Vitellogenesis in Blue Gourami is Accompanied by Brain Transcriptome Changes
Abstract
:1. Introduction
2. Results
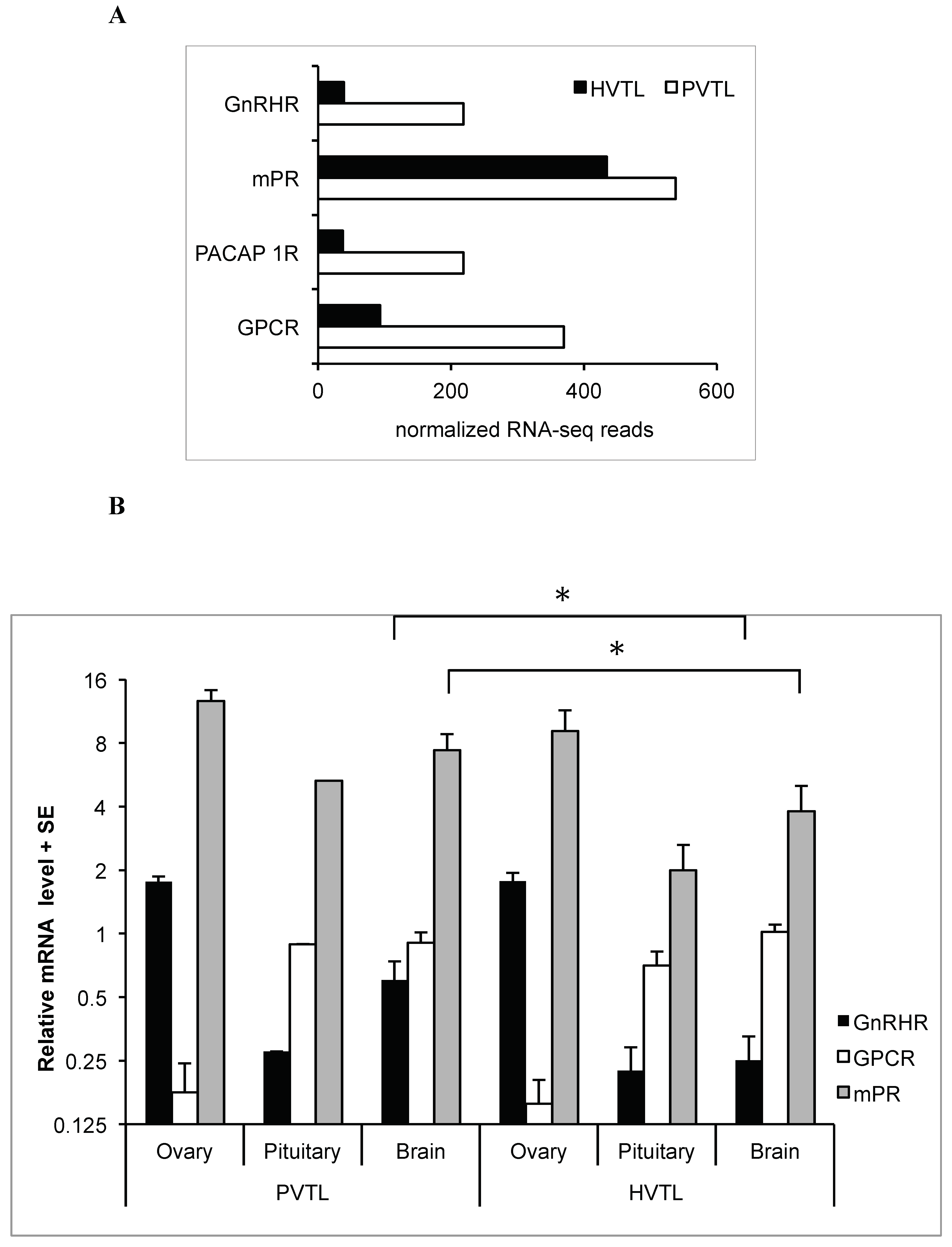
2.1. RNA-Seq
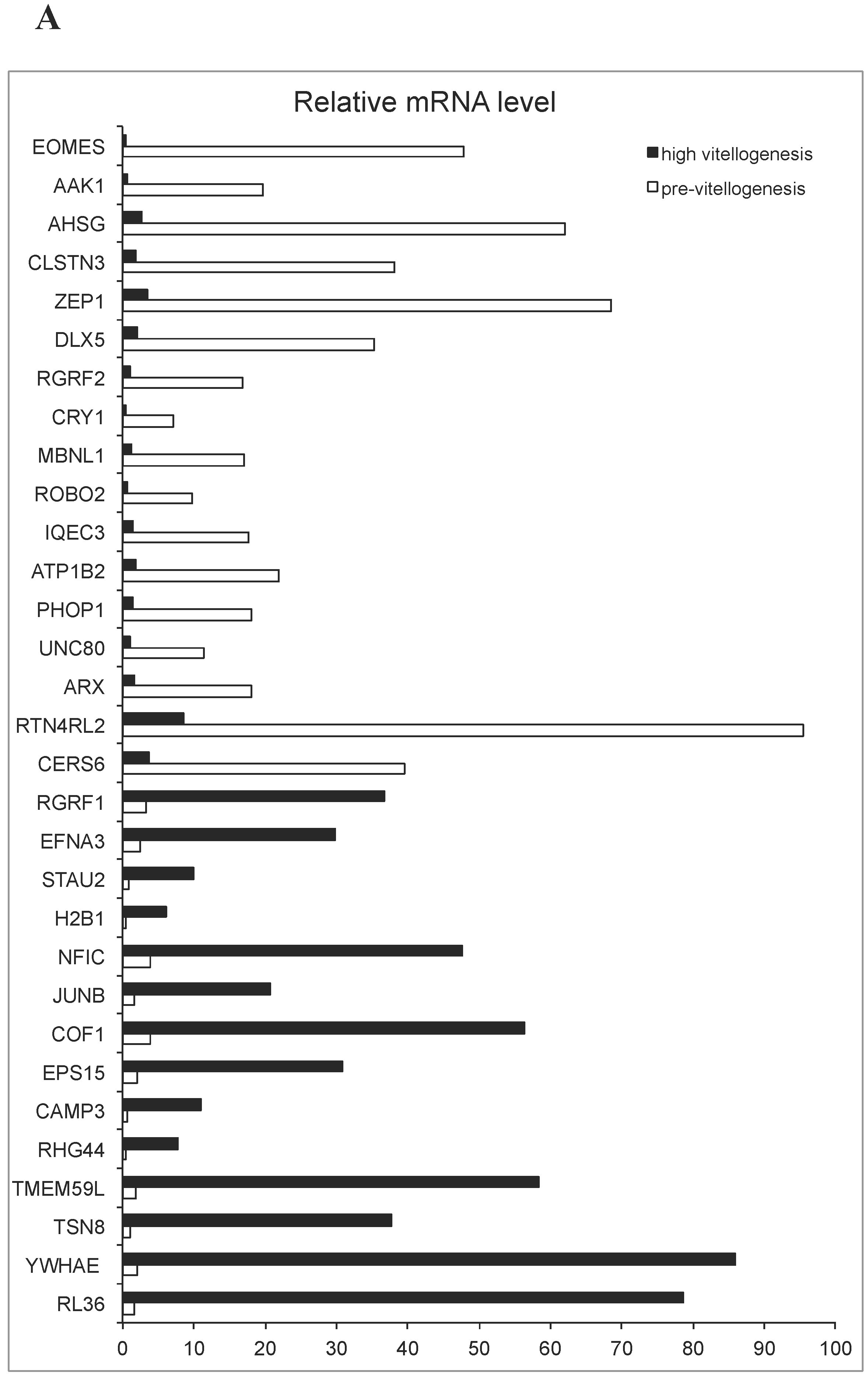
2.2. qRT-PCR
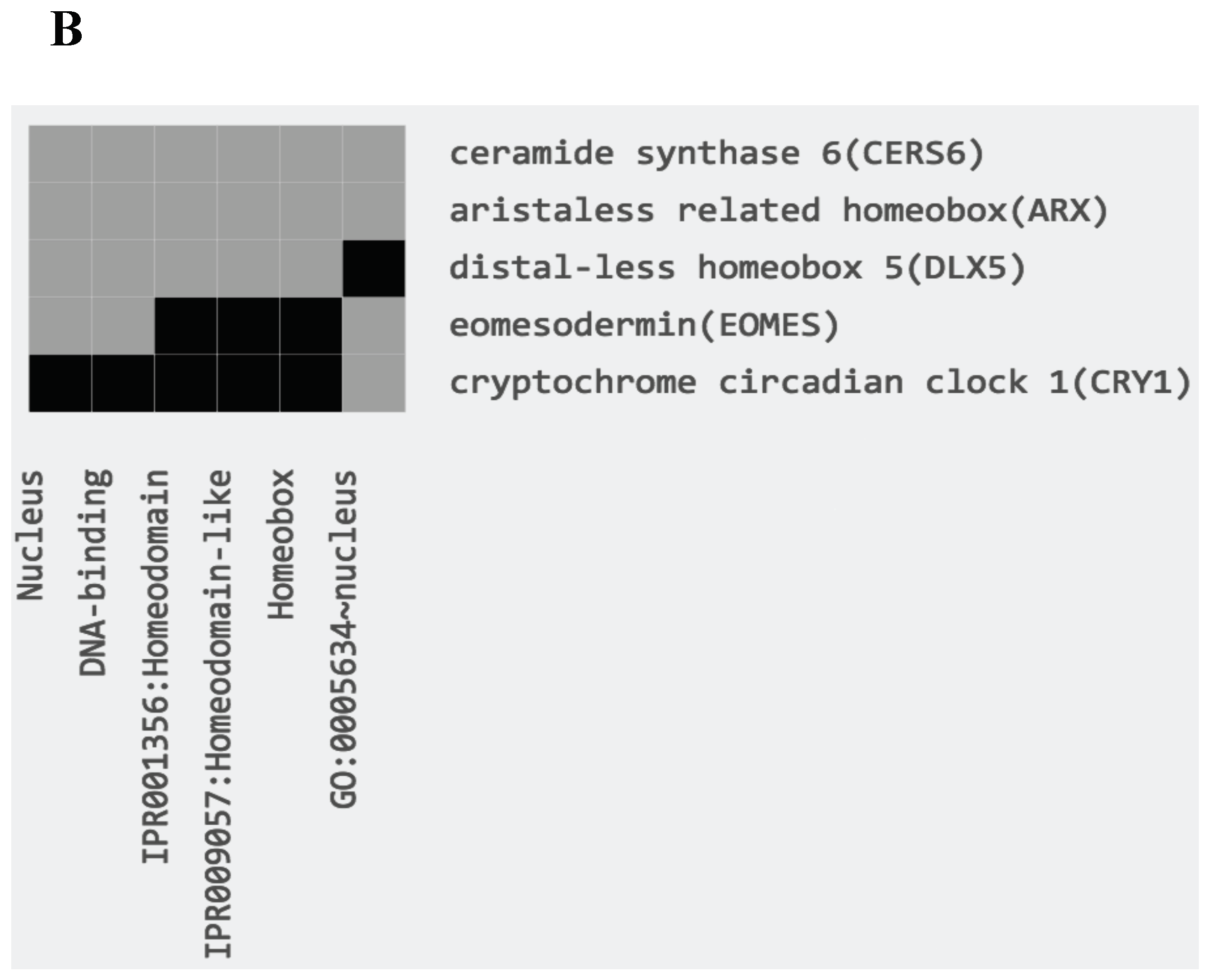
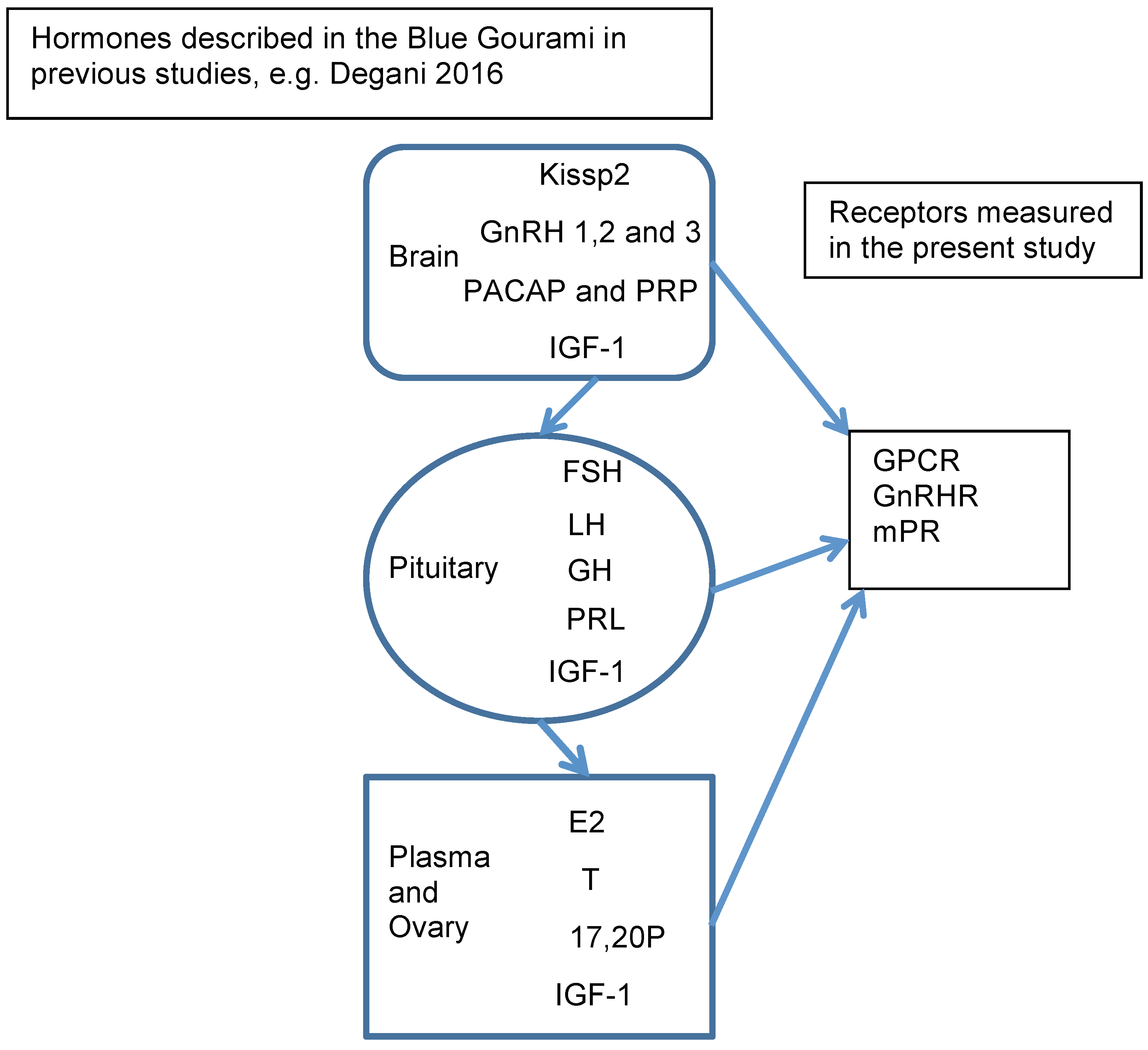
3. Discussion
4. Materials and Methods
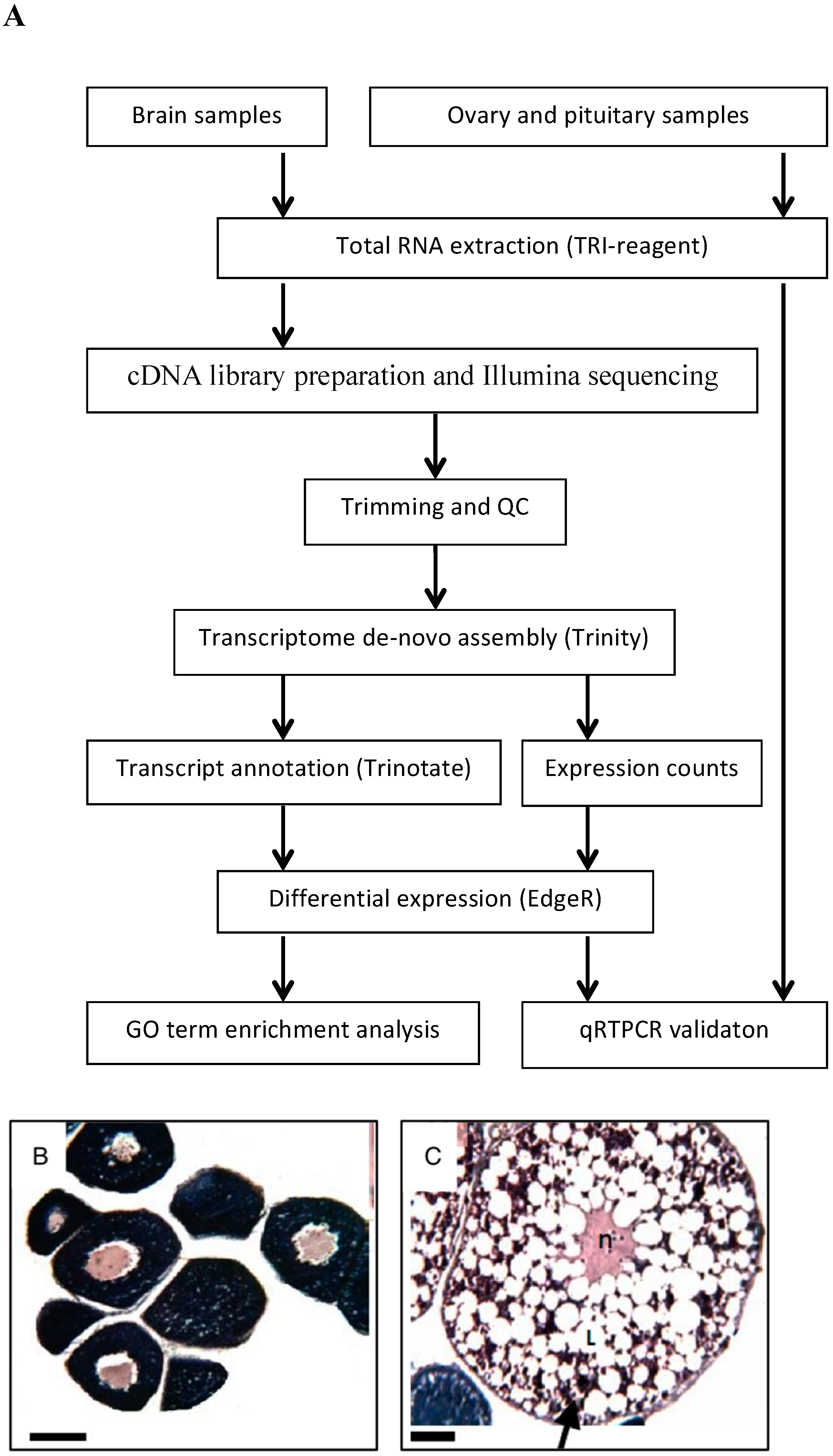
4.1. Fish and Sampling Procedure
4.2. Histological Analysis
4.3. RNA Extraction
4.4. Library Construction, Illumina Sequencing and Transcriptome Assembly
4.5. Functional Annotation
4.6. Gene Expression and Differentially Expressed Genes
4.7. Quantitative RT-PCR
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Degani, G. Oogenesis control in multi-spawning blue gourami (Trichogaster trichopterus) as a model for the Anabantidae family. Int. J. Sci. Res. 2016, 5, 179–184. [Google Scholar]
- Yaron, Z.; Levavi-Sivan, B. Endocrine Regulation of Fish Reproduction. In Encyclopedia of Fish Physiology: From Genome to Environment; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Degani, G. The effects of human chorionic gonadotropin on steroid changes in Trichogaster trichopterus (B & S 1801). Comp. Biochem. Physiol. A Physiol. 1990, 96, 525–528. [Google Scholar]
