RNA Helicase Mediates Competitive Fitness of Listeria monocytogenes on the Surface of Cantaloupe
Abstract
:1. Introduction
2. Results and Discussion
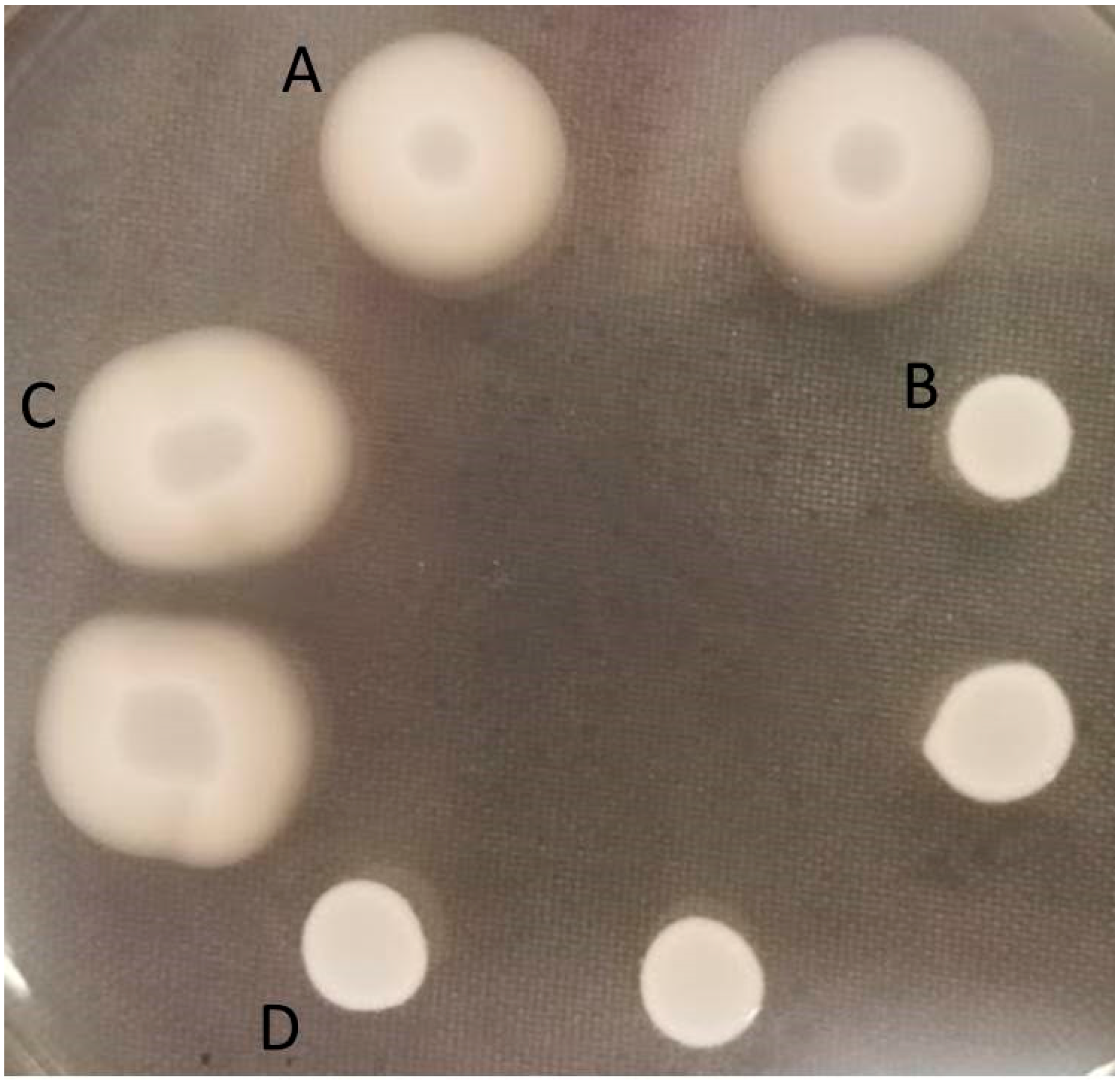
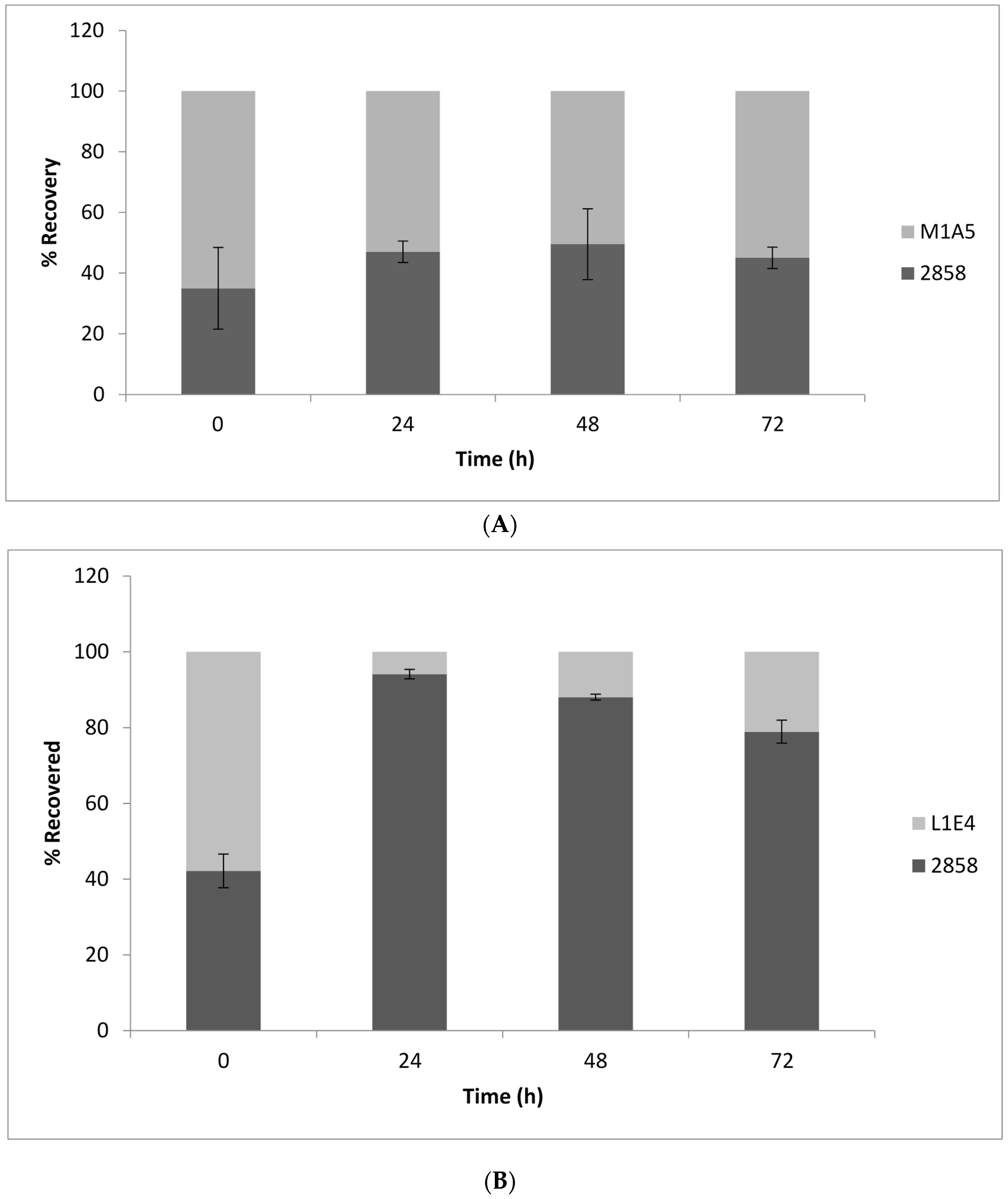
2.1. Impact of the Mutations on Growth and Competitive Fitness of L. monocytogenes on Cantaloupe Rind
2.2. Genetic Complementation of Mutant L1E4 Confirms Impact of the DEAD-Box RNA Helicase Encoded by lmo0866 in Competitive Fitness on Cantaloupe
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
3.2. Cantaloupe Adherence, Growth and Survival Assay
3.3. Competitive Fitness Assessments
3.4. Genetic Complementation of L1E4
3.5. In Vitro Growth Comparison of Strain 2858 and Motility Impaired Mutants
3.6. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McCollum, J.T.; Cronquist, A.B.; Silk, B.J.; Jackson, K.A.; O’Connor, K.A.; Cosgrove, S.; Gossack, J.P.; Parachini, S.S.; Jain, N.S.; Ettestad, P.; et al. Multistate outbreak of listeriosis associated with cantaloupe. N. Engl. J. Med. 2013, 369, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Lemon, K.P.; Higgins, D.E.; Kolter, R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 2007, 189, 4418–4424. [Google Scholar] [CrossRef] [PubMed]
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991, 55, 476–511. [Google Scholar] [PubMed]
- Chae, M.S.; Schraft, H. Comparative evaluation of adhesion and biofilm formation of different Listeria monocytogenes strains. Int. J. Food Microbiol. 2000, 62, 103–111. [Google Scholar] [CrossRef]
- Chae, M.S.; Schraft, H.; Truelstrup Hansen, L.; Mackereth, R. Effects of physicochemical surface characteristics of Listeria monocytogenes strains on attachment to glass. Food Microbiol. 2006, 23, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Todhanakasem, T.; Young, G.M. Loss of flagellum-based motility by Listeria monocytogenes results in formation of hyperbiofilms. J. Bacteriol. 2008, 190, 6030–6034. [Google Scholar] [CrossRef] [PubMed]
- Ukuku, D.O.; Fett, W. Behavior of Listeria monocytogenes Inoculated on cantaloupe surfaces and efficacy of washing treatments to reduce transfer from rind to fresh-cut pieces. J. Food Prot. 2002, 65, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Rakic-Martinez, M.; Osborne, J.; Jayeola, V.O.; Katic, V.; Kathariou, S. Capacity of Listeria monocytogenes