Adhesins of Yeasts: Protein Structure and Interactions
Abstract
:1. Introduction
2. The Structure of Yeast Adhesins
2.1. Architectures of Yeast Adhesins
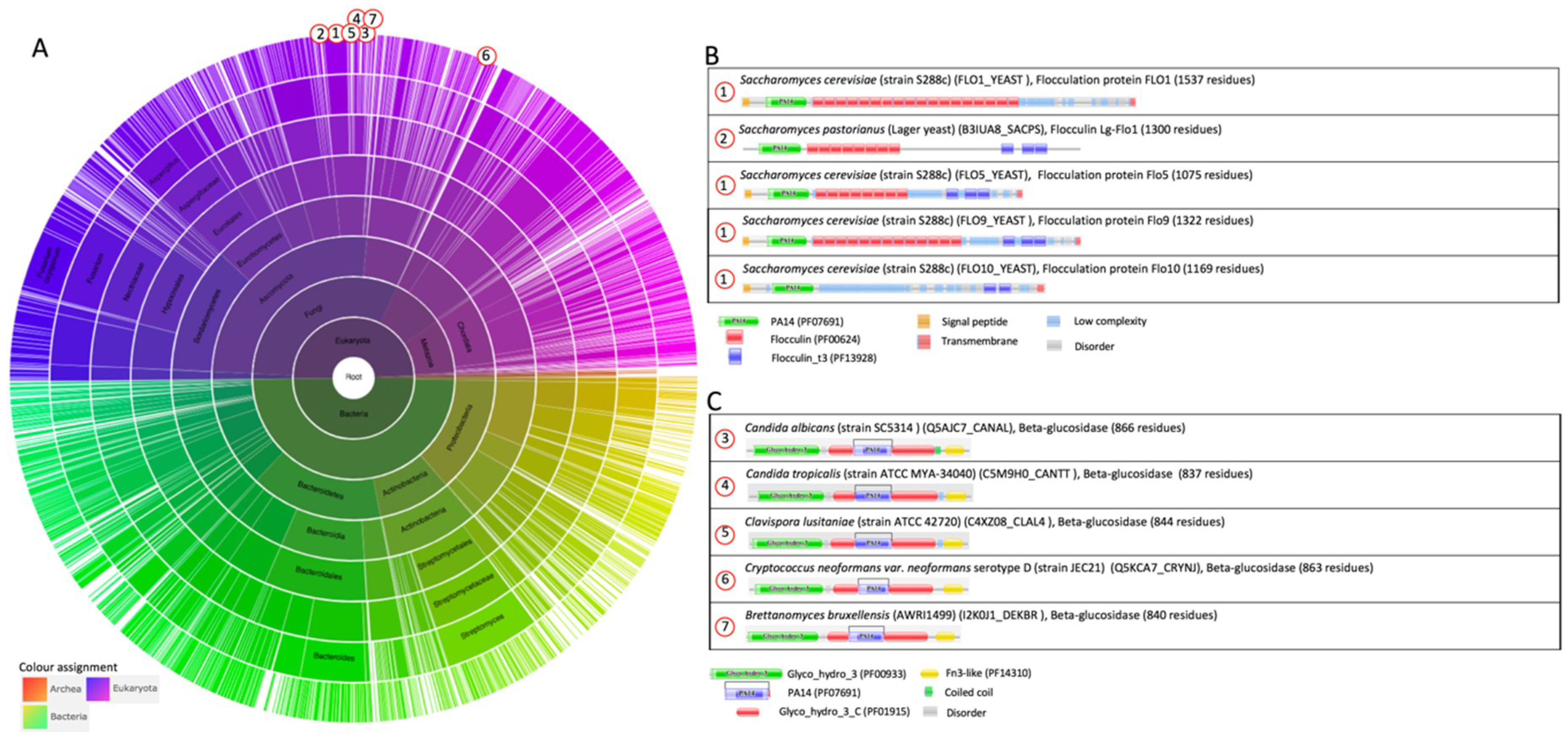
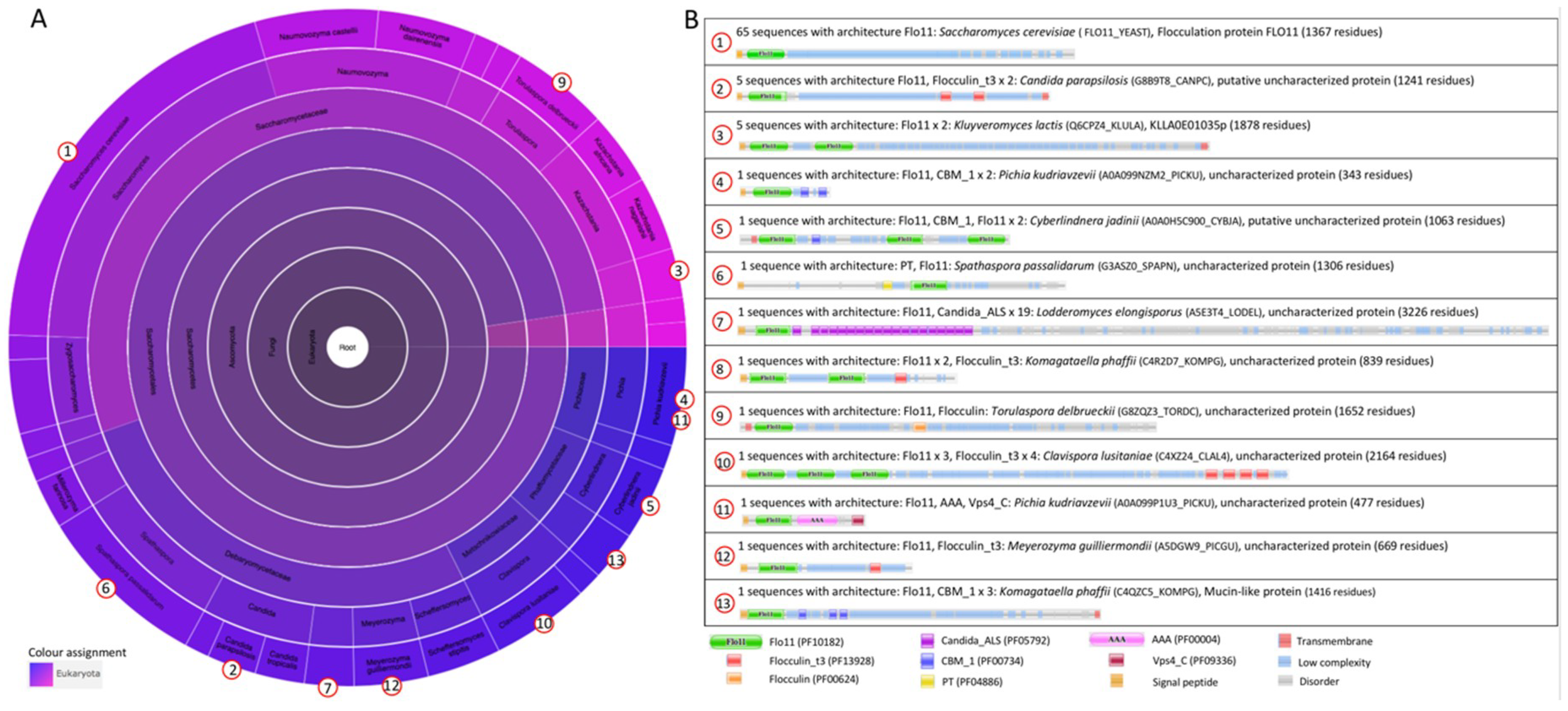
2.1.1. The S. cerevisiae Flocculation Protein Family
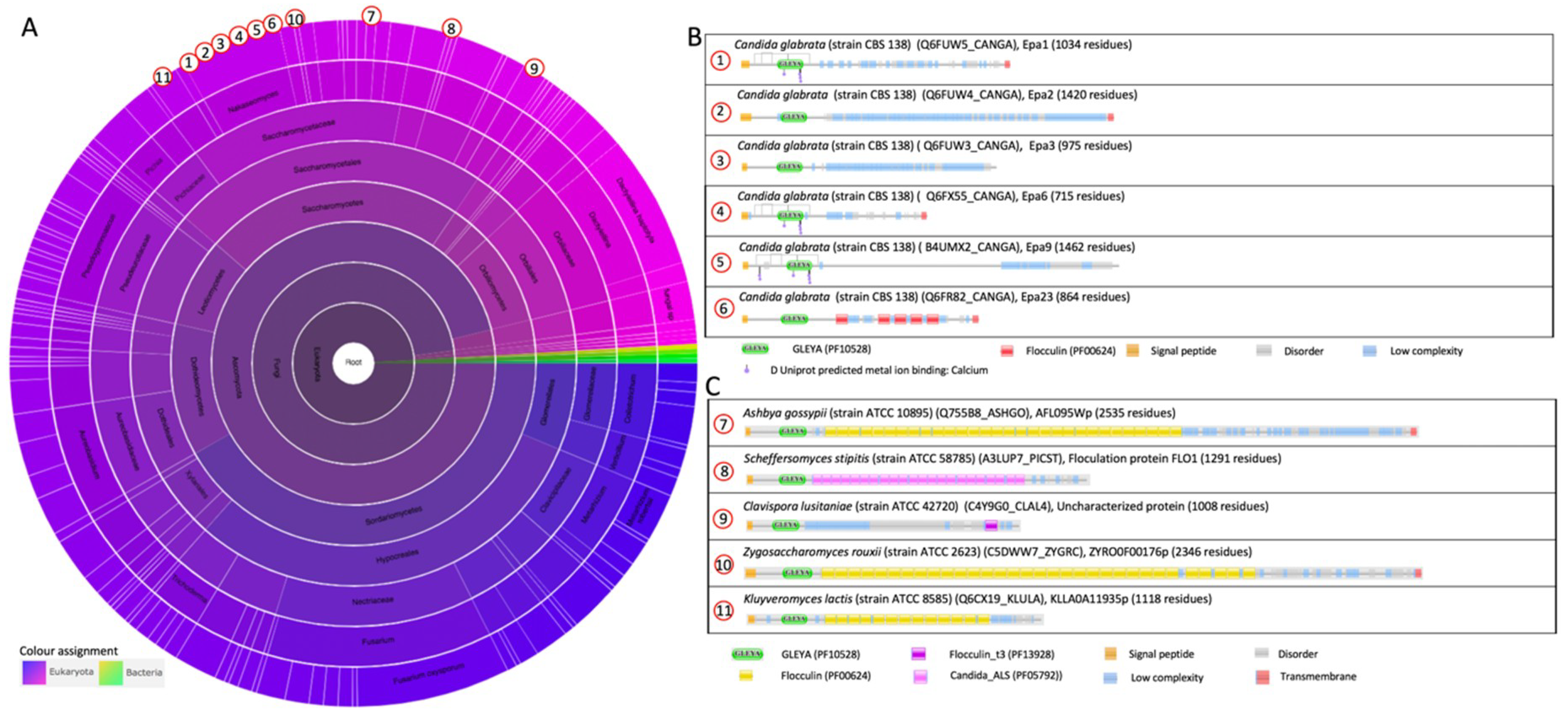
2.1.2. The Epithelial Adhesin Family
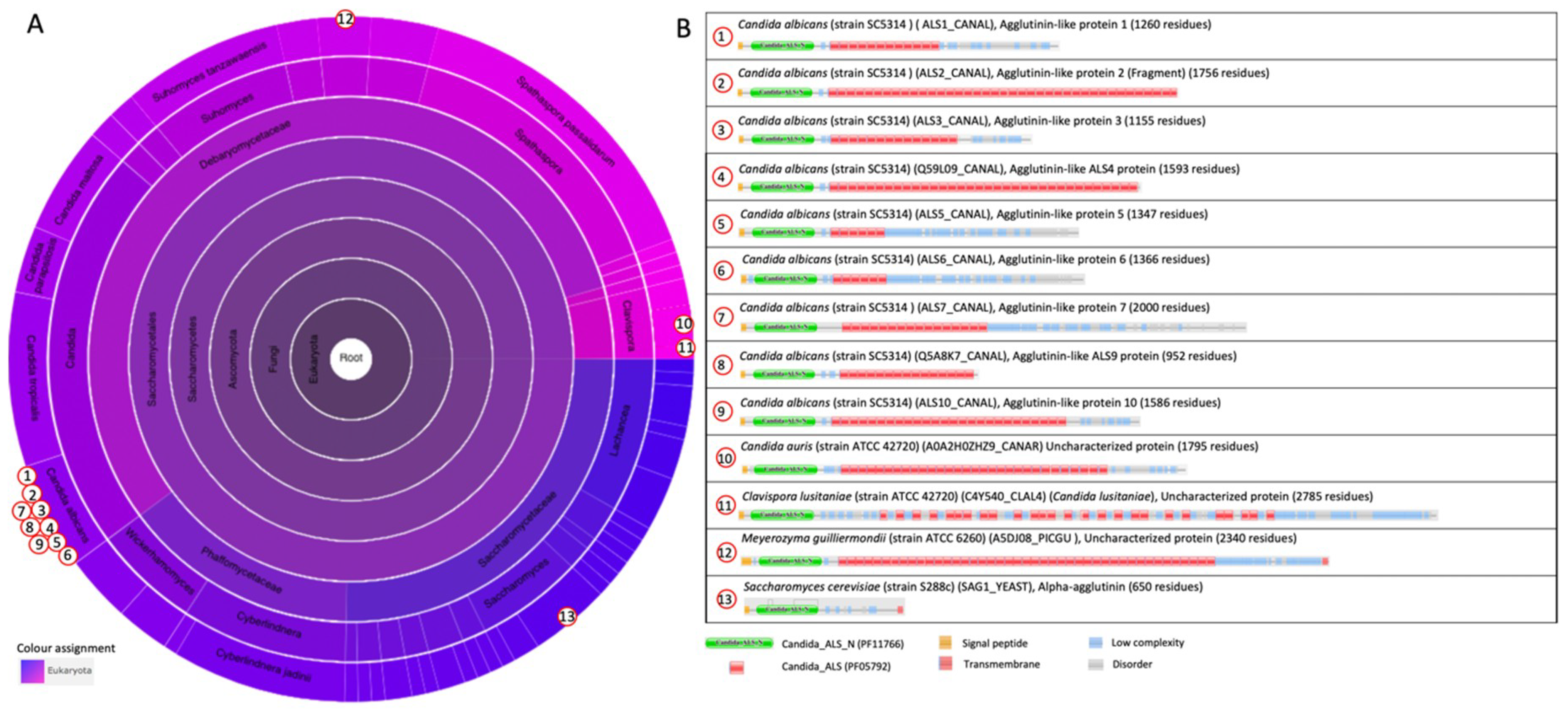
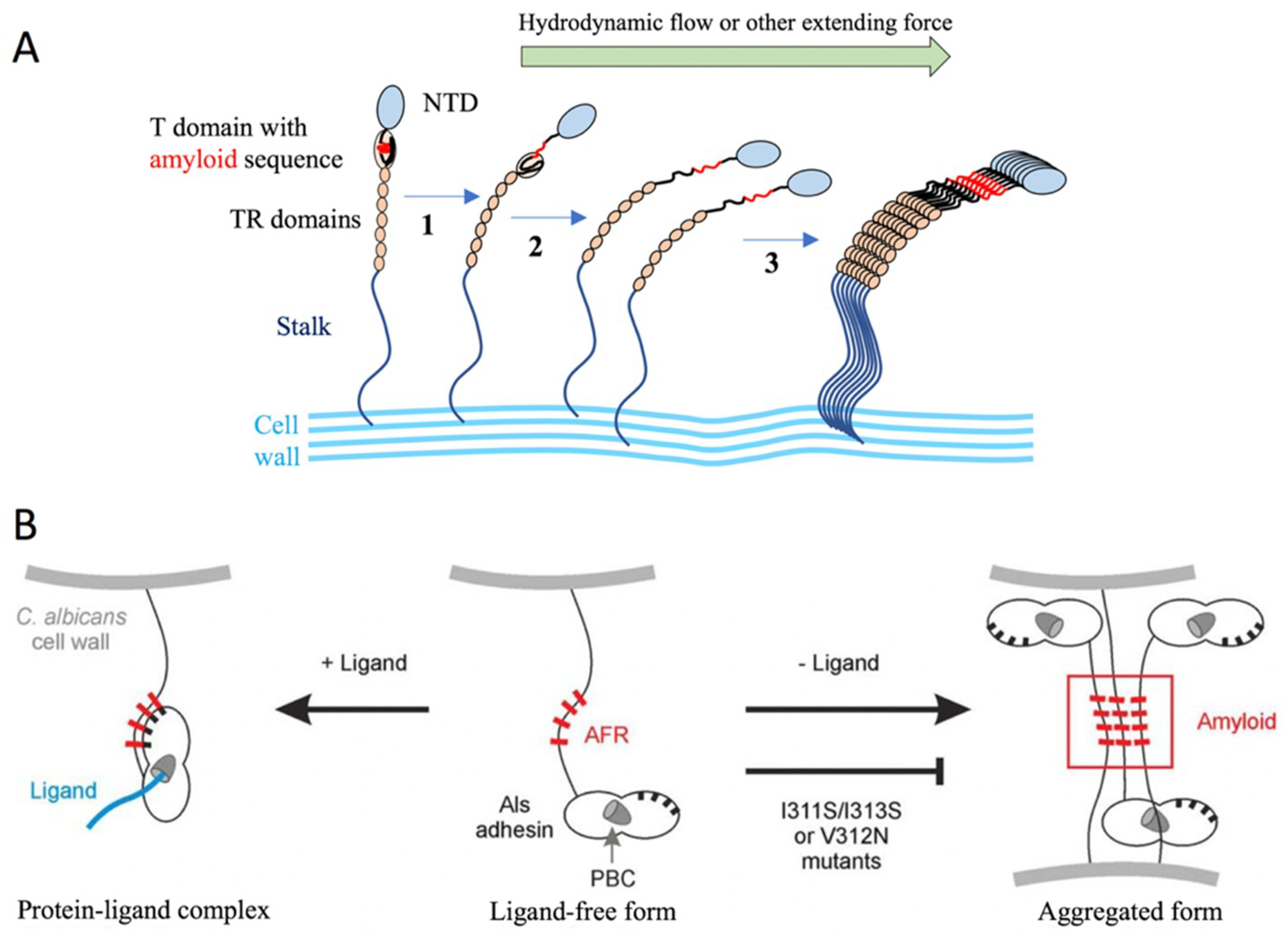
2.1.3. The Agglutinin-like Sequence Protein Family
2.2. Protein Structures of Yeast Adhesins
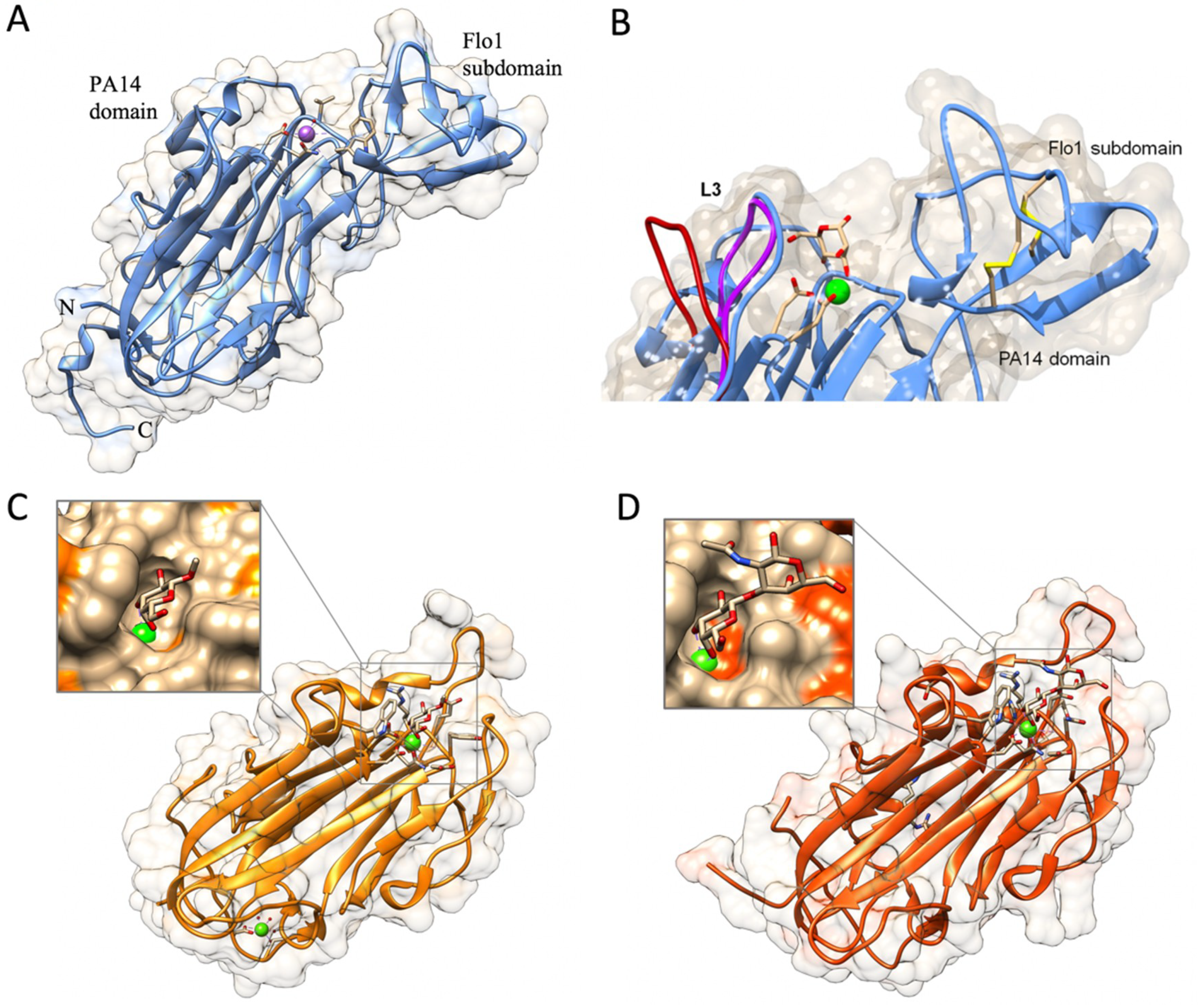
2.2.1. The PA14 Fold in Flocculation Protein and Epithelial Adhesin Family
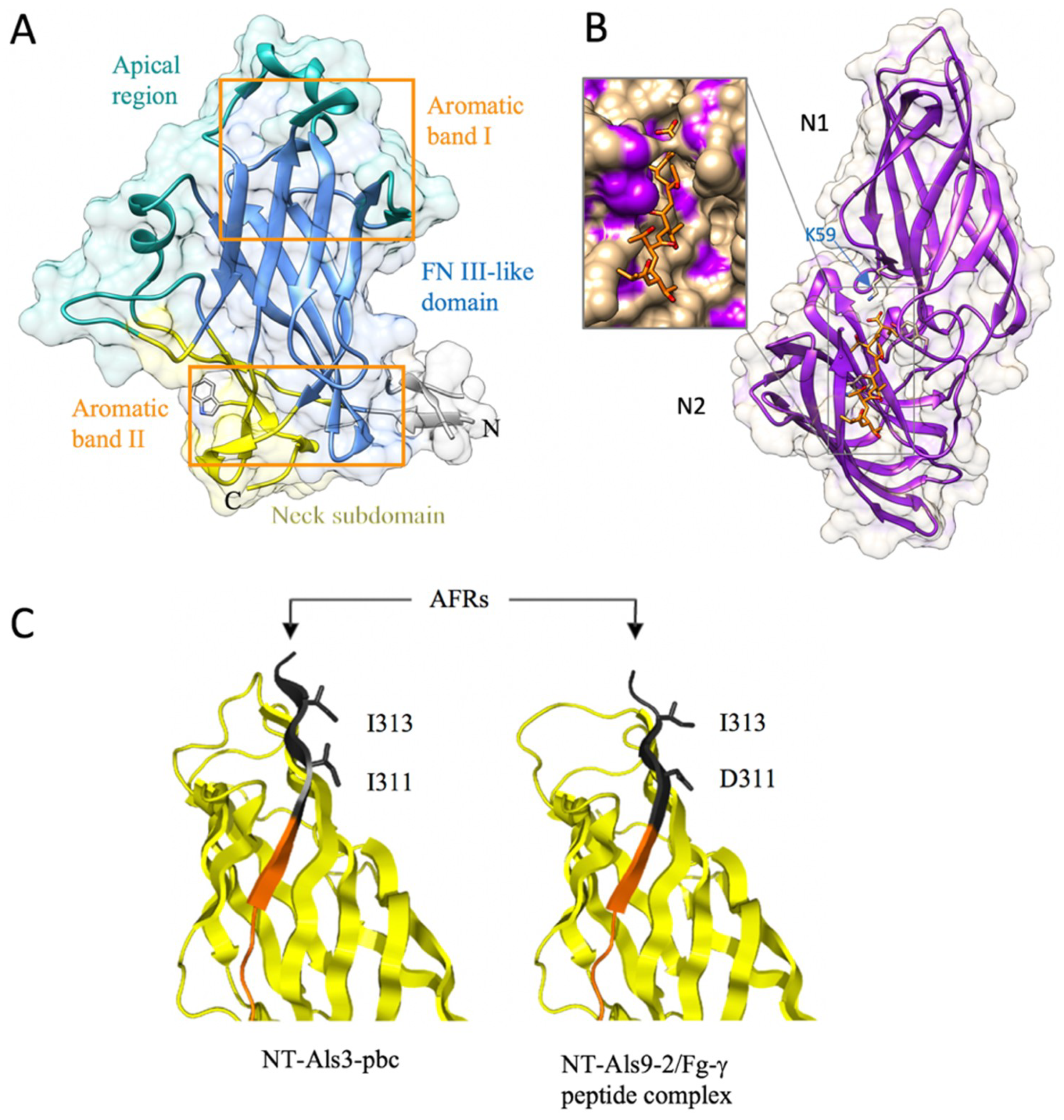
2.2.2. The Flo11 Fold
2.2.3. The Ig-like Fold in Agglutinin-like Sequence Protein (Als) Family
3. Yeast-Yeast and Yeast-Host Cell Interactions
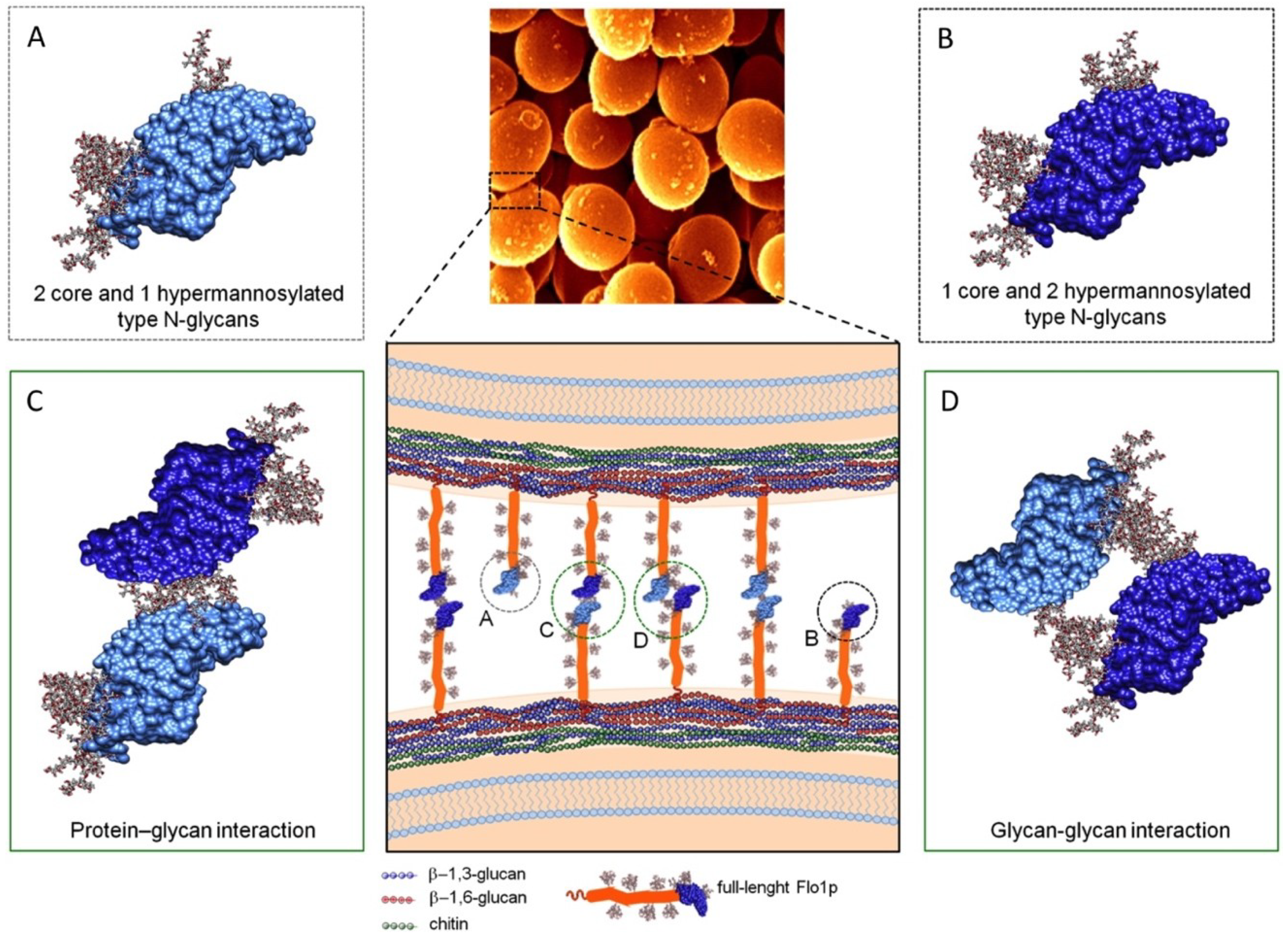
3.1. Flo Proteins Interactions
3.1.1. Cell-Cell Binding Based on S. cerevisiae-Lectin-Flocculin Interaction
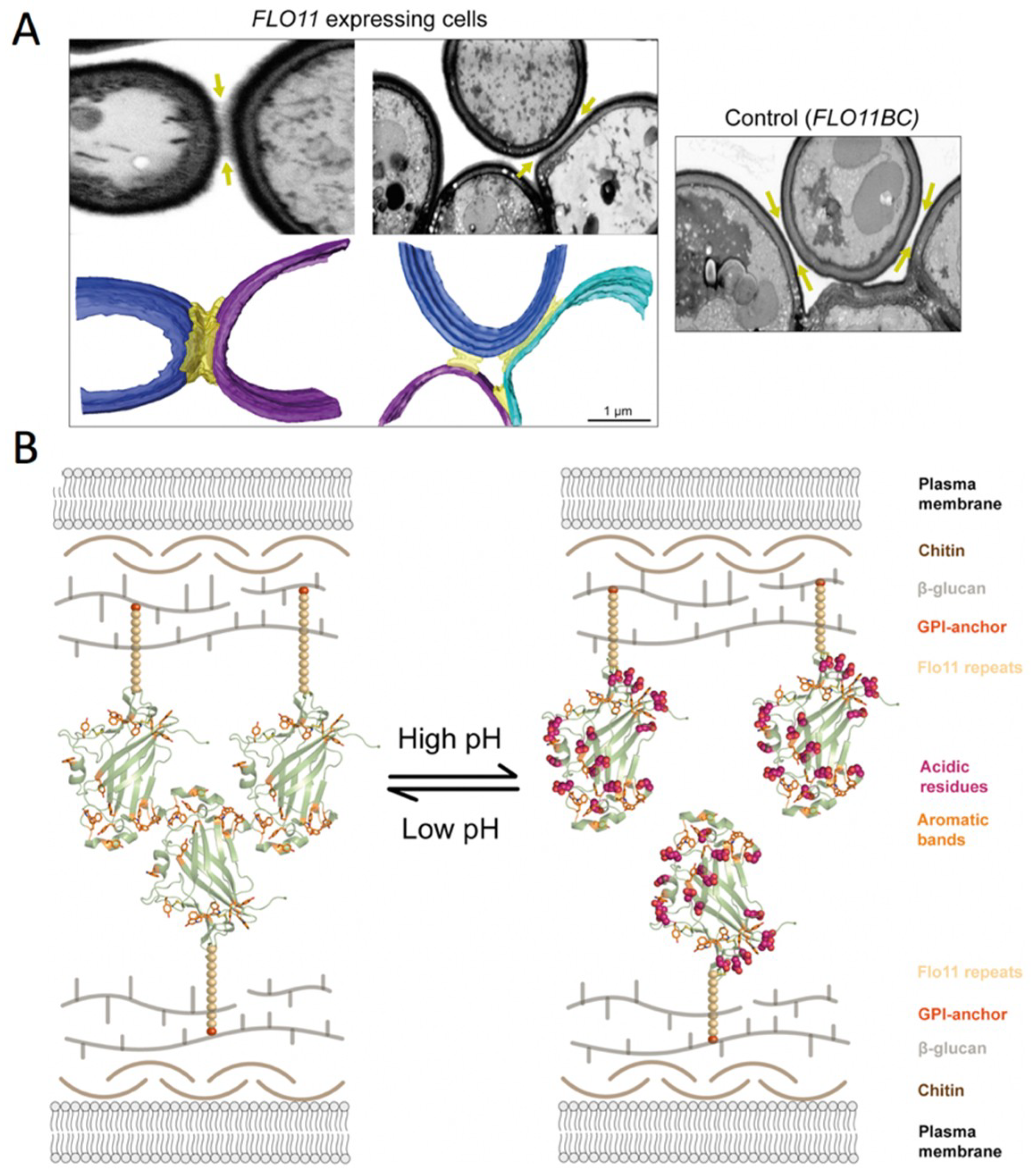
3.1.2. Cell-Cell Binding Based on S. cerevisiae-Flo11 Protein Interaction
3.2. C. albicans Als Protein Interactions
3.3. C. glabrata Epa Protein Interactions
