Neonatal alloimmune thrombocytopenia (NAIT) results from the maternal immune response against fetal-specific antigens inherited from the father. NAIT is the most common cause of severe neonatal thrombocytopenia in maternity wards. The diagnosis is ascertained only when the maternal alloantibody and the offending antigen present in the newborn are identified [
1,
2]. Platelet phenotyping is the best way to identify potentially immunogenic peptides. The main limitation for platelet phenotyping is the scarcity of specific, good-quality typing sera, especially for rare platelet antigens [
3,
4,
5]. Consequently, genotyping is the routine method for platelet typing. Up until now most laboratories perform DNA extraction from newborn blood samplings. However, this invasive strategy is slightly traumatic for the newborn who is already admitted in the intensive care unit due to severe thrombocytopenia. Therefore, a non-invasive strategy has been set up for extracting DNA from buccal swabs.
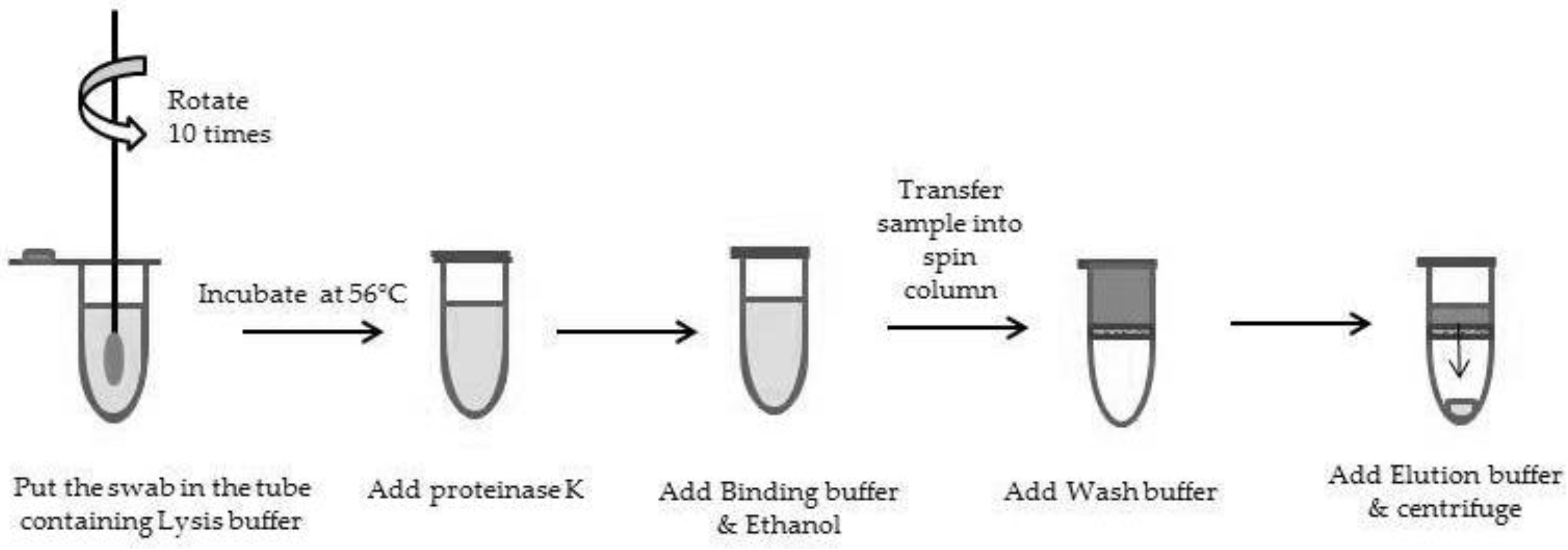
To demonstrate the reliability of such methodology for the genotyping of newborns, both EDTA-blood samplings and buccal swabs from thrombocytopenic newborns were collected in 2012 and 2013. The ethical approval of parents was obtained for all samples included in the study. Two distinct methods have been evaluated. The first method was manual, based on DNA extraction with the QIAamp mini blood kit (Qiagen, Courtaboeuf, France;
Figure 1). The manufacturers’ instructions were followed except for a slight modification of a longer swab pre-incubation in 400 µL PBS (overnight instead of a 10 min preincubation at 56 °C), which allows testing samples collected a few days before delivery in our laboratory. The second method was an automated one (MagNA Pure Compact instrument, Roche) based on the MagNA Pure Compact Nucleic Acid Kit I. The methodology is as follows: (1) a first incubation of the buccal swab in 500 µL PBS over 30 min at 56 °C after a vortex of 15 s (2 mL microtube); (2) centrifugation of the microtube 1 min at 600 g to collect as much volume as possible; (3) DNA extraction by the MagNA Pure Compact instrument (protocol: “Total_NA_Plasma_100_400”; sample volume of 400 µL; elution volume of 50 µL). DNA was quantified using Qubit technology (Thermoscientific, Villebon sur Yvette, France). Both DNA extracted from buccal swabs and from blood samplings were genotyped using BioArray technology (HPA BeadChip, Immucor).
