Full-Length Transcriptome Characterization and Molecular Analysis of the Longfin Batfish (Platax teira)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and RNA Extraction
2.2. PacBio Iso-Seq Library Preparation and Sequencing
2.3. Sequencing Data Processing
2.4. Functional Annotation
2.5. Gene Structure Analysis
2.6. Protein–Protein Interaction Analysis and Molecular Characterization of Immune Genes
3. Results
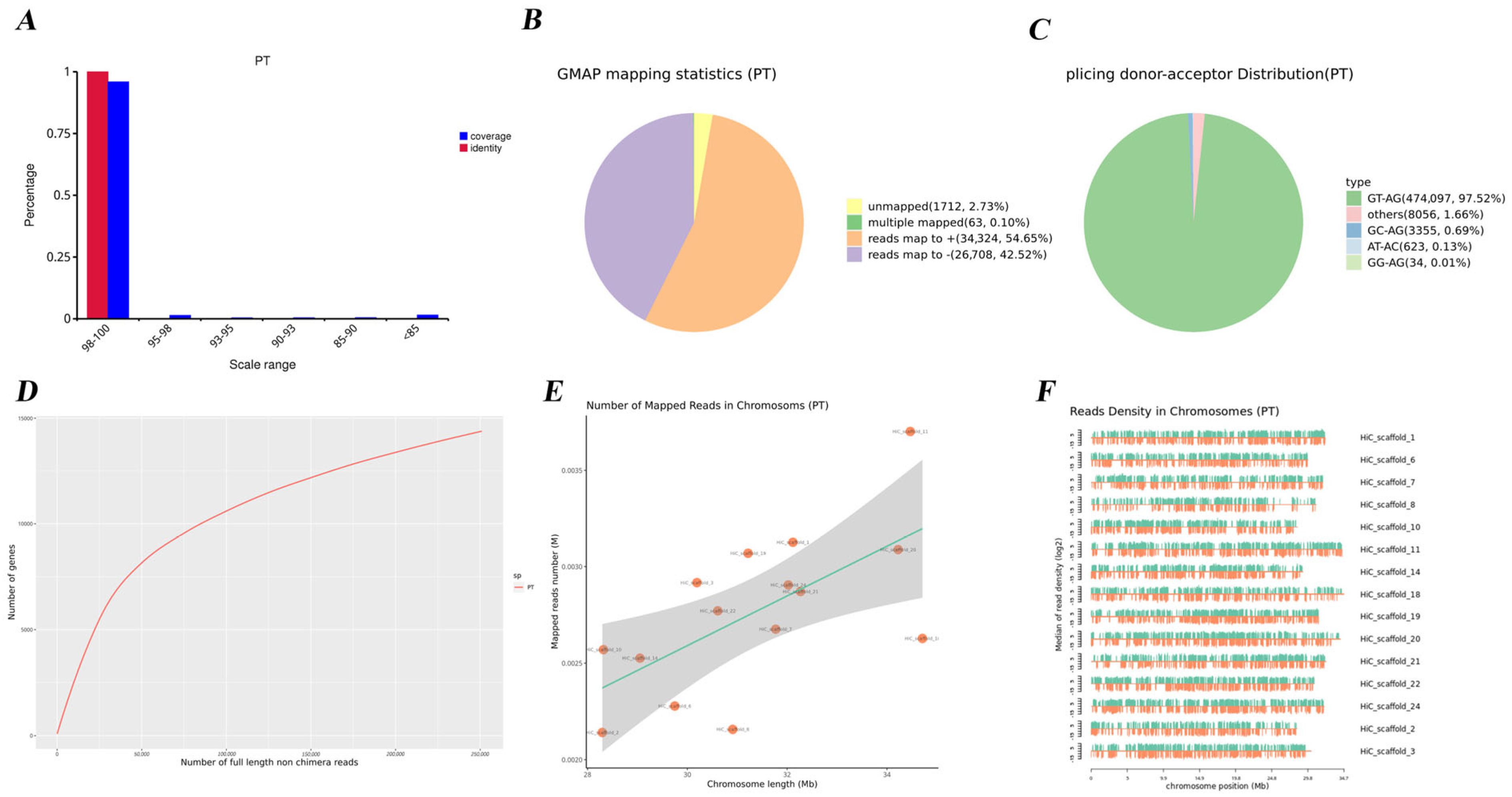
3.1. Quality Control of the Full-Length Transcriptomes
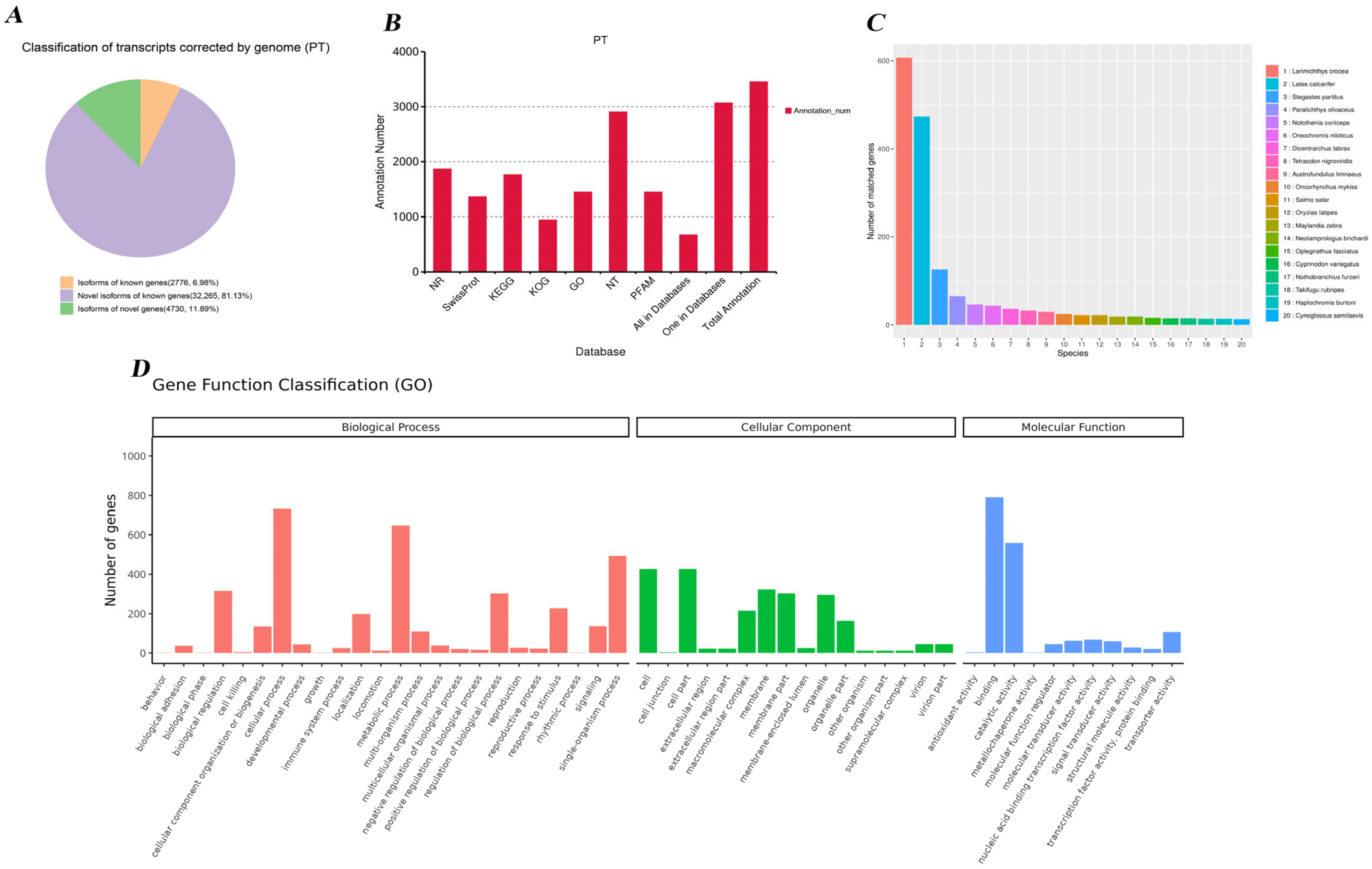
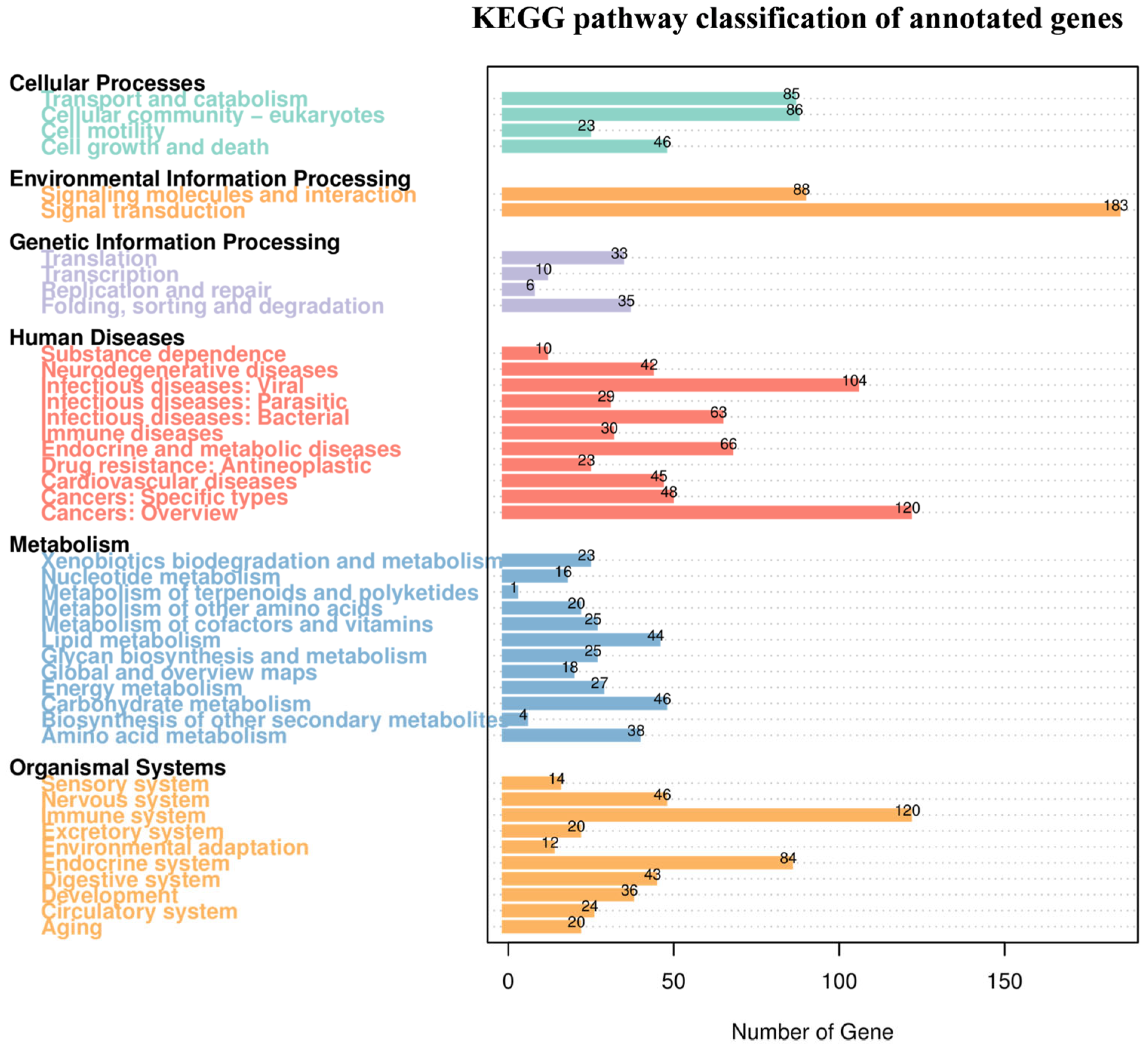
3.2. Transcript Identification and Gene Functional Annotation
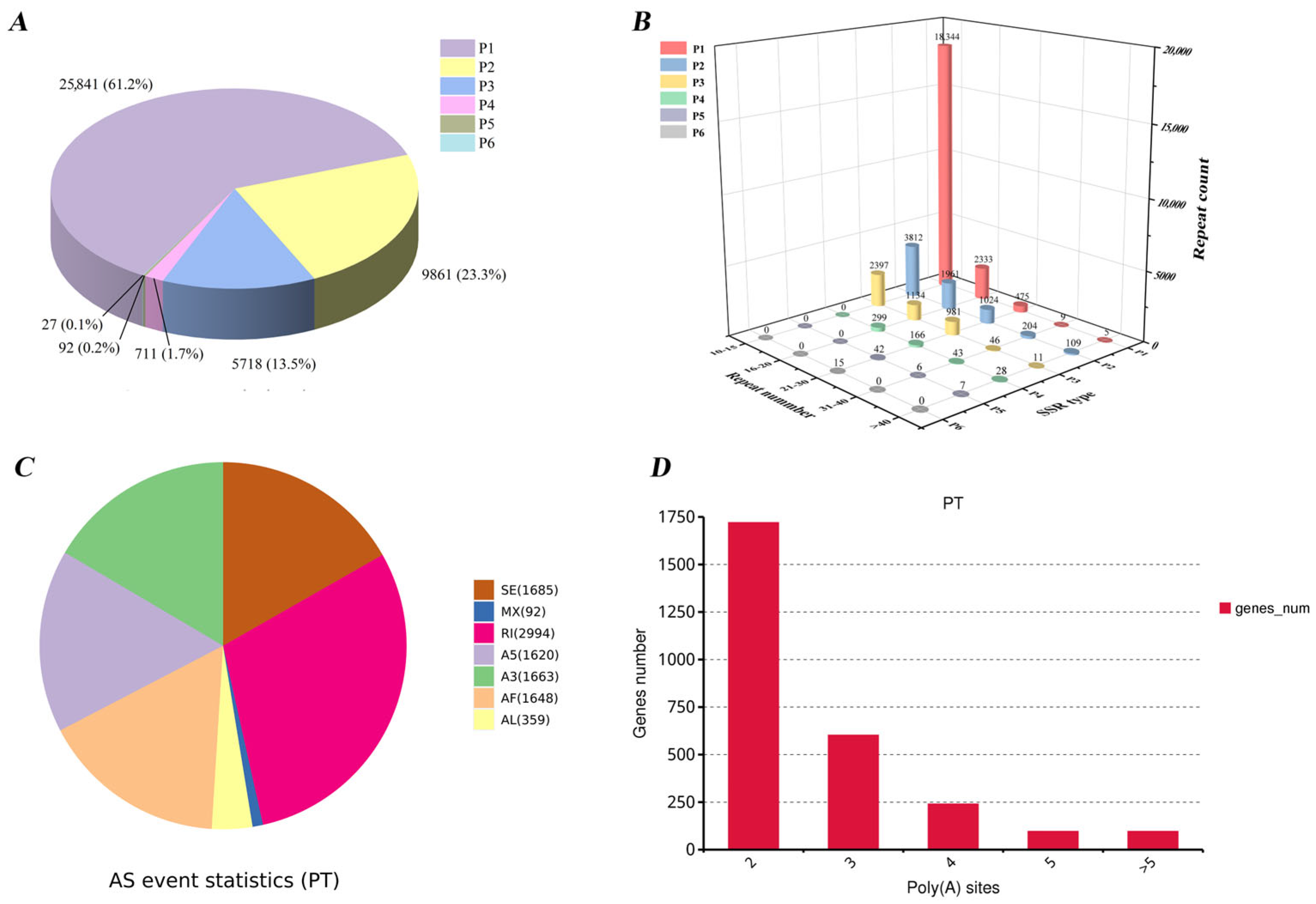
3.3. SSR Detection, Alternative Splicing (AS) Analysis, and Alternative Polyadenylation (APA) Events
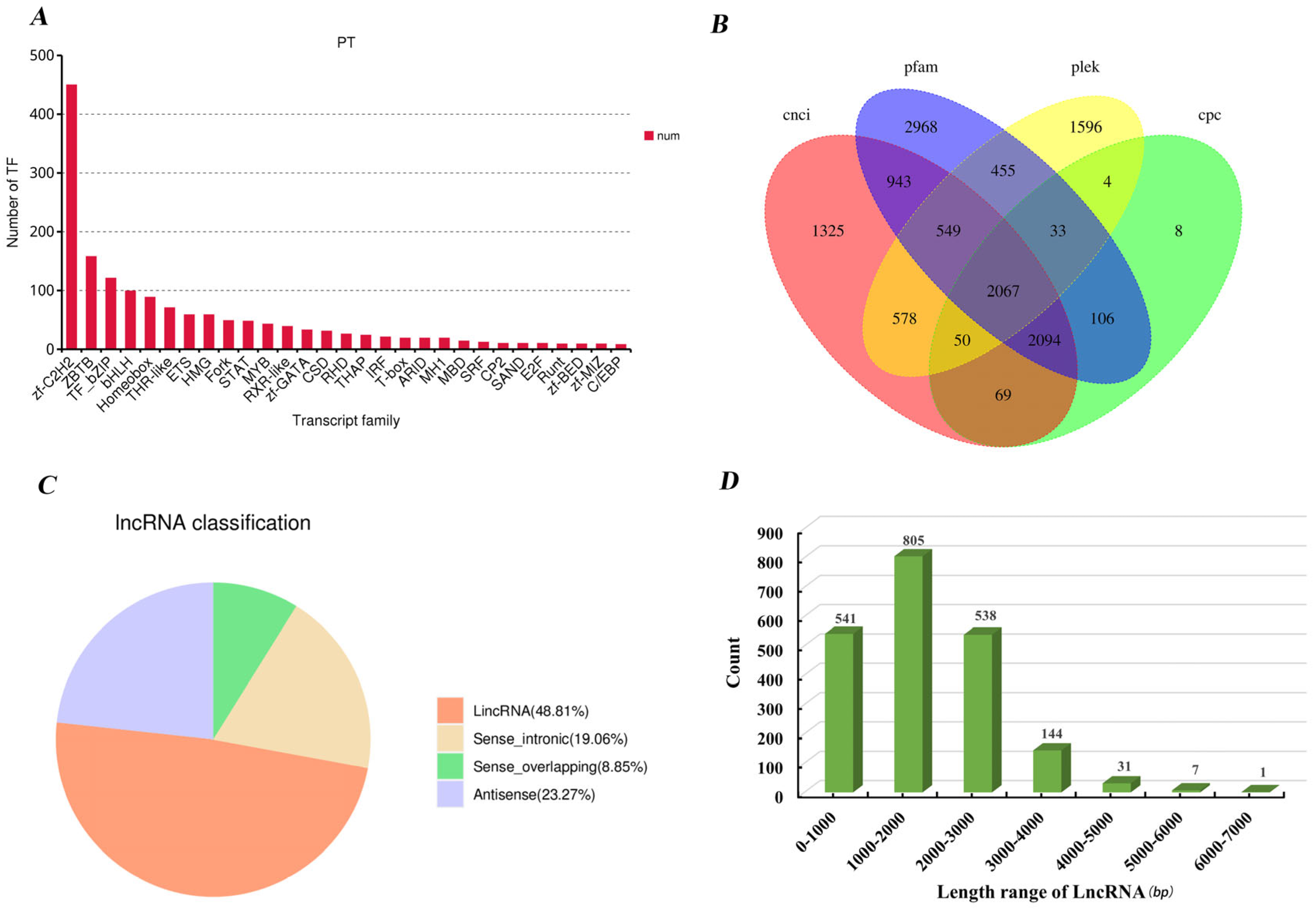
3.4. Transcription Factors (TFs) and Long Non-Coding RNAs
3.5. Screening Key Genes by PPI and Molecular Analysis of Immune Genes
4. Discussion
4.1. Transcriptome Completeness and Annotation Quality
4.2. Functional Implications of Novel Genes
4.3. Simple Sequence Repeats, Transcription Factors, and lncRNAs
4.4. Alternative Splicing and Immune Regulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Priyashadi, M.S.V.H.; Deepananda, K.H.M.A.; Jayasinghe, A. Socio-Economic Development of Marine Ornamental Reef Fish Fishers in Eastern Sri Lanka through the Lenses of Human Development Index. Mar. Policy 2022, 143, 105136. [Google Scholar] [CrossRef]
- Liu, B.; Guo, H.-Y.; Zhu, K.-C.; Liu, B.-S.; Guo, L.; Zhang, N.; Jiang, S.-G.; Zhang, D.-C. Nutritional Compositions in Different Parts of Muscle in the Longfin Batfish, Platax Teira (Forsskål, 1775). J. Appl. Anim. Res. 2019, 47, 403–407. [Google Scholar] [CrossRef]
- Leu, M.-Y.; Tai, K.-Y.; Meng, P.-J.; Tang, C.-H.; Wang, P.-H.; Tew, K.S. Embryonic, Larval and Juvenile Development of the Longfin Batfish, Platax teira (Forsskål, 1775) under Controlled Conditions with Special Regard to Mitigate Cannibalism for Larviculture. Aquaculture 2018, 493, 204–213. [Google Scholar] [CrossRef]
- Li, S.; Xie, Z.; Chen, P.; Tang, J.; Tang, L.; Chen, H.; Wang, D.; Zhang, Y.; Lin, H. The Complete Mitochondrial Genome of the Platax teira (Osteichthyes: Ephippidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 796–797. [Google Scholar] [CrossRef]