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Teunissen, A.W.R.H.; Steensma, H.Y. The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 1995, 11, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- de Groot, P.W.J.; Bader, O.; de Boer, A.D.; Weig, M.; Chauhan, N. Adhesins in Human Fungal Pathogens: Glue with Plenty of Stick. Eukaryot. Cell 2013, 12, 470–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reitsma, S.; Slaaf, D.W.; Vink, H.; van Zandvoort, M.A.M.J.; oude Egbrink, M.G.A. The endothelial glycocalyx: Composition, functions, and visualization. Pflugers Arch. 2007, 454, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Critchley, I.A.; Douglas, L.J. Role of Glycosides as Epithelial Cell Receptors for Candida albicans. Microbiology 1987, 133, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Vardar-Ünlü, G.; McSharry, C.; Douglas, L.J. Fucose-specific adhesins on germ tubes of Candida albicans. FEMS Immunol. Med. Microbiol. 2006, 20, 55–67. [Google Scholar] [CrossRef] [Green Version]
- Brückner, S.; Mösch, H.-U. Choosing the right lifestyle: Adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2012, 36, 25–58. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, C.J.; Ljungdahl, P.O.; Styles, C.A.; Fink, G.R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 1992, 68, 1077–1090. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F.J. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 13619–13624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palková, Z. Multicellular microorganisms: Laboratory versus nature. EMBO Rep. 2004, 5, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.; Flaxman, A.D.; Naghavi, M.; Lozano, R.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; Aboyans, V.; et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling Human Fungal Infections. Science 2012, 336, 647. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [PubMed]
- Black, R.E.; Victora, C.G.; Walker, S.P.; Bhutta, Z.A.; Christian, P.; de Onis, M.; Ezzati, M.; Grantham-McGregor, S.; Katz, J.; Martorell, R.; et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet 2013, 382, 427–451. [Google Scholar] [CrossRef]
- Rahman, S.A.; Adjeroh, D. Surface-Based Body Shape Index and Its Relationship with All-Cause Mortality. PLoS ONE 2015, 10, e0144639. [Google Scholar] [CrossRef] [PubMed]
- Bourke, C.D.; Berkley, J.A.; Prendergast, A.J. Immune Dysfunction as a Cause and Consequence of Malnutrition. Trends Immunol. 2016, 37, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Katona, P.; Katona Apte, J. The Interaction between Nutrition and Infection. Clin. Infect. Dis. 2008, 46, 1582–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olofin, I.; McDonald, C.M.; Ezzati, M.; Flaxman, S.; Black, R.E.; Fawzi, W.W.; Caulfield, L.E.; Danaei, G.; Nutrition Impact Model Study (anthropometry cohort pooling). Associations of