A total of 31 newborns were included in the study. All DNAs extracts from buccal swabs gave very low concentrations, below the recommended concentration given by the instructions for use of BioArray technology (<10 ng/µL). Genotyping was performed without diluting the DNA. Using the Qiagen protocol, 14 DNAs were extracted from buccal swabs. In 13 of 14 cases, concordant genotyping results were obtained between DNA extracted from buccal swabs and DNA extracted from EDTA-blood samplings. In only one case out of 14 samplings, HPA genotyping failed due to a high background with the BioArray assay and a low DNA amplification during the polymerase chain reaction (PCR). With the automated protocol (Roche Diagnostic, Meylan, France), 15 DNAs were extracted from swabs, and genotypings were concordant with those from EDTA-blood samplings. For the last two newborns, two buccal swabs were referred to our laboratory in parallel with blood sampling, which allowed extracting DNA with both manual and automated methods of DNA extraction. Genotyping results were concordant.
Taking into account these results, the automated DNA extraction from newborn buccal swabs with the MagNA Pure Compact instrument was chosen as the first-line strategy, with a significant gain of time in processing buccal swabs (30 min of incubation, compared with overnight for manual DNA extraction). This strategy allows HPA genotyping within one day. In the case of genotyping failure, a neonatal blood sampling is requested from the intensive care unit.
This non-invasive strategy for HPA genotyping is also suitable for parental genotyping during platelet immunological investigations for NAIT. However, the identification of the maternal alloantibody requires testing the maternal serum not only against a platelet panel but also against paternal platelets to detect the involvement of private or rare antigens.
To conclude, our results are in favor of DNA extraction from buccal swabs allowing non-invasive HPA genotyping of thrombocytopenic newborns. This strategy can be offered to other pathologies where diagnosis requires newborn DNA.
Acknowledgments
This work has been done at Institut National de Transfusion Sanguine, Paris, France. We greatly thank T. Beranger, F. Bianchi, C. Chenet, V. Dufour, N. Ferre, N. Giuliani, J. Quesne and S. Philippe for technical assistance, and E. Le Toriellec and C. Martageix for sample collection.
Author Contributions
Gérald Bertrand designed the protocols, performed the experiments, analyzed data and wrote the manuscript; Cécile Kaplan designed the study and wrote the manuscript.
Conflict of Interest
The authors declare no conflict of interest.
References
- Kaplan, C.; Daffos, F.; Forestier, F.; Morel, M.C.; Chesnel, N.; Tchernia, G. Current trends in neonatal alloimmune thrombocytopenia diagnosis and therapy. In Platelet Immunology: Fundamental and Clinical Aspects; John Libbey-Eurotex/ed.INSERM: Paris, France, 1991; pp. 267–278. [Google Scholar]
- Bertrand, G.; Kaplan, C. How do we treat fetal and neonatal alloimmune thrombocytopenia? Transfusion 2014, 54, 1698–1703. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, G.; Bianchi, F.; Alexandre, M.; Quesne, J.; Chenet, C.; Martageix, C.; Jallu, V.; Kaplan, C. HPA-13bw neonatal alloimmune thrombocytopenia and low frequency alloantigens: Case report and review of the literature. Transfusion 2007, 47, 1510–1513. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, G.; Bianchi, F.; Quesne, J.; Philippe, S.; Chenet, C.; Martageixm, C.; Kaplan, C. A severe case of neonatal thrombocytopenia with a fatal outcome, implicating HPA-12bw fetomaternal platelet alloimmunization. Transfusion 2013, 53, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, C.; Porcelijn, L.; Vanlieferinghen, P.; Julien, E.; Bianchi, F.; Martageix, C.; Bertrand, G.; Jallu, V. Anti-HPA-9bw (Maxa) fetomaternal alloimmunization, a clinically severe neonatal thrombocytopenia: Difficulties in diagnosis and therapy and report on eight families. Transfusion 2005, 45, 1799–1803. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).