- Parker, J.; Roth, O. Comparative Assessment of Immunological Tolerance in Fish with Natural Immunodeficiency. Dev. Comp. Immunol. 2022, 132, 104393. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, W.; Li, Y.; Wang, B.; Shao, Y.; Di, X.; Zhang, H.; Cai, W.; Wei, Y.; Ma, X. Construction of a Full-Length Transcriptome Resource for the African Sharptooth Catfish (Clarias Gariepinus), a Prototypical Air-Breathing Fish, Based on Isoform Sequencing (Iso-Seq). Gene 2024, 930, 148802. [Google Scholar] [CrossRef]
- Chen, X.; Tang, Y.Y.; Yin, H.; Sun, X.; Zhang, X.; Xu, N. A Survey of the Full-Length Transcriptome of Gracilariopsis lemaneiformis Using Single-Molecule Long-Read Sequencing. BMC Plant Biol. 2022, 22, 597. [Google Scholar] [CrossRef]
- Liao, X.; Zhang, L.; Tian, H.; Yang, B.; Wang, E.; Zhu, B. Transcript Annotation of Chinese Sturgeon (Acipenser sinensis) Using Iso-Seq and RNA-Seq Data. Sci. Data 2023, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Zhang, F.; Smith, J.; Kuo, R.; Hou, Z.-C. Full-Length Transcriptome Sequencing from Multiple Tissues of Duck, Anas Platyrhynchos. Sci. Data 2019, 6, 275. [Google Scholar] [CrossRef]
- Tian, K.; Zhang, C.; Gao, C.; Shi, J.; Xu, C.; Xie, W.; Yan, S.; Xiao, C.; Jia, X.; Tian, Y.; et al. Full-Length Transcriptome Sequencing of Seven Tissues of GuShi Chickens. Poult. Sci. 2024, 104, 104697. [Google Scholar] [CrossRef]
- Liu, M.-J.; Gao, J.; Guo, H.-Y.; Zhu, K.-C.; Liu, B.-S.; Zhang, N.; Sun, J.-H.; Zhang, D.-C. Transcriptomics Reveal the Effects of Breeding Temperature on Growth and Metabolism in the Early Developmental Stage of Platax Teira. Biology 2023, 12, 1161. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Watanabe, C.K. GMAP: A Genomic Mapping and Alignment Program for mRNA and EST Sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef]
- Yan, O.; Pan, J.; Lin, X.; Liu, B.; Guo, H.; Zhu, T.; Zhang, N.; Zhu, K.; Zhang, D. Chromosome-Level Genome and Characteristic Analysis of Platax Teira. South China Fish. Sci. 2024, 20, 31–42. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Rogers, M.F.; Thomas, J.; Reddy, A.S.; Ben-Hur, A. SpliceGrapher: Detecting Patterns of Alternative Splicing from RNA-Seq Data in the Context of Gene Models and EST Data. Genome Biol. 2012, 13, R4. