 52, 3303–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, M.; Joshi, S.N.; Mbambo, G.; Mu, A.Z.; Roemmich, S.M.; Shrestha, B.; Strauss, K.A.; Johnson, N.E.; Oo, K.Z.; Hlaing, T.M.; et al. An ultrasensitive reverse transcription polymerase chain reaction assay to detect asymptomatic low-density Plasmodium falciparum and Plasmodium vivax infections in small volume blood samples. Malar. J. 2015, 14, 520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apinjoh, T.O.; Ouattara, A.; Titanji, V.P.K.; Djimde, A.; Amambua-Ngwa, A. Genetic diversity and drug resistance surveillance of Plasmodium falciparum for malaria elimination: Is there an ideal tool for resource-limited sub-Saharan Africa? Malar. J. 2019, 18, 217. [Google Scholar] [CrossRef] [PubMed]
- Tripura, R.; Peto, T.J.; Chea, N.; Chan, D.; Mukaka, M.; Sirithiranont, P.; Dhorda, M.; Promnarate, C.; Imwong, M.; von Seidlein, L.; et al. A Controlled Trial of Mass Drug Administration to Interrupt Transmission of Multidrug-Resistant Falciparum Malaria in Cambodian Villages. Clin. Infect. Dis. 2018, 67, 817–826. [Google Scholar] [CrossRef]
- von Seidlein, L.; Peto, T.J.; Landier, J.; Nguyen, T.-N.; Tripura, R.; Phommasone, K.; Pongvongsa, T.; Lwin, K.M.; Keereecharoen, L.; Kajeechiwa, L.; et al. The impact of targeted malaria elimination with mass drug administrations on falciparum malaria in Southeast Asia: A cluster randomised trial. PLoS Med. 2019, 16, e1002745. [Google Scholar] [CrossRef]
- Mwesigwa, J.; Achan, J.; Affara, M.; Wathuo, M.; Worwui, A.; Mohammed, N.I.; Kanuteh, F.; Prom, A.; Dierickx, S.; di Tanna, G.L.; et al. Mass Drug Administration With Dihydroartemisinin-piperaquine and Malaria Transmission Dynamics in The Gambia: A Prospective Cohort Study. Clin. Infect. Dis. 2019, 69, 278–286. [Google Scholar] [CrossRef]
- Roth, J.M.; de Bes, L.; Sawa, P.; Omweri, G.; Osoti, V.; Oberheitmann, B.; Schallig, H.D.F.H.; Mens, P.F. Plasmodium Detection and Differentiation by Direct-on-Blood PCR Nucleic Acid Lateral Flow Immunoassay. J. Mol. Diagn. 2018, 20, 78–86. [Google Scholar] [CrossRef] [Green Version]
- Kolluri, N.; Klapperich, C.M.; Cabodi, M. Towards lab-on-a-chip diagnostics for malaria elimination. Lab Chip 2017, 18, 75–94. [Google Scholar] [CrossRef]
- Snounou, G.; Singh, B. Nested PCR analysis of Plasmodium parasites. Methods Mol. Med. 2002, 72, 189–203. [Google Scholar]
- Schneider, P.; Schoone, G.; Schallig, H.; Verhage, D.; Telgt, D.; Eling, W.; Sauerwein, R. Quantification of Plasmodium falciparum gametocytes in differential stages of development by quantitative nucleic acid sequence-based amplification. Mol. Biochem. Parasitol. 2004, 137, 35–41. [Google Scholar] [CrossRef]
- Chenet, S.M.; Schneider, K.A.; Villegas, L.; Escalante, A.A. Local population structure of Plasmodium: Impact on malaria control and elimination. Malar. J. 2012, 11, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batista, C.L.; Barbosa, S.; Da Silva Bastos, M.; Viana, S.A.S.; Ferreira, M.U. Genetic diversity of Plasmodium vivax over time and space: A community-based study in rural Amazonia. Parasitology 2015, 142, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Roh, M.E.; Tessema, S.K.; Murphy, M.; Nhlabathi, N.; Mkhonta, N.; Vilakati, S.; Ntshalintshali, N.; Saini, M.; Maphalala, G.; Chen, A.; et al. High Genetic Diversity of Plasmodium falciparum in the Low-Transmission Setting of the Kingdom of Eswatini. J. Infect. Dis. 2019, 220, 1346–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obaldia, N.; Baro, N.K.; Calzada, J.E.; Santamaria, A.M.; Daniels, R.; Wong, W.; Chang, H.-H.; Hamilton, E.J.; Arevalo-Herrera, M.; Herrera, S.; et al. Clonal Outbreak of Plasmodium falciparum Infection in Eastern Panama. J. Infect. Dis. 2015, 211, 1087–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyebola, K.M.; Aina, O.O.; Idowu, E.T.; Olukosi, Y.A.; Ajibaye, O.S.; Otubanjo, O.A.; Awolola, T.S.; Awandare, G.A.; Amambua-Ngwa, A. A barcode of multilocus nuclear DNA identifies genetic relatedness in pre- and post-Artemether/Lumefantrine treated Plasmodium falciparum in Nigeria. BMC Infect. Dis. 2018, 18, 392. [Google Scholar] [CrossRef]