- Degani, G.; Boker, R. Sensitivity to maturation-inducing steroids and gonadotropin in the oocytes of blue gourami Trichogaster trichopterus (Anabantidae, Pallas, 1770). Gen. Comp. Endocrinol. 1992, 85, 430–439. [Google Scholar] [CrossRef]
- Degani, G.; Jackson, K.; Marmelstein, G. The effect of LHRH analogue on sex steroid profiles in female Trichogaster trichopterus (Anabantidae, Pallas). J. Aquac. Trop. 1995, 10, 297–307. [Google Scholar]
- Degani, G. 11-ketotesterone (KT-11), Estradiol estradiol (E2) level and cytochrome P450 (bgCYP19a) Ttranscription of in the testis of male blue gourami (Trichogaster trichopterus). Int. J. Sci. Res. 2015, 4, 641–643. [Google Scholar]
- Levy, G.; Degani, G. Involvement of GnRH, PACAP and PRP in the Reproduction of Blue Gourami Females (Trichogaster trichopterus). J. Mol. Neurosci. 2012, 48, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Levy, G.; Degani, G. Evidence of a reproduction-related function for pituitary adenylate cyclase-activating polypeptide-related peptide in an Anabantidae fish. J. Mol. Endocrinol. 2011, 46, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Oakley, A.E.; Clifton, D.K.; Steiner, R.A. Kisspeptin Signaling in the Brain. Endocr. Rev. 2009, 30, 713–743. [Google Scholar] [CrossRef]
- Zohar, Y.; Muñoz-Cueto, J.A.; Elizur, A.; Kah, O. Neuroendocrinology of reproduction in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 438–455. [Google Scholar] [CrossRef]
- Servili, A.; Le Page, Y.; Leprince, J.; Caraty, A.; Escobar, S.; Parhar, I.S.; Seong, J.Y.; Vaudry, H.; Kah, O. Organization of Two Independent Kisspeptin Systems Derived from Evolutionary-Ancient Kiss Genes in the Brain of Zebrafish. Endocrinology 2011, 152, 1527–1540. [Google Scholar] [CrossRef] [Green Version]
- Shahjahan, M.; Kitahashi, T.; Parhar, I.S. Central Pathways Integrating Metabolism and Reproduction in Teleosts. Front. Endocrinol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Degani, G.; Alon, A.; Stoler, A.; Berkovitch, D. Evidence of a reproduction-related function for brine kisspeptin 2 and its receptors in anabantidae fish (trichogaster trichopterus). Int. J. Zool. Investig. 2017, 3, 106–122. [Google Scholar]