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, Accurate, and Uncertainty-Aware Differential Splicing Analysis across Multiple Conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Ben-Hur, A.; Reddy, A.S.N. A Survey of the Sorghum Transcriptome Using Single-Molecule Long Reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef]
- Zhang, H.-M.; Liu, T.; Liu, C.-J.; Song, S.; Zhang, X.; Liu, W.; Jia, H.; Xue, Y.; Guo, A.-Y. AnimalTFDB 2.0: A Resource for Expression, Prediction and Functional Study of Animal Transcription Factors. Nucleic Acids Res. 2015, 43, D76–D81. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the Protein-Coding Potential of Transcripts Using Sequence Features and Support Vector Machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A Tool for Predicting Long Non-Coding RNAs and Messenger RNAs Based on an Improved k-Mer Scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, S.; Shi, Y.; Yan, Y.; Liu, Q. Full-Length Transcriptome of Anadromous Coilia Nasus Using Single Molecule Real-Time (SMRT) Sequencing. Aquac. Fish. 2022, 7, 420–426. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, L.; Hui, M.; Li, Y.; Sha, Z. Insights into Adaptive Divergence of Japanese Mantis Shrimp Oratosquilla Oratoria Inferred from Comparative Analysis of Full-Length Transcriptomes. Front. Mar. Sci. 2022, 9, 975686. [Google Scholar] [CrossRef]
- Wang, A.; Sha, Z.; Hui, M. Full-Length Transcriptome Comparison Provides Novel Insights into the Molecular Basis of Adaptation to Different Ecological Niches of the Deep-Sea Hydrothermal Vent in Alvinocaridid Shrimps. Diversity 2022, 14, 371. [Google Scholar] [CrossRef]
- Fu, Q.; Zhang, P.; Zhao, S.; Li, Y.; Li, X.; Cao, M.; Yang, N.; Li, C. A Novel Full-Length Transcriptome Resource from Multiple Immune-Related Tissues in Turbot (Scophthalmus maximus) Using Pacbio SMART Sequencing. Fish Shellfish. Immunol. 2022, 129, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Zhang, M.; Yang, N.; Fu, Q.; Su, B.; Zhang, X.; Li, Q.; Yan, X.; Thongda, W.; Li, C. Full Length Transcriptome Profiling Reveals Novel Immune-Related Genes in Black Rockfish (Sebastes schlegelii). Fish Shellfish. Immunol. 2020, 106, 1078–1086. [Google Scholar] [CrossRef]