- Nkhoma, S.C.; Nair, S.; Cheeseman, I.H.; Rohr-Allegrini, C.; Singlam, S.; Nosten, F.; Anderson, T.J.C. Close kinship within multiple-genotype malaria parasite infections. Proc. R. Soc. B Biol. Sci. 2012, 279, 2589–2598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campino, S.; Auburn, S.; Kivinen, K.; Zongo, I.; Ouedraogo, J.-B.; Mangano, V.; Djimde, A.; Doumbo, O.K.; Kiara, S.M.; Nzila, A.; et al. Population Genetic Analysis of Plasmodium falciparum Parasites Using a Customized Illumina GoldenGate Genotyping Assay. PLoS ONE 2011, 6, e20251. [Google Scholar] [CrossRef] [PubMed]
- Zhong, D.; Koepfli, C.; Cui, L.; Yan, G. Molecular approaches to determine the multiplicity of Plasmodium infections. Malar. J. 2018, 17, 172. [Google Scholar] [CrossRef]
- Lerch, A.; Koepfli, C.; Hofmann, N.E.; Kattenberg, J.H.; Rosanas-Urgell, A.; Betuela, I.; Mueller, I.; Felger, I. Longitudinal tracking and quantification of individual Plasmodium falciparum clones in complex infections. Sci. Rep. 2019, 9, 3333. [Google Scholar] [CrossRef]
- Koepfli, C.; Mueller, I. Malaria Epidemiology at the Clone Level. Trends Parasitol. 2017, 33, 974–985. [Google Scholar] [CrossRef]
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Langlois, A.-C.; Khim, N.; Kim, S.; Duru, V.; Bouchier, C.; Ma, L.; et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014, 505, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Cowell, A.; Winzeler, E. Exploration of the Plasmodium falciparum Resistome and Druggable Genome Reveals New Mechanisms of Drug Resistance and Antimalarial Targets. Microbiol. Insights 2018, 11, 1178636118808529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howes, R.E.; Dewi, M.; Piel, F.B.; Monteiro, W.M.; Battle, K.E.; Messina, J.P.; Sakuntabhai, A.; Satyagraha, A.W.; Williams, T.N.; Baird, J.K.; et al. Spatial distribution of G6PD deficiency variants across malaria-endemic regions. Malar. J. 2013, 12, 418. [Google Scholar] [CrossRef] [PubMed]
- Talundzic, E.; Ndiaye, Y.D.; Deme, A.B.; Olsen, C.; Patel, D.S.; Biliya, S.; Daniels, R.; Vannberg, F.O.; Volkman, S.K.; Udhayakumar, V.; et al. Molecular Epidemiology of Plasmodium falciparum kelch13 Mutations in Senegal Determined by Using Targeted Amplicon Deep Sequencing. Antimicrob. Agents Chemother. 2017, 61, e02116-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyce, R.M.; Hathaway, N.; Fulton, T.; Reyes, R.; Matte, M.; Ntaro, M.; Mulogo, E.; Waltmann, A.; Bailey, J.A.; Siedner, M.J.; et al. Reuse of malaria rapid diagnostic tests for amplicon deep sequencing to estimate Plasmodium falciparum transmission intensity in western Uganda. Sci. Rep. 2018, 8, 10159. [Google Scholar] [CrossRef] [PubMed]
- Lerch, A.; Koepfli, C.; Hofmann, N.E.; Messerli, C.; Wilcox, S.; Kattenberg, J.H.; Betuela, I.; O’Connor, L.; Mueller, I.; Felger, I. Development of amplicon deep sequencing markers and data analysis pipeline for genotyping multi-clonal malaria infections. BMC Genom. 2017, 18, 864. [Google Scholar] [CrossRef] [Green Version]
- Rabinovich, R.N.; Drakeley, C.; Djimde, A.A.; Hall, B.F.; Hay, S.I.; Hemingway, J.; Kaslow, D.C.; Noor, A.; Okumu, F.; Steketee, R.; et al. malERA: An updated research agenda for malaria elimination and eradication. PLoS Med. 2017, 14, e1002456. [Google Scholar] [CrossRef]
