Protein Extract of Tobacco Expressing Solanum torvum PP5-Encoding Gene Inhibits Verticillium dahliae Proliferation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Isolation of Full-Length cDNA of StPP5
2.3. Bioinformatics Analysis
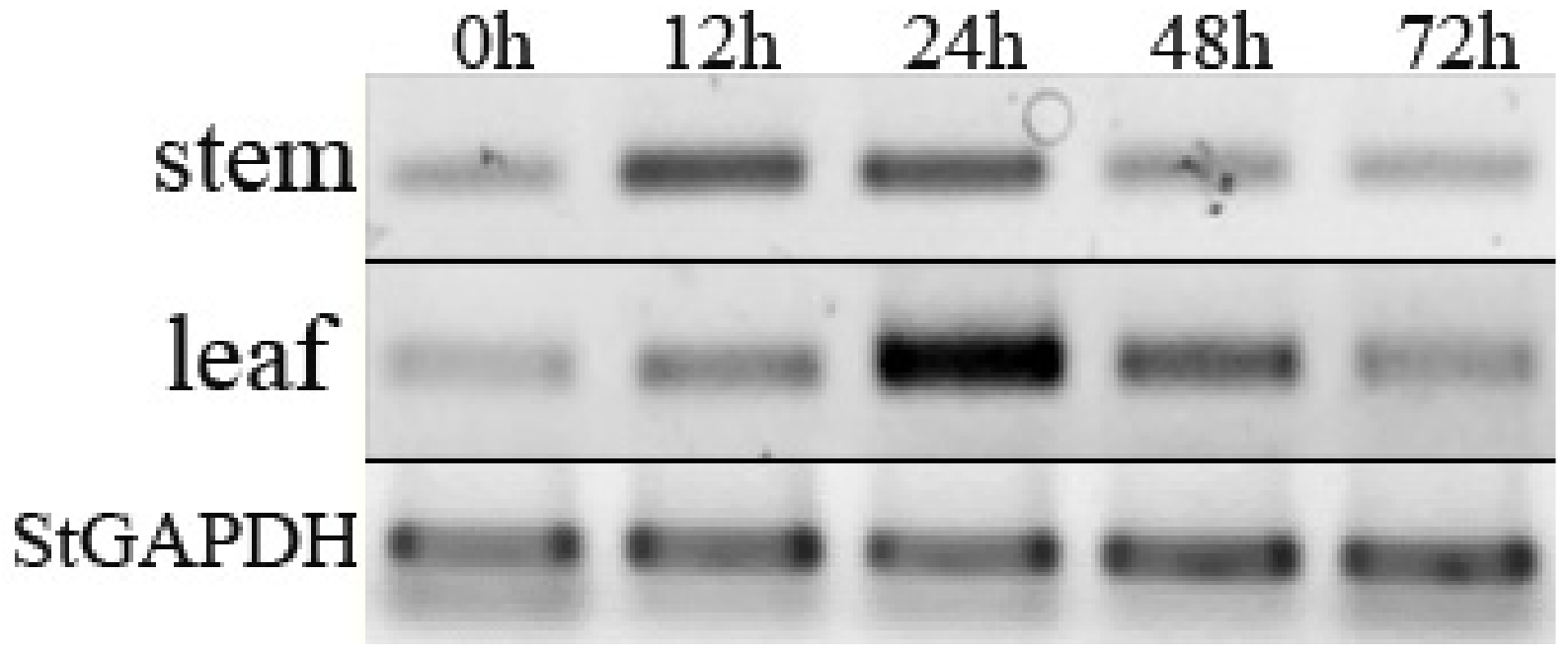
2.4. Expression Analysis of StPP5 Gene
2.4.1. RT-PCR Analysis of StPP5 Expression
2.4.2. RT-qPCR Analysis of StPP5 Expression
2.5. Construction of Overexpression Vector
2.6. Genetic Transformation and Identification of Transgenic Plants
2.7. Antifungal Assay
3. Results
3.1. Isolation of Full-Length cDNA of StPP5 Gene
3.2. Phylogenetic Analysis of StPP5
3.3. Expression of StPP5 Gene under V. dahliae Infection
3.4. Genetic Transformation and Plant Regeneration
3.5. Inhibitory Effect of Transgenic Tobacco Protein Extracts on V. dahliae
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weese, T.L.; Bohs, L. Eggplant origins: Out of Africa, into the Orient. Taxon 2010, 59, 49–56. [Google Scholar] [CrossRef]
- Liu, S.P.; Zhu, Y.P.; Xie, C.; Jue, D.W.; Hong, Y.B.; Chen, M.; Hubdar, A.K.; Yang, Q. Transgenic potato plants expressing Sto Ve1 exhibit enhanced resistance to Verticillium dahliae. Plant Mol. Biol. Rep. 2012, 30, 1032–1039. [Google Scholar] [CrossRef]
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. alboatrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.A. Mechanisms of disease resistance in Gossypium species and variation in Verticillium dahliae. Proc. World Cotton Res. Conf. 1994, 1, 225–235. [Google Scholar]
- Liu, H.; Zhang, N.; Yin, C.H.; Wang, J.H.; Ma, Z.H.; Zhou, L.G. Comparison between radial growth assay and liquid suspension culture assay for screening of antifungal natural compounds. Nat. Prod. Res. Dev. 2009, 21, 577–580. [Google Scholar]
- Parkhi, V.; Kumar, V.; Campbell, L.M.; Bell, A.A.; Shah, J.; Rathore, K.S. Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res. 2010, 19, 959–975. [Google Scholar] [CrossRef] [PubMed]
- Kawchuk, L.M.; Hachey, J.; Lynch, D.R.; Kulcsar, F.; van Rooijen, G.; Waterer, D.R.; Robertson, A.; Kokko, E.; Byers, R.; Howard, R.J.; et al. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 6511–6515. [Google Scholar] [CrossRef] [Green Version]