- Chang, J.P.; Pemberton, J.G. Comparative aspects of GnRH-Stimulated signal transduction in the vertebrate pituitary—Contributions from teleost model systems. Mol. Cell. Endocrinol. 2018, 463, 142–167. [Google Scholar] [CrossRef] [PubMed]
- Degani, G. Effect of Sexual Behavior on Oocyte Development and Steroid Changes in Trichogaster trichopterus (Pallas). Copeia 1993, 1993, 1091–1096. [Google Scholar] [CrossRef]
- Degani, G. Blue Gourami (Trichogaster trichopterus) Model for Labyrinth Fish; Laser Pages Publishing: Jerusalem, Israel, 2001; ISBN 978-965-90044-6-1. [Google Scholar]
- Jackson, K.; Abraham, M.; Degani, G. Oocyte maturation triggered by the presence of male in the Blue Gourami, Trichogaster trichopterus. J. Morphol. 1994, 220, 1–9. [Google Scholar] [CrossRef]
- Becker, D.; Galili, N.; Degani, G. GCMS-indentified steroids and steroid glucoronides in gonads and holding water of Trichogaster trichopterus (Anabantidae, Pallas 1770). Comp. Biochem. Physiol. Part B Comp. Biochem. 1992, 103, 15–19. [Google Scholar] [CrossRef]