- Ge, H.; Zhang, H.; Yang, L.; Wang, H.; Tu, L.; Jiang, Z.; Zheng, J.; Chen, B.; Chen, J.; Li, Y.; et al. Full-Length Transcriptome Sequencing from the Longest-Lived Freshwater Bony Fish of the World: Bigmouth Buffalo (Ictiobus cyprinellus). Front. Mar. Sci. 2021, 8, 736188. [Google Scholar] [CrossRef]
- Luo, W.; Wu, Q.; Wang, T.; Xu, Z.; Wang, D.; Wang, Y.; Yang, S.; Long, Y.; Du, Z. Full-Length Transcriptome Analysis of Misgurnus anguillicaudatus. Mar. Genom. 2020, 54, 100785. [Google Scholar] [CrossRef]
- Ao, J.; Li, J.; You, X.; Mu, Y.; Ding, Y.; Mao, K.; Bian, C.; Mu, P.; Shi, Q.; Chen, X. Construction of the High-Density Genetic Linkage Map and Chromosome Map of Large Yellow Croaker (Larimichthys crocea). Int. J. Mol. Sci. 2015, 16, 26237–26248. [Google Scholar] [CrossRef]
- Vij, S.; Kuhl, H.; Kuznetsova, I.S.; Komissarov, A.; Yurchenko, A.A.; Van Heusden, P.; Singh, S.; Thevasagayam, N.M.; Prakki, S.R.S.; Purushothaman, K.; et al. Chromosomal-Level Assembly of the Asian Seabass Genome Using Long Sequence Reads and Multi-Layered Scaffolding. PLoS Genet. 2016, 12, e1005954. [Google Scholar] [CrossRef]
- Gratten, J.; Beraldi, D.; Lowder, B.V.; McRae, A.F.; Visscher, P.M.; Pemberton, J.M.; Slate, J. Compelling Evidence That a Single Nucleotide Substitution in TYRP1 Is Responsible for Coat-Colour Polymorphism in a Free-Living Population of Soay Sheep. Proc. Biol. Sci. 2007, 274, 619–626. [Google Scholar] [CrossRef]
- Fujii, R. The Regulation of Motile Activity in Fish Chromatophores. Pigment. Cell Res. 2000, 13, 300–319. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, S.; Xu, J.; Feng, J.; Mahboob, S.; Al-Ghanim, K.A.; Sun, X.; Xu, P. Comparative Transcriptome Analysis Reveals the Genetic Basis of Skin Color Variation in Common Carp. PLoS ONE 2014, 9, e108200. [Google Scholar] [CrossRef]
- Bhatt, P.; Kumaresan, V.; Palanisamy, R.; Ravichandran, G.; Mala, K.; Amin, S.M.N.; Arshad, A.; Yusoff, F.M.; Arockiaraj, J. A Mini Review on Immune Role of Chemokines and Its Receptors in Snakehead Murrel Channa striatus. Fish Shellfish. Immunol. 2018, 72, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chen, C.; Lin, F.; Xu, X.; Zhang, X.; Liu, Y.; Li, C.; Cui, P.; Fu, Q. Characterization, Immune Response and Antibacterial Mechanism of CXCL12 in Black Rockfish (Sebastes schlegelii). Int. J. Biol. Macromol. 