- Fradin, E.F.; Zhang, Z.; Juarez Ayala, J.C.; Castroverde, C.D.; Nazar, R.N.; Robb, J.; Liu, C.M.; Thomma, B.P. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009, 150, 320–332. [Google Scholar] [CrossRef] [Green Version]
- Fradin, E.F.; Abd-El-Haliem, A.; Masini, L.; van den Berg, G.C.; Joosten, M.H.; Thomma, B.P. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 2011, 156, 2255–2265. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.J.; Yin, Y.; Wang, Z.; Chen, M.; Huang, Q.S.; Huang, L.P.; Yang, Q. Cloning and analysis of full-length cDNA of Sto Ve1 gene from Solanum torvum. Plant Physiol. Commun. 2006, 42, 638–642. [Google Scholar]
- Miao, W.; Wang, X.; Li, M.; Song, C.; Wang, Y.; Hu, D.; Wang, J.S. Genetic transformation of cotton with a harpin-encoding gene hpaXoo confers an enhanced defense response against different pathogens through a priming mechanism. BMC Plant Biol. 2010, 10, 67–80. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, X.; Yang, S.; Chi, J.; Zhang, G.; Ma, Z. Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 2011, 30, 2085–2096. [Google Scholar] [CrossRef]
- Stone, J.M.; Collinge, M.A.; Smith, R.D.; Horn, M.A.; Walker, J.C. Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 1994, 266, 793–795. [Google Scholar] [CrossRef]
- Meskiene, I.; Bögre, L.; Glaser, W.; Balog, J.; Brandstötter, M.; Zwerger, K.; Ammerer, G.; Hirt, H. MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc. Natl. Acad. Sci. USA 1998, 95, 1938–1943. [Google Scholar] [CrossRef] [Green Version]
- Kerk, D.; Bulgrien, J.; Smith, D.W.; Barsam, B.; Veretnik, S.; Gribskov, M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002, 129, 908–925. [Google Scholar] [CrossRef] [Green Version]
- Mumby, M.C.; Walter, G. Protein-serine threonine phosphatases-structure, regulation, and functions in cell-growth. Physiol. Rev. 1993, 73, 673–699. [Google Scholar] [CrossRef]