- Jackson, K.Z.; Goldberg, D.; Ofir, M.; Abraham, M.; Degani, G. Blue gourami (Trichogaster trichopterus) gonadotropic beta subunits (I and II) cDNA sequences and expression during oogenesis. J. Mol. Endocrinol. 1999, 23, 177–187. [Google Scholar] [CrossRef]
- Degani, G.; Jackson, K.; Goldberg, D.; Sarfati, R.; Avtalion, R.R. βFSH, βLH and Growth Hormone Gene Expression in Blue Gourami (Trichogaster trichopterus, Pallas 1770) During Spermatogenesis and Male Sexual Behavior. Zool. Sci. 2003, 20, 737–743. [Google Scholar] [CrossRef]
- Goldberg, D.; Jackson, K.; Yom-Din, S.; Degani, G. Growth Hormone of Trichogaster trichopterus: cDNA Cloning, Sequencing and Analysis of mRNA Expression during Oogenesis. J. Aquac. Trop. 2004, 19, 215–229. [Google Scholar]
- Degani, G.; Yom-Din, S.; Goldberg, D.; Jackson, K. cDNA cloning of blue gourami (Trichogaster trichopterus) prolactin and its expression during the gonadal cycles of males and females. J. Endocrinol. Investig. 2010, 33, 7–12. [Google Scholar] [CrossRef]
- Drew, R.E.; Settles, M.L.; Churchill, E.J.; Williams, S.M.; Balli, S.; Robison, B.D. Brain transcriptome variation among behaviorally distinct strains of zebrafish (Danio rerio). BMC Genom. 2012, 13, 323. [Google Scholar] [CrossRef] [PubMed]