- Wijesundere, D.A.; Ramasamy, R. Analysis of Historical Trends and Recent Elimination of Malaria from Sri Lanka and Its Applicability for Malaria Control in Other Countries. Front. Public Health 2017, 5, 212. [Google Scholar] [CrossRef] [Green Version]
- Mogeni, P.; Williams, T.N.; Omedo, I.; Kimani, D.; Ngoi, J.M.; Mwacharo, J.; Morter, R.; Nyundo, C.; Wambua, J.; Nyangweso, G.; et al. Detecting Malaria Hotspots: A Comparison of Rapid Diagnostic Test, Microscopy, and Polymerase Chain Reaction. J. Infect. Dis. 2017, 216, 1091–1098. [Google Scholar] [CrossRef] [Green Version]
- Gruenberg, M.; Moniz, C.A.; Hofmann, N.E.; Wampfler, R.; Koepfli, C.; Mueller, I.; Monteiro, W.M.; Lacerda, M.; de Melo, G.C.; Kuehn, A.; et al. Plasmodium vivax molecular diagnostics in community surveys: Pitfalls and solutions. Malar. J. 2018, 17, 55. [Google Scholar] [CrossRef]
- Roth, J.M.; Sawa, P.; Omweri, G.; Osoti, V.; Makio, N.; Bradley, J.; Bousema, T.; Schallig, H.D.F.H.; Mens, P.F. Plasmodium falciparum gametocyte dynamics after pyronaridine–artesunate or artemether–lumefantrine treatment. Malar. J. 2018, 17, 223. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, B.P.; Tiono, A.B.; Ouédraogo, A.; Guelbéogo, W.M.; Bradley, J.; Nebie, I.; Siaka, D.; Lanke, K.; Eziefula, A.C.; Diarra, A.; et al. Single low dose primaquine to reduce gametocyte carriage and Plasmodium falciparum transmission after artemether-lumefantrine in children with asymptomatic infection: A randomised, double-blind, placebo-controlled trial. BMC Med. 2016, 14, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, N.F.; Bastos, M.S.; Ferreira, M.U. Plasmodium vivax: Reverse transcriptase real-time PCR for gametocyte detection and quantitation in clinical samples. Exp. Parasitol. 2012, 132, 348–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghansah, A.; Kamau, E.; Amambua-Ngwa, A.; Ishengoma, D.S.; Maiga-Ascofare, O.; Amenga-Etego, L.; Deme, A.; Yavo, W.; Randrianarivelojosia, M.; Ochola-Oyier, L.I.; et al. Targeted Next Generation Sequencing for malaria research in Africa: Current status and outlook. Malar. J. 2019, 18, 324. [Google Scholar] [CrossRef]
- Daniels, R.; Ndiaye, D.; Wall, M.; McKinney, J.; Séne, P.D.; Sabeti, P.C.; Volkman, S.K.; Mboup, S.; Wirth, D.F. Rapid, Field-Deployable Method for Genotyping and Discovery of Single-Nucleotide Polymorphisms Associated with Drug Resistance in Plasmodium falciparum. Antimicrob. Agents Chemother. 2012, 56, 2976–2986. [Google Scholar] [CrossRef] [Green Version]
- Ndiaye, Y.D.; Diédhiou, C.K.; Bei, A.K.; Dieye, B.; Mbaye, A.; Mze, N.P.; Daniels, R.F.; Ndiaye, I.M.; Déme, A.B.; Gaye, A.; et al. High resolution melting: A useful field-deployable method to measure dhfr and dhps drug resistance in both highly and lowly endemic Plasmodium populations. Malar. J. 2017, 16, 153. [Google Scholar] [CrossRef] [Green Version]
- Wesolowski, A.; Taylor, A.R.; Chang, H.-H.; Verity, R.; Tessema, S.; Bailey, J.A.; Alex Perkins, T.; Neafsey, D.E.; Greenhouse, B.; Buckee, C.O. Mapping malaria by combining parasite genomic and epidemiologic data. BMC Med. 2018, 16, 190. [Google Scholar] [CrossRef] [Green Version]
- Imai, K.; Tarumoto, N.; Runtuwene, L.R.; Sakai, J.; Hayashida, K.; Eshita, Y.; Maeda, R.; Tuda, J.; Ohno, H.; Murakami, T.; et al. An innovative diagnostic technology for the codon mutation C580Y in kelch13 of Plasmodium falciparum with MinION nanopore sequencer. Malar. J. 2018, 17, 217. [Google Scholar] [CrossRef]
- Runtuwene, L.R.; Tuda, J.S.B.; Mongan, A.E.; Makalowski, W.; Frith, M.C.; Imwong, M.; Srisutham, S.; Nguyen Thi, L.A.; Tuan, N.N.; Eshita, Y.; et al. Nanopore sequencing of drug-resistance-associated genes in malaria parasites, Plasmodium falciparum. Sci. Rep. 2018, 8, 8286. [Google Scholar] [CrossRef]