2025, 309, 143153. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chai, Y.; Gao, A.; Han, J.; Guo, Y.; Lin, Y.; Wu, L.; Ye, J. CXCL13/CXCR5 Axis Mediates IgM+ B Cell Migration through AKT and STAT3 Signaling Pathways in Nile Tilapia (Oreochromis niloticus). Aquaculture 2024, 591, 741109. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, L.; Wang, H.-Q.; Luo, X.-C.; Dan, X.-M.; Li, Y.-W. Molecular Cloning and Expression Analysis of CCL25 and Its Receptor CCR9s from Epinephelus coioides Post Cryptocaryon irritans Infection. Fish Shellfish. Immunol. 2017, 67, 402–410. [Google Scholar] [CrossRef]
- Han, Z.; Xiao, S.; Li, W.; Ye, K.; Wang, Z.Y. The Identification of Growth, Immune Related Genes and Marker Discovery through Transcriptome in the Yellow Drum (Nibea albiflora). Genes Genom. 2018, 40, 881–891. [Google Scholar] [CrossRef]
- Li, C.; Teng, T.; Shen, F.; Guo, J.; Chen, Y.; Zhu, C.; Ling, Q. Transcriptome Characterization and SSR Discovery in Squaliobarbus Curriculus. J. Ocean. Limnol. 2019, 37, 235–244. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, P.; Mo, Q.; Luo, W.; Du, Z.; Jiang, J.; Yang, S.; Zhao, L.; Gong, Q.; Wang, Y. Comprehensive Analysis of Full-Length Transcriptomes of Schizothorax prenanti by Single-Molecule Long-Read Sequencing. Genomics 2022, 114, 456–464. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, H.; Chen, Y.; Song, M.; Liu, B.; Song, J.; Liu, X.; Li, H. Full-Length Transcriptome Sequencing and Identification of Immune-Related Genes in the Critically Endangered Hucho bleekeri. Dev. Comp. Immunol. 2021, 116, 103934. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, L.; Huang, S.; Wang, G. Full-Length Transcriptome Sequencing of Heliocidaris crassispina Using PacBio Single-Molecule Real-Time Sequencing. Fish Shellfish. Immunol. 2022, 120, 507–514. [Google Scholar] [CrossRef]
- Wu, X.; Gong, Q.; Chen, Y.; Liu, Y.; Song, M.; Li, F.; Li, P.; Lai, J. Full-Length Transcriptome and Analysis of Bmp-Related Genes in Platypharodon extremus. Heliyon 2022, 8, e10783. [Google Scholar] [CrossRef] [PubMed]
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription Factors: Bridge between Cell Signaling and Gene Regulation. Proteomics 2021, 21, e2000034. [Google Scholar] [CrossRef] [PubMed]
