Automatic Discrimination between Scomber japonicus and Scomber australasicus by Geometric and Texture Features
Abstract
:1. Introduction
Related Work
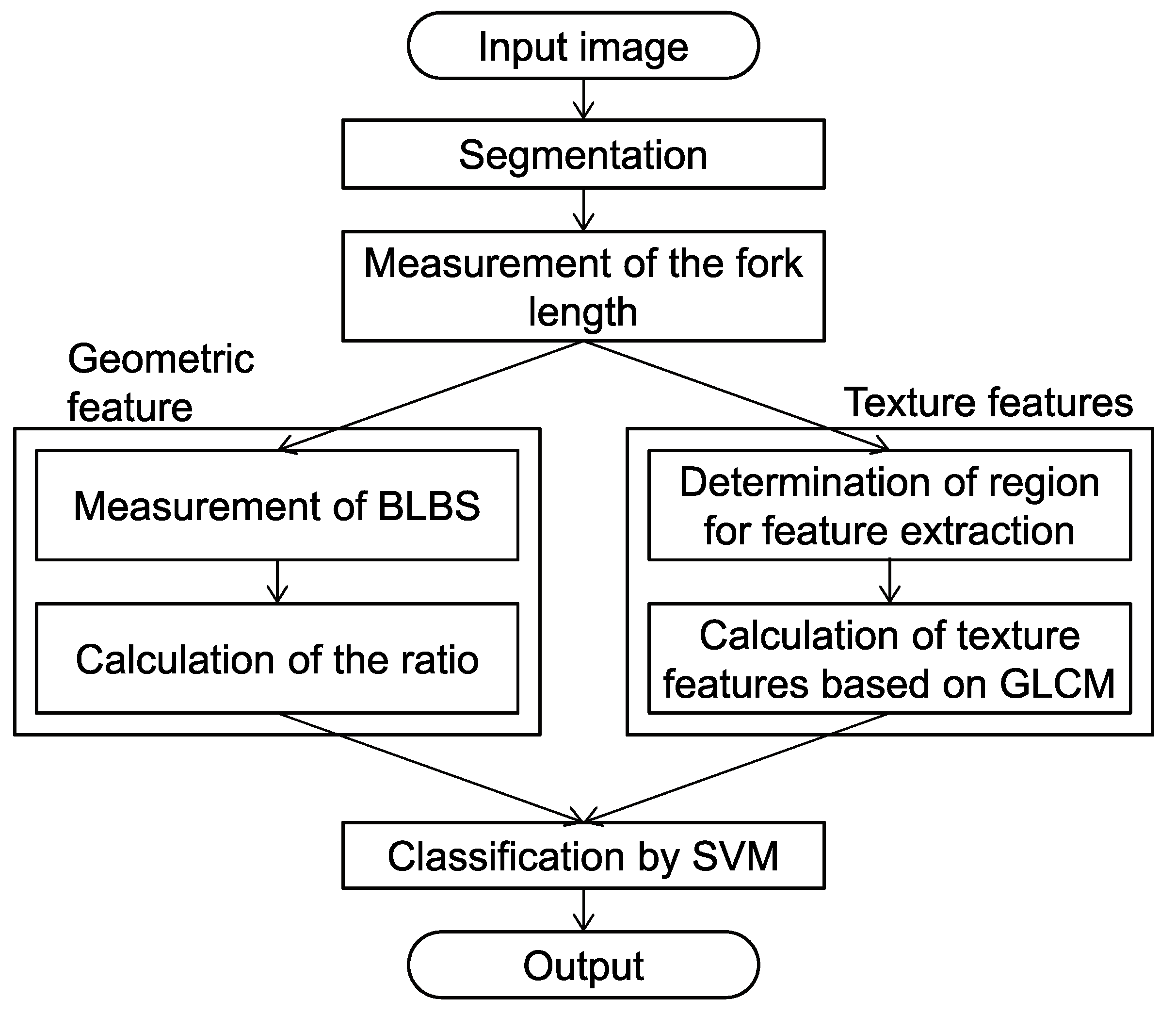
2. Materials and Methods
2.1. Input
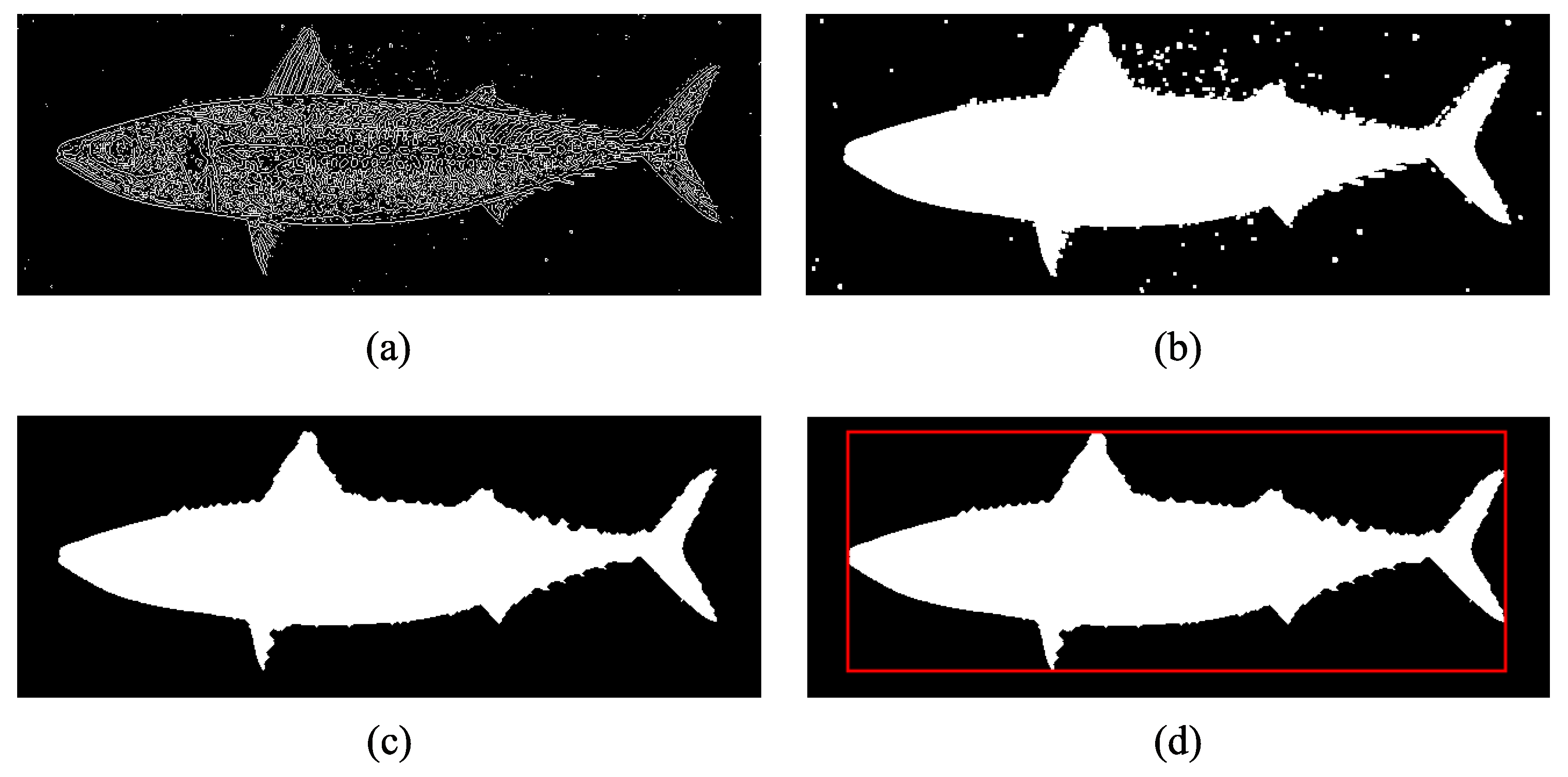
2.2. Segmentation
2.3. Fork-Length Measurement
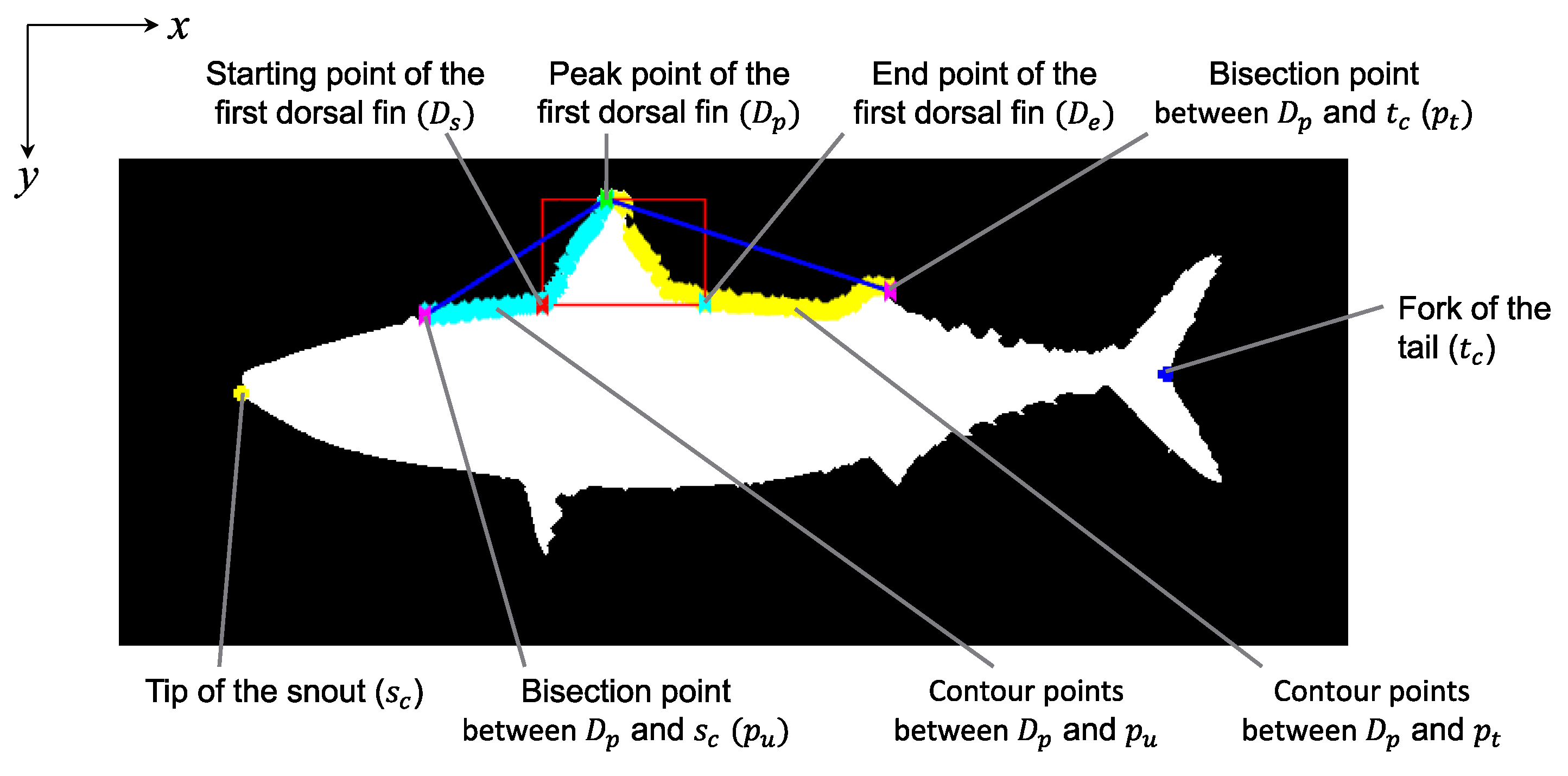
2.4. Measurement of Base Length between Spines
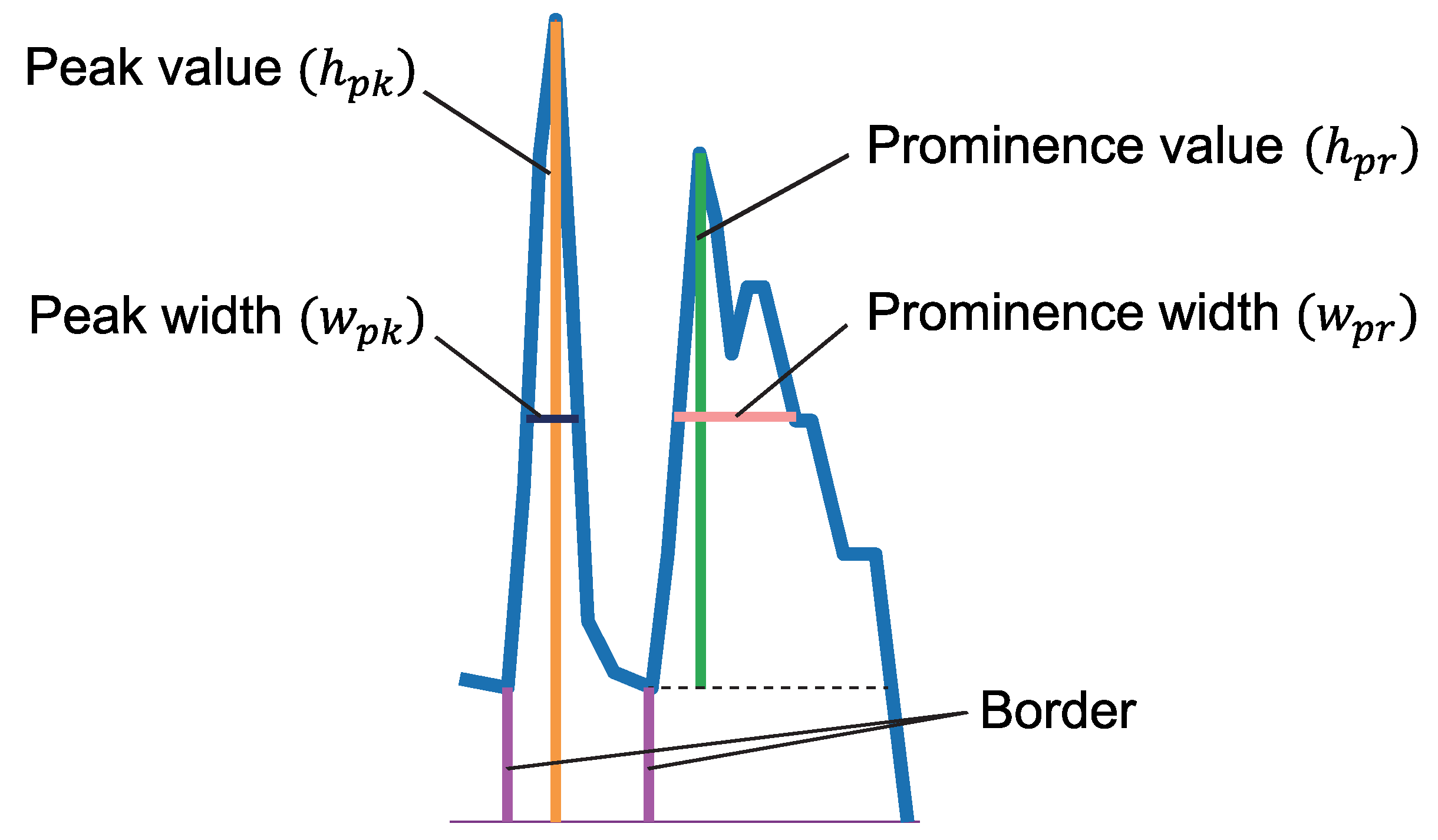
2.4.1. Rough Detection of First Dorsal Fin
2.4.2. Detection of Spine Positions
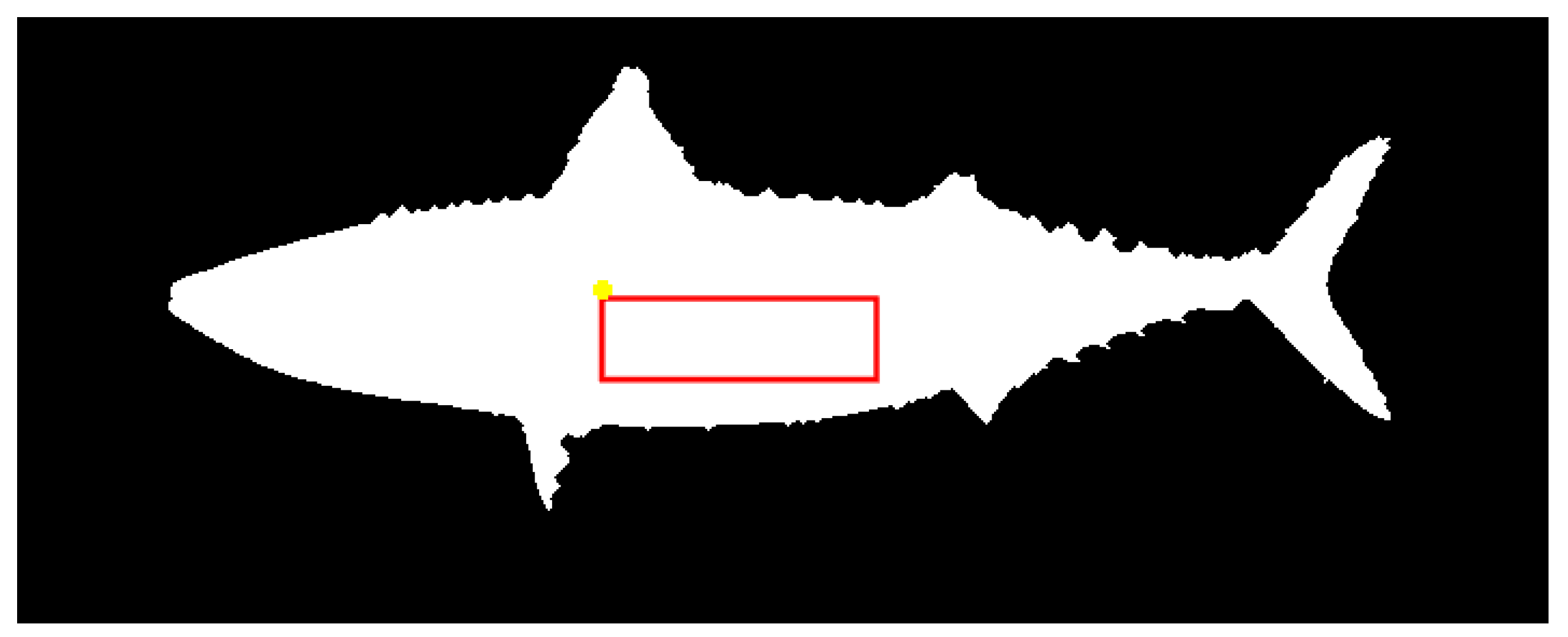
2.5. Texture Feature Extraction
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sassa, C.; Tsukamoto, Y. Distribution and growth of Scomber japonicus and S. australasicus larvae in the southern East China Sea in response to oceanographic conditions. Mar. Ecol. Prog. Ser. 2010, 419, 185–199. [Google Scholar] [CrossRef]
- Baker, E.A.; Collette, B.B. Mackerel from the northern Indian Ocean and the Red Sea are Scomber australasicus, not Scomber japonicus. Ichthyol. Res. 1998, 45, 29–33. [Google Scholar] [CrossRef]