Suboptimal Growth with All-Cause and Cause-Specific Mortality in Children under Five Years: A Pooled Analysis of Ten Prospective Studies. PLoS ONE 2013, 8, e64636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rytter, M.J.H.; Kolte, L.; Briend, A.; Friis, H.; Christensen, V.B. The Immune System in Children with Malnutrition—A Systematic Review. PLoS ONE 2014, 9, e105017. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.K.; Zambruni, M.; Melby, C.L.; Melby, P.C. Impact of Childhood Malnutrition on Host Defense and Infection. Clin. Microbiol. Rev. 2017, 30, 919–971. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, R.; Syrjänen, J. Obesity and the risk and outcome of infection. Int. J. Obes. 2012, 37, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Lamps, L.W.; Lai, K.K.T.; Milner, D.A. Fungal Infections of the Gastrointestinal Tract in the Immunocompromised Host. Adv. Anat. Pathol. 2014, 21, 217–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antinori, S.; Milazzo, L.; Sollima, S.; Galli, M.; Corbellino, M. Candidemia and invasive candidiasis in adults: A narrative review. Eur. J. Intern. Med. 2016, 34, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. North. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Krcmery, V.; Barnes, A.J. Non-albicans Candida spp. causing fungaemia: Pathogenicity and antifungal resistance. J. Hosp. Infect. 2002, 50, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011, 11, 142–151. [Google Scholar] [CrossRef]