- Levi, L.; Pekarski, I.; Gutman, E.; Fortina, P.; Hyslop, T.; Biran, J.; Levavi-Sivan, B.; Lubzens, E. Revealing genes associated with vitellogenesis in the liver of the zebrafish (Danio rerio) by transcriptome profiling. BMC Genom. 2009, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.M.; Kille, P.; Workman, V.L.; Paull, G.C.; Tyler, C.R. Sexually dimorphic gene expression in the brains of mature zebrafish. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2008, 149, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Nakajo, M.; Kanda, S.; Karigo, T.; Takahashi, A.; Akazome, Y.; Uenoyama, Y.; Kobayashi, M.; Oka, Y. Evolutionally Conserved Function of Kisspeptin Neuronal System Is Nonreproductive Regulation as Revealed by Nonmammalian Study. Endocrinology 2018, 159, 163–183. [Google Scholar] [CrossRef]
- Tang, H.; Liu, Y.; Luo, D.; Ogawa, S.; Yin, Y.; Li, S.; Zhang, Y.; Hu, W.; Parhar, I.S.; Lin, H.; et al. The kiss/kissr systems are dispensable for zebrafish reproduction: Evidence from gene knockout studies. Endocrinology 2015, 156, 589–599. [Google Scholar] [CrossRef]