- Gousset, C.; Collonnier, C.; Mulya, K.; Mariska, I.; Rotino, G.L.; Besse, P.; Servaes, A.; Sihachakr, D. Solanum torvum, as a useful source of resistance against bacterial and fungal diseases for improvement of eggplant (S. melongena L.). Plant Sci. 2005, 168, 319–327. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, J.L.; Zhang, F.; Huang, Q.S.; Huang, L.P.; Yang, Q. Differential expression analysis by cDNA-AFLP of Solanum torvum upon Verticillium dahliae infection. Russ. J. Plant Physiol. 2010, 57, 676–684. [Google Scholar] [CrossRef]
- Meng, X.; Li, F.; Liu, C.; Zhang, C.; Wu, Z.; Chen, Y. Isolation and characterization of an ERF Transcription factor gene from cotton (Gossypium barbadense L.). Plant Mol. Biol. Rep. 2010, 28, 176–183. [Google Scholar] [CrossRef]
- Quiroga, E.N.; Sampietro, A.R.; Vattuone, M.A. Screening antifungal activities of selected medicinal plants. J. Ethnopharmacol. 2001, 74, 89–96. [Google Scholar] [CrossRef]
- Wu, B.M.; Subbarao, K.V. A model for multiseasonal spread of verticillium wilt of lettuce. Phytopathology 2014, 104, 908–917. [Google Scholar] [CrossRef]
- Short, D.P.G.; Gurung, S.; Koike, S.T.; Klosterman, S.J.; Subbarao, K.V. Frequency of Verticillium species in commercial spinach fields and transmission of V. dahliae from spinach to subsequent Lettuce crops. Phytopathology 2015, 105, 80–90. [Google Scholar] [CrossRef] [Green Version]
- Aguero, C.B.; Uratsu, S.L.; Greve, C.; Powell, A.L.; Labavitch, J.M.; Meredith, C.P.; Dandekar, A.M. Evaluation of tolerance to pierce′s disease and botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 2005, 6, 43–51. [Google Scholar] [CrossRef]
- Cohen, P.T.W. Overview of protein serine/threonine phosphatases. In Protein Phosphatases; Springer: Berlin/Heidelberg, Germany, 2004; pp. 1–20. [Google Scholar]
- Mochida, S.; Maslen, S.L.; Skehel, M.; Hunt, T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 2010, 330, 1670–1673. [Google Scholar] [CrossRef]
- Das, I.; Dennis, J.E. Normal-boundary Intersection: A new method for generating the pareto surface in nonlinear multicriteria optimization problems. SIAM J. Optim. 1998, 8, 631–657. [Google Scholar] [CrossRef] [Green Version]