- Dara, A.; Travassos, M.A.; Adams, M.; Schaffer DeRoo, S.; Drábek, E.F.; Agrawal, S.; Laufer, M.K.; Plowe, C.V.; Silva, J.C. A new method for sequencing the hypervariable Plasmodium falciparum gene var2csa from clinical samples. Malar. J. 2017, 16, 343. [Google Scholar] [CrossRef] [Green Version]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; AbuOun, M.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb Genom. 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Baird, J.K. 8-Aminoquinoline Therapy for Latent Malaria. Clin. Microbiol. Rev. 2019, 32, e00011-19. [Google Scholar] [CrossRef] [PubMed]
- WHO. Considerations for Implementation of G6PD Testing and Radical Curein P. vivax Endemic Countries; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Larocca, A.; Moro Visconti, R.; Marconi, M. Malaria diagnosis and mapping with m-Health and geographic information systems (GIS): Evidence from Uganda. Malar. J. 2016, 15, 520. [Google Scholar] [CrossRef] [PubMed]
- Abaza, H.; Marschollek, M. mHealth Application Areas and Technology Combinations. Methods Inf. Med. 2017, 56, e105–e122. [Google Scholar] [CrossRef]
- Singh, Y.; Jackson, D.; Bhardwaj, S.; Titus, N.; Goga, A. National surveillance using mobile systems for health monitoring: Complexity, functionality and feasibility. BMC Infect. Dis. 2019, 19, 786. [Google Scholar] [CrossRef] [Green Version]
- Dowell, S.F.; Blazes, D.; Desmond-Hellmann, S. Four steps to precision public health. Nature 2016, 540, 189–191. [Google Scholar] [CrossRef]
- Ishengoma, D.S.; Saidi, Q.; Sibley, C.H.; Roper, C.; Alifrangis, M. Deployment and utilization of next-generation sequencing of Plasmodium falciparum to guide anti-malarial drug policy decisions in sub-Saharan Africa: Opportunities and challenges. Malar. J. 2019, 18, 267. [Google Scholar] [CrossRef] [Green Version]
- Tessema, S.K.; Raman, J.; Duffy, C.W.; Ishengoma, D.S.; Amambua-Ngwa, A.; Greenhouse, B. Applying next-generation sequencing to track falciparum malaria in sub-Saharan Africa. Malar. J. 2019, 18, 268. [Google Scholar] [CrossRef] [Green Version]
- Achidi, E.A.; Agbenyega, T.; Allen, S.; Amodu, O.; Bojang, K.; Conway, D.; Corran, P.; Deloukas, P.; Djimde, A.; Dolo, A.; et al. A global network for investigating the genomic epidemiology of malaria. Nature 2008, 456, 732–737. [Google Scholar]
- Ghansah, A.; Amenga-Etego, L.; Amambua-Ngwa, A.; Andagalu, B.; Apinjoh, T.; Bouyou-Akotet, M.; Cornelius, V.; Golassa, L.; Andrianaranjaka, V.H.; Ishengoma, D.; et al. Monitoring parasite diversity for malaria elimination in sub-Saharan Africa. Science (80-) 2014, 345, 1297–1298. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, J.G.; Mather, F.J.; Wele, M.; Li, J.; Tangara, C.O.; Kassogue, Y.; Srivastav, S.K.; Thiero, O.; Diakite, M.; Sangare, M.; et al. Expanding Research Capacity in Sub-Saharan Africa Through Informatics, Bioinformatics, and Data Science Training Programs in Mali. Front. Genet. 2019, 10, 331. [Google Scholar] [CrossRef] [PubMed]
- Nsanzabana, C.; Djalle, D.; Guérin, P.J.; Ménard, D.; González, I.J. Tools for surveillance of anti-malarial drug resistance: An assessment of the current landscape. Malar. J. 2018, 17, 75. [Google Scholar] [CrossRef] [PubMed]
- Nsanzabana, C.; Ariey, F.; Beck, H.-P.; Ding, X.C.; Kamau, E.; Krishna, S.; Legrand, E.; Lucchi, N.; Miotto, O.; Nag, S.; et al. Molecular assays for antimalarial drug resistance surveillance: A target product profile. PLoS ONE 2018, 13, e0204347. [Google Scholar] [CrossRef] [PubMed]