- Rosewell Shaw, A.; Bennett, A.; Fan, D.; Ai, W. Alternative Splicing of KLF4 in Myeloid Cells: Implications for Cellular Plasticity and Trained Immunity in Cancer and Inflammatory Disease. Front. Immunol. 2025, 16, 1585528. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.M.; Adams, H.; Magor, G.W.; Perkins, A.C. KLF Feedback Loops in Innate Immunity. Front. Immunol. 2025, 16, 1606277. [Google Scholar] [CrossRef]
- Huang, Y.; Ren, Q. A Kruppel-like Factor from Macrobrachium rosenbergii (MrKLF) Involved in Innate Immunity against Pathogen Infection. Fish Shellfish. Immunol. 2019, 95, 519–527. [Google Scholar] [CrossRef]
- Ren, Q.; Liu, Z.; Wu, L.; Yin, G.; Xie, X.; Kong, W.; Zhou, J.; Liu, S. C/EBPβ: The Structure, Regulation, and Its Roles in Inflammation-Related Diseases. Biomed. Pharmacother. 2023, 169, 115938. [Google Scholar] [CrossRef]
- Özdemir, S.; Aydın, Ş.; Laçin, B.B.; Arslan, H. Identification and Characterization of Long Non-Coding RNA (lncRNA) in Cypermethrin and Chlorpyrifos Exposed Zebrafish (Danio rerio) Brain. Chemosphere 2023, 344, 140324. [Google Scholar] [CrossRef]
- Ren, Y.; Li, J.; Guo, L.; Liu, J.N.; Wan, H.; Meng, Q.; Wang, H.; Wang, Z.; Lv, L.; Dong, X.; et al. Full-Length Transcriptome and Long Non-Coding RNA Profiling of Whiteleg Shrimp Penaeus vannamei Hemocytes in Response to Spiroplasma eriocheiris Infection. Fish Shellfish. Immunol. 2020, 106, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, W.; Jiang, L.; Lü, Z.; Liu, M.; Gong, L.; Liu, B.; Liu, L.; Yin, X. Immunity-Associated Long Non-Coding RNA and Expression in Response to Bacterial Infection in Large Yellow Croaker (Larimichthys crocea). Fish Shellfish. Immunol. 2019, 94, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Zhang, A.; Yang, S.; Li, H. Role of Alternative Splicing in Fish Immunity. Fish Shellfish. Immunol. 2024, 149, 109601. [Google Scholar] [CrossRef]
- Zhang, H.; Jia, J.; Zhai, J. Plant Intron-Splicing Efficiency Database (PISE): Exploring Splicing of ∼1,650,000 Introns in Arabidopsis, Maize, Rice, and Soybean from ~57,000 Public RNA-Seq Libraries. Sci. China Life Sci. 2023, 66, 602–611. [Google Scholar] [CrossRef]