- National Research Institute of Fisheries Science. Manual for Discrimination of Scomber Japonicus and Scomber Australasicus (Masaba Gomasaba Hanbetsu Manyuaru); National Research Institute of Fisheries Science: Yokohama, Japan, 1999. (In Japanese)
- Kitasato, A.; Miyazaki, T.; Sugaya, Y.; Omachi, S. Discrimination of Scomber japonicus and Scomber australasicus by dorsal fin length and fork length. In Proceedings of the 22nd Korea-Japan joint Workshop on Frontiers of Computer Vision, Takayama, Japan, 17–19 February 2016; pp. 338–341. [Google Scholar]
- Rova, A.; Mori, G.; Dill, L.M. One fish, two fish, butterfish, trumpeter: Recognizing fish in underwater video. In Proceedings of the IAPR Conference on Machine Vision Applications, Nagoya, Japan, 8–12 May 2017; pp. 404–407. [Google Scholar]
- Belongie, S.; Malik, J.; Puzicha, J. Shape matching and object recognition using shape contexts. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 509–522. [Google Scholar] [CrossRef] [Green Version]
- Felzenszwalb, P.F.; Huttenlocher, D.P. Pictorial structures for object recognition. Int. J. Comput. Vis. 2005, 61, 55–79. [Google Scholar] [CrossRef]