- Perlroth, J.; Choi, B.; Spellberg, B. Nosocomial fungal infections: Epidemiology, diagnosis, and treatment. Med. Mycol. 2007, 45, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Hube, B. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell. Microbiol. 2013, 15, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A Saccharomyces gene family involved in invasive growth, cell–cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Bojsen, R.K.; Andersen, K.S.; Regenberg, B. Saccharomyces cerevisiae—a model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 2012, 65, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Dougherty, S.D.; Erdman, S.E. Conserved WCPL and CX4C Domains Mediate Several Mating Adhesin Interactions in Saccharomyces cerevisiae. Genetics 2009, 182, 173–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipke, P.N.; Wojciechowicz, D.; Kurjan, J. AG alpha 1 is the structural gene for the Saccharomyces cerevisiae alpha-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 1989, 9, 3155–3165. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Hauser, K.; Mrśa, V.; Watzele, M.; Watzele, G.; Gruber, C.; Tanner, W. Saccharomyces cerevisiae a- and alpha-agglutinin: Characterization of their molecular interaction. EMBO J. 1991, 10, 4081–4088. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Baldermann, C.; Rachel, R.; Tanner, W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: Cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 1994, 13, 4737–4744. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Lu, C.F.; Marykwas, D.L.; Lipke, P.N.; Kurjan, J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 1991, 11, 4196–4206. [Google Scholar] [CrossRef] [PubMed]
- Erdman, S.; Lin, L.; Malczynski, M.; Snyder, M. Pheromone-regulated Genes Required for Yeast Mating Differentiation. J. Cell. Biol. 1998, 140, 461–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.; Willaert, R. Flocculation protein structure and cell–cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 2010, 32, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.; Ielasi, F.S.; Nookaew, I.; Stals, I.; Alonso-Sarduy, L.; Daenen, L.; Van Mulders, S.E.; Stassen, C.; van Eijsden, R.G.E.; Siewers, V.; et al. Molecular Mechanism of Flocculation Self-Recognition in Yeast and Its Role in Mating and Survival. MBio 2015, 6, e00427-15. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.S.; Dranginis, A.M. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 1996, 178, 7144–7151. [Google Scholar] [CrossRef] [PubMed]