strains from the 2011 cantaloupe outbreak to adhere, survive, and grow on cantaloupe. J. Food Prot. 2016, 79, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Gorski, L.; Duhé, J.M.; Flaherty, D. The use of flagella and motility for plant colonization and fitness by different strains of the foodborne pathogen Listeria monocytogenes. PLoS ONE 2009, 4, e5142. [Google Scholar] [CrossRef] [PubMed]
- Gorski, L.; Palumbo, J.D.; Mandrell, R.E. Attachment of Listeria monocytogenes to radish tissue is dependent upon temperature and flagellar motility. Appl. Environ. Microbiol. 2003, 69, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Markkula, A.; Mattila, M.; Lindström, M.; Korkeala, H. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3 °C. Environ. Microbiol. 2012, 14, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Azizoglu, R.O.; Kathariou, S. Inactivation of a cold-induced putative rna helicase gene of Listeria monocytogenes is accompanied by failure to grow at low temperatures but does not affect freeze-thaw tolerance. J. Food Prot. 2010, 73, 1474–1479. [Google Scholar] [CrossRef] [PubMed]
- Netterling, S.; Bäreclev, C.; Vaitkevicius, K.; Johansson, J. RNA Helicase important for Listeria monocytogenes hemolytic activity and virulence factor expression. Infect. Immun. 2016, 84, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Markkula, A.; Lindström, M.; Johansson, P.; Björkroth, J.; Korkeala, H. Roles of four putative DEAD-box RNA helicase genes in growth of Listeria monocytogenes EGD-e under heat, pH, osmotic, ethanol, and oxidative stress conditions. Appl. Environ. Microbiol. 2012, 78, 6875–6882. [Google Scholar] [CrossRef] [PubMed]
- Bäreclev, C.; Vaitkevicius, K.; Netterling, S.; Johansson, J. DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression. RNA Biol. 2014, 11, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Bitar, A.P.; Marquis, H. A mariner-based transposition system for Listeria monocytogenes. Appl. Environ. Microbiol. 2007, 73, 2758–2761. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Lee, S.; Jayeola, V.; Kathariou, S. Novel cadmium resistance determinant in Listeria monocytogenes. Appl. Environ. Microbiol. 2017, 83, e02580-16. [Google Scholar] [CrossRef] [PubMed]