- Spampinato, C.; Giordano, D.; Di Salvo, R.; Chen-Burger, Y.; Fisher, R.B.; Nadarajan, G. Automatic fish classification for underwater species behavior understanding. In Proceedings of the First ACM International Workshop on Analysis and Retrieval of Tracked Events and Motion in Imagery Streams, Firenze, Italy, 25–29 October 2010; pp. 45–50. [Google Scholar]
- Haralick, R.M.; Shanmugan, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Mokhtarian, F.; Abbasi, S. Shape similarity retrieval under affine transforms. Pattern Recognit. 2002, 35, 31–41. [Google Scholar] [CrossRef]
- Fabic, J.N.; Turla, I.E.; Capacillo, J.A.; David, L.T.; Naval, P.C. Fish population estimation and species classification from underwater video sequences using blob counting and shape analysis. In Proceedings of the 2013 IEEE International Underwater Technology Symposium, Tokyo, Japan, 5–8 March 2013. [Google Scholar]
- Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI-8, 679–698. [Google Scholar] [CrossRef]
- Khotanzad, A.; Hong, Y.H. Invariant image recognition by Zernike moments. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 489–497. [Google Scholar] [CrossRef]
- Hossain, E.; Alam, S.M.S.; Ali, A.A.; Amin, M.A. Fish activity tracking and species identification in underwater video. In Proceedings of the 5th International Conference on Informatics, Electronics and Vision, Dhaka, Bangladesh, 13–14 May 2016. [Google Scholar]
- Bosch, A.; Zisserman, A.; Muñoz, X. Image classification using random forests and ferns. In Proceedings of the IEEE 11th International Conference on Computer Vision, Rio de Janeiro, Brazil, 14–21 October 2007. [Google Scholar]
- Hasija, S.; Buragohain, M.J.; Indu, S. Fish species classification using graph embedding discriminant analysis. In Proceedings of the International Conference on Machine Vision and Information Technology, Singapore, 7–19 February 2017; pp. 81–86. [Google Scholar]
- Fouad, M.M.M.; Zawbaa, H.M.; El-Bendary, N.; Hassanien, A.E. Automatic Nile Tilapia fish classification approach using machine learning techniques. In Proceedings of the 13th International Conference on Hybrid Intelligent Systems, Gammarth, Tunisia, 4–6 December 2013; pp. 173–178. [Google Scholar]