- Schmitz, U.; Pinello, N.; Jia, F.; Alasmari, S.; Ritchie, W.; Keightley, M.-C.; Shini, S.; Lieschke, G.J.; Wong, J.J.-L.; Rasko, J.E.J. Intron Retention Enhances Gene Regulatory Complexity in Vertebrates. Genome Biol. 2017, 18, 216. [Google Scholar] [CrossRef]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.N.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative Splicing and Protein Diversity: Plants Versus Animals. Front. Plant Sci. 2019, 10, 708. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative Isoform Regulation in Human Tissue Transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Ren, F.; Zhang, N.; Zhang, L.; Miller, E.; Pu, J.J. Alternative Polyadenylation: A New Frontier in Post Transcriptional Regulation. Biomark. Res. 2020, 8, 67. [Google Scholar] [CrossRef]
- Tan, S.; Zhang, J.; Peng, Y.; Du, W.; Yan, J.; Fang, Q. Integrative Transcriptome Analysis Reveals Alternative Polyadenylation Potentially Contributes to GCRV Early Infection. Front. Microbiol. 2023, 14, 1269164. [Google Scholar] [CrossRef]
- Liu, D.; Yu, H.; Xue, N.; Bao, H.; Gao, Q.; Tian, Y. Alternative Splicing Patterns of Hnrnp Genes in Gill Tissues of Rainbow Trout (Oncorhynchus mykiss) during Salinity Changes. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2024, 271, 110948. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Xiong, F.; Wu, X.M.; Hu, Y.W. The Expanding and Function of NLRC3 or NLRC3-like in Teleost Fish: Recent Advances and Novel Insights. Dev. Comp. Immunol. 2021, 114, 103859. [Google Scholar] [CrossRef]
- Wang, T.; Yan, B.; Lou, L.; Lin, X.; Yu, T.; Wu, S.; Lu, Q.; Liu, W.; Huang, Z.; Zhang, M.; et al. Nlrc3-like Is Required for Microglia Maintenance in Zebrafish. J. Genet. Genom. 2019, 46, 291–299. [Google Scholar] [CrossRef]
- Fang, H.; Wu, X.M.; Hu, Y.W.; Song, Y.J.; Zhang, J.; Chang, M.X. NLRC3-like 1 Inhibits NOD1-RIPK2 Pathway via Targeting RIPK2. Dev. Comp. Immunol. 2020, 112, 103769. [Google Scholar] [CrossRef]
- Li, F.-L.; Wu, X.-J.; Feng, H.-Y.; Liu, X.-S.; Pan, X.-Y.; Chen, Q.-L.; Li, R.; Wang, C. Harnessing Tyrosine Kinase 2: Breakthroughs in Autoimmune Disease and Cancer Therapy. Bioorganic Chem. 2025, 164, 108802. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Zhu, Y.; Yan, J.; Zhang, L.; Zhu, S.; An, L.; Meng, Q.; Zhang, Z.; Wang, X. Full-Length Transcriptome Sequencing Analysis Reveals Differential Skin Color Regulation in Snakeheads Fish Channa argus. Aquac. Fish. 2024, 9, 590–596. [Google Scholar] [CrossRef]