- Millar, R.P. GnRHs and GnRH receptors. Anim. Reprod. Sci. 2005, 88, 5–28. [Google Scholar] [CrossRef]
- Degani, G.; Hurvitz, A.; Eliraz, Y.; Meerson, A. Sex-related gonadal gene expression differences in the Russian sturgeon (Acipenser gueldenstaedtii) grown in stable aquaculture conditions. Anim. Reprod. Sci. 2019, 200, 75–85. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-Seq: Reference generation and analysis with Trinity. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degani, G.; Alon, A.; Hajouj, A.; Meerson, A. Vitellogenesis in Blue Gourami is Accompanied by Brain Transcriptome Changes. Fishes 2019, 4, 54. https://doi.org/10.3390/fishes4040054
Degani G, Alon A, Hajouj A, Meerson A. Vitellogenesis in Blue Gourami is Accompanied by Brain Transcriptome Changes. Fishes. 2019; 4(4):54. https://doi.org/10.3390/fishes4040054
Chicago/Turabian StyleDegani, Gad, Amir Alon, Akram Hajouj, and Ari Meerson. 2019. "Vitellogenesis in Blue Gourami is Accompanied by Brain Transcriptome Changes" Fishes 4, no. 4: 54. https://doi.org/10.3390/fishes4040054