- Lowe, D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Bay, H.; Ess, A.; Tuytelaars, T.; Van Gool, L. Speeded-up robust features. Comput. Vis. Image Underst. 2008, 110, 346–359. [Google Scholar] [CrossRef]
- Vapnik, V.N. Statistical Learning Theory; John Wiley & Sons: Hoboken, NJ, USA, 1998. [Google Scholar]
- Rodrigues, M.T.A.; Freitas, M.H.G.; Pádua, F.L.C.; Gomes, R.M.; Carrano, E.G. Evaluating cluster detection algorithms and feature extraction techniques in automatic classification of fish species. Pattern Anal. Appl. 2015, 18, 783–797. [Google Scholar] [CrossRef]
- Jégou, H.; Douze, M.; Schmid, C.; Pérez, P. Aggregating local descriptors into a compact image representation. In Proceedings of the 2010 IEEE Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010; pp. 3304–3311. [Google Scholar]
- De Castro, L.N.; Von Zuben, F.J. aiNet: An Artificial Immune Network for Data Analysis. In Data Mining: A Heuristic Approach; Idea Group Inc.: London, UK, 2002; pp. 231–260. [Google Scholar]
- Bezerra, G.B.; Barra, T.V.; de Castro, L.N.; Von Zuben, F.J. Adaptive radius immune algorithm for data clustering. In Lecture Notes in Computer Science; Springer: Heidelberg/Berlin, Germany, 2005; pp. 290–303. [Google Scholar]