- Li, F.H.; Fu, F.L.; Sha, L.N.; Liang, H.; Li, W.C. Differential expression of serine/threonine protein phosphatase type-2c under drought stress in Maize. Plant Mol. Biol. Rep. 2009, 27, 29–37. [Google Scholar] [CrossRef]
- Sontag, E. Protein phosphatase 2A: The Trojan Horse of cellular signaling. Cell Signal. 2001, 13, 7–16. [Google Scholar] [CrossRef]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef]
- Liu, S.P.; Hong, Y.B.; Wu, Z.; Ma, Y.S.; Jue, D.W.; Xie, C.; Zhu, Y.P.; Chen, M.; Yang, Q. Protein extract of tobacco expressing Sto Ve1 gene inhibits Verticillium dahliae proliferation. Czech J. Genet. Plant Breed. 2013, 49, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Vranová, E.; Langebartels, C.; Van Montagu, M.; Inzé, D.; Camp, W.V. Oxidative stress, heat shock and drought differentially affect expression of a tobacco protein phosphatase 2C. J. Exp. Bot. 2000, 51, 1763–1764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.J.; Peng, Y.; Chen, X.; Dardick, C.; Ronald, P.C. Rice XB15, a Protein Phosphatase 2C, Negatively Regulates Cell Death and XA21-Mediated Innate Immunity. PLoS Biol. 2008, 6, 1910–1926. [Google Scholar]





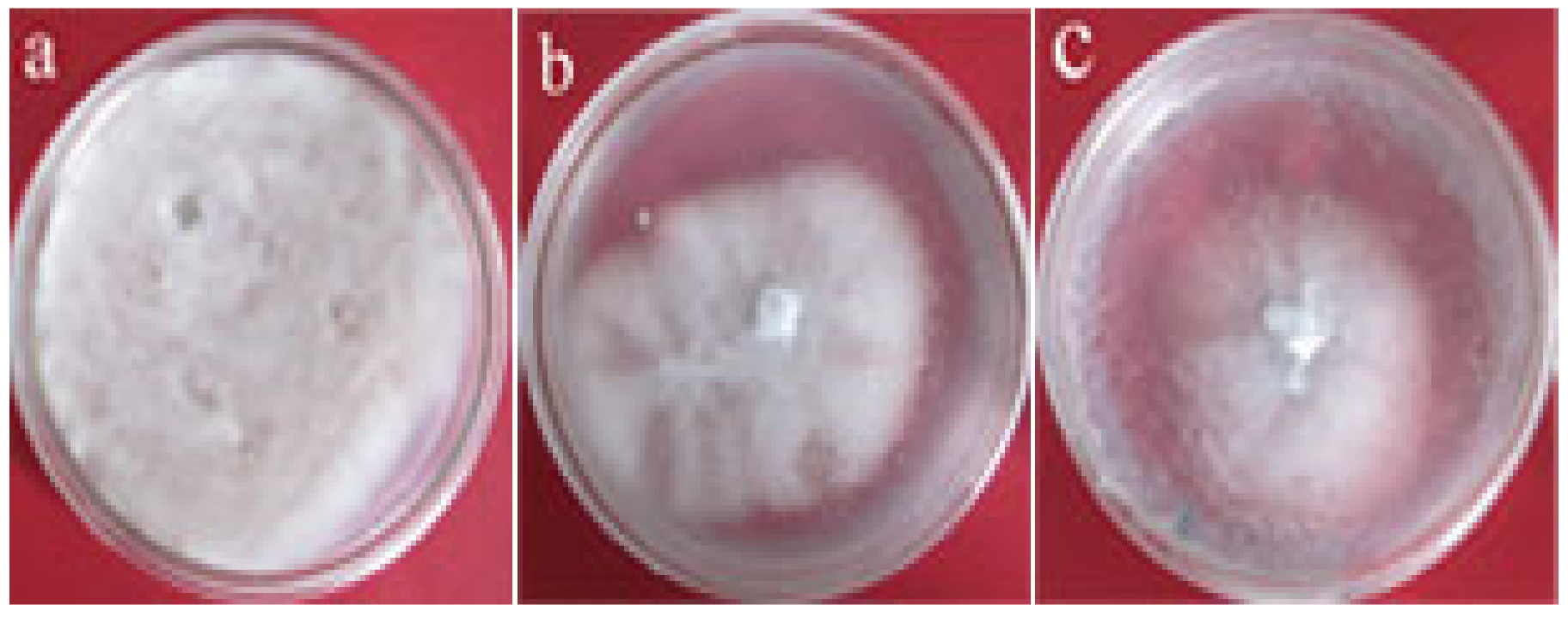
| Treatment | Average Inhibition Zone Diameter (mm) | Inhibition Rate (%) |
|---|---|---|
| Wild-type tobacco Transgenic line P6 | 62.0 40.3 | 31.1 55.2 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Xie, C.; Yang, Q.; Zhu, W.; Wang, M. Protein Extract of Tobacco Expressing Solanum torvum PP5-Encoding Gene Inhibits Verticillium dahliae Proliferation. Horticulturae 2022, 8, 252. https://doi.org/10.3390/horticulturae8030252
Chen M, Xie C, Yang Q, Zhu W, Wang M. Protein Extract of Tobacco Expressing Solanum torvum PP5-Encoding Gene Inhibits Verticillium dahliae Proliferation. Horticulturae. 2022; 8(3):252. https://doi.org/10.3390/horticulturae8030252
Chicago/Turabian StyleChen, Min, Chao Xie, Qing Yang, Wenjiao Zhu, and Man Wang. 2022. "Protein Extract of Tobacco Expressing Solanum torvum PP5-Encoding Gene Inhibits Verticillium dahliae Proliferation" Horticulturae 8, no. 3: 252. https://doi.org/10.3390/horticulturae8030252