- Vajjhala, P.R.; Wong, J.S.; To, H.-Y.; Munn, A.L. The β domain is required for Vps4p oligomerization into a functionally active ATPase. FEBS J. 2006, 273, 2357–2373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linder, T.; Gustafsson, C.M. Molecular phylogenetics of ascomycotal adhesins—A novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 2008, 45, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Decanniere, K.; Willaert, R.G. The epithelial adhesin 1 (Epa1p) from the human-pathogenic yeast Candida glabrata: Structural and functional study of the carbohydrate-binding domain. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Reyna, M.; Diderrich, R.; Veelders, M.S.; Eulenburg, G.; Kalugin, V.; Brückner, S.; Keller, P.; Rupp, S.; Mösch, H.-U.; Essen, L.-O. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc. Natl. Acad. Sci. USA 2012, 109, 16864–16869. [Google Scholar] [CrossRef] [PubMed]
- Bidaud, A.L.; Chowdhary, A.; Dannaoui, E. Candida auris: An emerging drug resistant yeast—A mini-review. J. Mycol. Med. 2018, 28, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida aurison 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Sears, D.; Schwartz, B.S. Candida auris: An emerging multidrug-resistant pathogen. Int. J. Infect. Dis. 2017, 63, 95–98. [Google Scholar] [CrossRef] [PubMed]