- Vandooren, J.; Itoh, Y. Alpha-2-Macroglobulin in Inflammation, Immunity and Infections. Front. Immunol. 2021, 12, 803244. [Google Scholar] [CrossRef]
- Ning, J.; Liu, Y.; Gao, F.; Song, C.; Cui, Z. Two Alpha-2 Macroglobulin from Portunus trituberculatus Involved in the Prophenoloxidase System, Phagocytosis and Regulation of Antimicrobial Peptides. Fish Shellfish. Immunol. 2019, 89, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Mohindra, V.; Dangi, T.; Chowdhury, L.M.; Jena, J.K. Tissue Specific Alpha-2-Macroglobulin (A2M) Splice Isoform Diversity in Hilsa Shad, Tenualosa Ilisha (Hamilton, 1822). PLoS ONE 2019, 14, e0216144. [Google Scholar] [CrossRef] [PubMed]
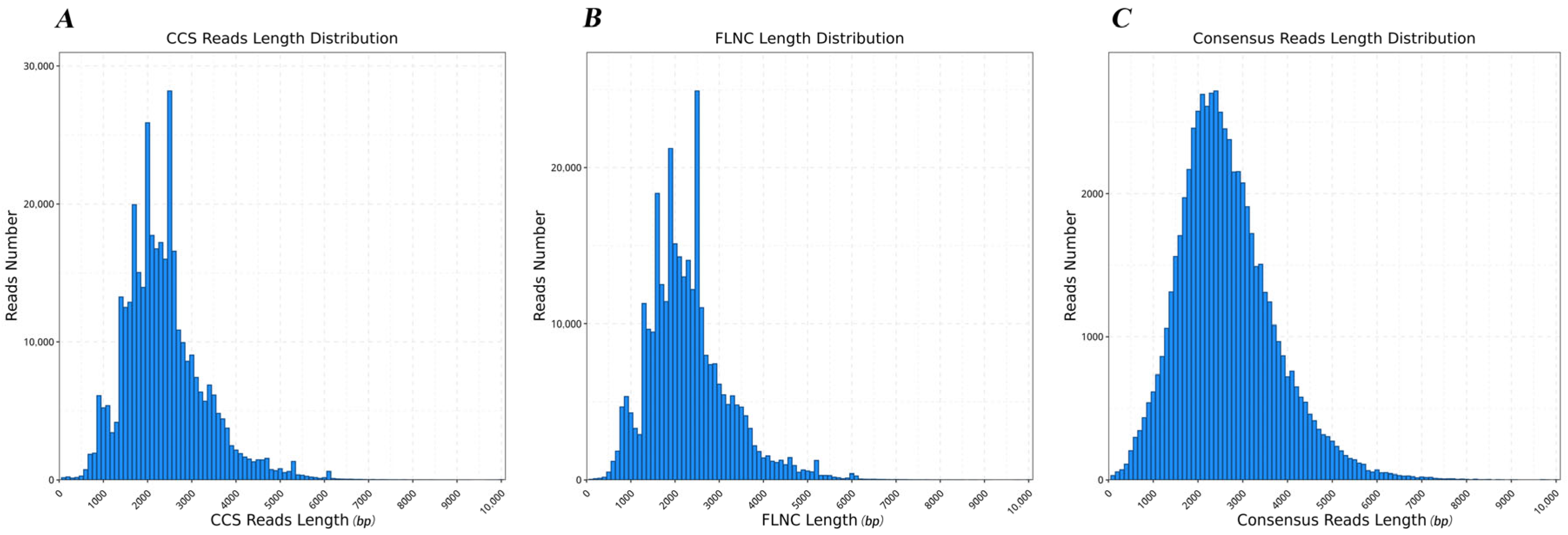


| Type | Total Number | Min Length | Average Length | Max Length | N50 |
|---|---|---|---|---|---|
| Subread | 25,608,838 | - | 2222 | - | 2448 |
| CCS | 362,543 | 50 | 2333 | 11,729 | 2496 |
| FLNC | 304,085 | 50 | 2250 | 10,126 | 2421 |
| Polished consensus | 62,807 | 50 | 2588 | 10,081 | 2881 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, L.; Liu, B.; Guo, H.; Zhu, K.; Zhang, N.; Sun, J.; Zhang, D. Full-Length Transcriptome Characterization and Molecular Analysis of the Longfin Batfish (Platax teira). Fishes 2025, 10, 575. https://doi.org/10.3390/fishes10110575
Lv L, Liu B, Guo H, Zhu K, Zhang N, Sun J, Zhang D. Full-Length Transcriptome Characterization and Molecular Analysis of the Longfin Batfish (Platax teira). Fishes. 2025; 10(11):575. https://doi.org/10.3390/fishes10110575
Chicago/Turabian StyleLv, Lingeng, Baosuo Liu, Huayang Guo, Kecheng Zhu, Nan Zhang, Jinhui Sun, and Dianchang Zhang. 2025. "Full-Length Transcriptome Characterization and Molecular Analysis of the Longfin Batfish (Platax teira)" Fishes 10, no. 11: 575. https://doi.org/10.3390/fishes10110575
APA StyleLv, L., Liu, B., Guo, H., Zhu, K., Zhang, N., Sun, J., & Zhang, D. (2025). Full-Length Transcriptome Characterization and Molecular Analysis of the Longfin Batfish (Platax teira). Fishes, 10(11), 575. https://doi.org/10.3390/fishes10110575