- Azizoglu, R.O.; Gorski, L.; Kathariou, S. Isolation of Listeria monocytogenes from food and water: Official and experimental protocols. In Current Protocols in Microbiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; Volume 33, pp. 9B.5.1–9B.5.19. [Google Scholar]
- Peel, M.; Donachie, W.; Shaw, A. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and western blotting. J. Gen. Microbiol. 1988, 134, 2171–2178. [Google Scholar] [CrossRef] [PubMed]
- Way, S.S.; Thompson, L.J.; Lopes, J.E.; Hajjar, A.M.; Kollmann, T.R.; Freitag, N.E.; Wilson, C.B. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell. Microbiol. 2004, 6, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Lauer, P.; Chow, M.Y.; Loessner, M.J.; Portnoy, D.A.; Calendar, R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 2002, 184, 4177–4186. [Google Scholar] [CrossRef] [PubMed]
- Breidt, F.; Romick, T.L.; Fleming, H.P. A rapid method for the determination of bacterial growth kinetics. J. Rapid Methods Autom. Microbiol. 1994, 3, 59–68. [Google Scholar] [CrossRef]
| Strains | Genotype and Features | Phenotype |
|---|---|---|
| 2858 | Wild-type strain from 2011 cantaloupe outbreak | EmS (Erythromycin susceptible) |
| L1E4 | Transposon mutant of 2858 DEAD-box RNA helicase in lmo0866 homolog (nt 981) | Non-motile, cold sensitive, EmR (Erythromycin resistant), reduced hemolysis on blood agar plates |
| M1A5 | Transposon mutant of 2858 in lmo0694 homolog (nt 161) (flagella biosynthesis and chemotaxis gene cluster) | Non-motile, EmR |
| L1E4-C | Complemented L1E4 mutant | Motile, cold growth proficient; EmR, CmR (chloramphenicol resistant) |
| L1E4-E | L1E4 mutant with empty vector (pPL2) | Non-motile, cold-sensitive; EmR, CmR |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Price, R.; Parsons, C.; Kathariou, S. RNA Helicase Mediates Competitive Fitness of Listeria monocytogenes on the Surface of Cantaloupe. Horticulturae 2018, 4, 40. https://doi.org/10.3390/horticulturae4040040
Price R, Parsons C, Kathariou S. RNA Helicase Mediates Competitive Fitness of Listeria monocytogenes on the Surface of Cantaloupe. Horticulturae. 2018; 4(4):40. https://doi.org/10.3390/horticulturae4040040
Chicago/Turabian StylePrice, Robert, Cameron Parsons, and Sophia Kathariou. 2018. "RNA Helicase Mediates Competitive Fitness of Listeria monocytogenes on the Surface of Cantaloupe" Horticulturae 4, no. 4: 40. https://doi.org/10.3390/horticulturae4040040