- Khotimah, W.N.; Arifin, A.Z.; Yuniarti, A.; Wijaya, A.Y.; Navastara, D.A.; Kalbuadi, M.A. Tuna fish classification using decision tree algorithm and image processing method. In Proceedings of the International Conference on Computer, Control, Informatics and Its Applications, Bandung, Indonesia, 5–7 October 2015; pp. 126–131. [Google Scholar]
- Najman, L.; Talbot, H. Mathematical Morphology; Wiley-ISTE: Washington, DC, USA, 2013. [Google Scholar]
- Szeliski, R. Computer Vision; Springer: Heidelberg/Berlin, Germany, 2010. [Google Scholar]
- Niblack, W. An Introduction to Digital Image Processing; Prentice Hall: Upper Saddle River, NJ, USA, 1986. [Google Scholar]
- FishPix. Available online: http://fishpix.kahaku.go.jp/fishimage-e/index.html (accessed on 27 June 2018).








| Feature | d | |
|---|---|---|
| Contrast | 4 | |
| Correlation | 4 | |
| Energy | 1 | |
| Homogeneity | 4 | |
| Entropy | 4 |
| Texture feature | RBF | Linear |
|---|---|---|
| Contrast | 70% | 70% |
| Correlation | 65% | 62% |
| Energy | 70% | 62% |
| Homogeneity | 81% | 81% |
| Entropy | 70% | 68% |
| Contrast + Homogeneity | 84% | 84% |
| All textures | 76% | 76% |
| Parameter | Value |
|---|---|
| 0.51 | |
| 0.65 | |
| 0.85 | |
| 1.626 |
| Method | Accuracy [%] |
|---|---|
| Proposed method | 97% |
| Ratio only | 89% |
| Texture only | 84% |
| Khotimah et al. [25] | 76% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitasato, A.; Miyazaki, T.; Sugaya, Y.; Omachi, S. Automatic Discrimination between Scomber japonicus and Scomber australasicus by Geometric and Texture Features. Fishes 2018, 3, 26. https://doi.org/10.3390/fishes3030026
Kitasato A, Miyazaki T, Sugaya Y, Omachi S. Automatic Discrimination between Scomber japonicus and Scomber australasicus by Geometric and Texture Features. Fishes. 2018; 3(3):26. https://doi.org/10.3390/fishes3030026
Chicago/Turabian StyleKitasato, Airi, Tomo Miyazaki, Yoshihiro Sugaya, and Shinichiro Omachi. 2018. "Automatic Discrimination between Scomber japonicus and Scomber australasicus by Geometric and Texture Features" Fishes 3, no. 3: 26. https://doi.org/10.3390/fishes3030026