- Mlotshwa, B.C.; Mwesigwa, S.; Mboowa, G.; Williams, L.; Retshabile, G.; Kekitiinwa, A.; Wayengera, M.; Kyobe, S.; Brown, C.W.; Hanchard, N.A.; et al. The collaborative African genomics network training program: A trainee perspective on training the next generation of African scientists. Genet. Med. 2017, 19, 826–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiffin, N.; George, A.; LeFevre, A.E. How to use relevant data for maximal benefit with minimal risk: Digital health data governance to protect vulnerable populations in low-income and middle-income countries. BMJ Glob. Health 2019, 4, e001395. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.; Kan, L. Digital Technology and the Future of Health Systems. Health Syst. Reform 2019, 5, 113–120. [Google Scholar] [CrossRef] [Green Version]
- WHO. WHO External Quality Assurance Scheme for Malaria Nucleic Acid Amplification Testing (NAAT EQA). Available online: https://www.who.int/malaria/areas/diagnosis/faq-nucleic-acid-amplification-tests/en/ (accessed on 25 September 2019).
- MalariaGEN Malaria Genomic Epidemiology Network. Available online: https://www.malariagen.net/about (accessed on 25 September 2019).
- MRC. Centre for Genomics and Global Health Plasmodium Diversity Network Africa. Available online: https://www.cggh.org/collaborations/plasmodium-diversity-network-africa (accessed on 25 September 2019).
- Lai, S.; Sun, J.; Ruktanonchai, N.W.; Zhou, S.; Yu, J.; Routledge, I.; Wang, L.; Zheng, Y.; Tatem, A.J.; Li, Z. Changing epidemiology and challenges of malaria in China towards elimination. Malar. J. 2019, 18, 107. [Google Scholar] [CrossRef]

| Assay | Limit of Detection (Parasites/µL) | Throughput | Cost/Sample Excluding Labor and Equipment (USD) | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|---|
| Nested PCR | 1 | moderate | <10 | Requires simple and cheap thermocycler Can be performed with low amount of DNA (e.g., from dried blood spots) | Moderately sensitive Requires good laboratory infrastructure and well-trained staff | [42] |
| qPCR (high blood volume) | 0.022 | high | <10 | Highly sensitive | Requires high blood volume Requires good laboratory infrastructure and well-trained staff | [34] |
| qPCR (low blood volume or dried blood spots) | 0.15 | high | <10 | Highly sensitive Can be performed with low amount of DNA (e.g., from dried blood spots) | Requires good laboratory infrastructure and well-trained staff | [33] |
| qRT-PCR | 0.002 | high | <20 | Highly sensitive Can be performed with low amount of DNA (e.g., from dried blood spots) Can detect and quantify gametocytes | Difficult to work with RNA Requires good laboratory infrastructure and well-trained staff | [14] |
| LAMP | 1 to 5 | moderate | <3 | Cheap Does not require laboratory infrastructure and well-trained staff Can be performed with low of DNA (e.g., from dried blood spots) or directly from blood sample Fast | Moderately sensitive Limited throughput | [31] |
| QT NASBA | <1 | high | Can be performed with low amount of DNA (e.g., from dried blood spots) Can detect and quantify gametocytes | Not as robust as qRT-PCR | [43] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nsanzabana, C. Strengthening Surveillance Systems for Malaria Elimination by Integrating Molecular and Genomic Data. Trop. Med. Infect. Dis. 2019, 4, 139. https://doi.org/10.3390/tropicalmed4040139
Nsanzabana C. Strengthening Surveillance Systems for Malaria Elimination by Integrating Molecular and Genomic Data. Tropical Medicine and Infectious Disease. 2019; 4(4):139. https://doi.org/10.3390/tropicalmed4040139
Chicago/Turabian StyleNsanzabana, Christian. 2019. "Strengthening Surveillance Systems for Malaria Elimination by Integrating Molecular and Genomic Data" Tropical Medicine and Infectious Disease 4, no. 4: 139. https://doi.org/10.3390/tropicalmed4040139