- de Cássia Orlandi Sardi, J.; Silva, D.R.; Soares Mendes-Giannini, M.J.; Rosalen, P.L. Candida auris: Epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb. Pathog. 2018, 125, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.; Woodworth, K.; Walters, M.; Berkow, E.L.; Jackson, B.; Chiller, T.; Vallabhaneni, S. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2018, 64, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Hecht, J.E. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast 2000, 18, 49–60. [Google Scholar] [CrossRef]
- Diderrich, R.; Kock, M.; Maestre-Reyna, M.; Keller, P.; Steuber, H.; Rupp, S.; Essen, L.-O.; Mösch, H.-U. Structural Hot Spots Determine Functional Diversity of the Candida glabrata Epithelial Adhesin Family. J. Biol. Chem. 2015, 290, 19597–19613. [Google Scholar] [CrossRef] [PubMed]
- Veelders, M.; Brückner, S.; Ott, D.; Unverzagt, C.; Mösch, H.-U.; Essen, L.-O. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 22511–22516. [Google Scholar] [CrossRef] [PubMed]
- Sim, L.; Groes, M.; Olesen, K.; Henriksen, A. Structural and biochemical characterization of the N-terminal domain of flocculin Lg-Flo1p from Saccharomyces pastorianus reveals a unique specificity for phosphorylated mannose. FEBS J. 2013, 280, 1073–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ielasi, F.S.; Verhaeghe, T.; Desmet, T.; Willaert, R.G. Engineering the carbohydrate-binding site of Epa1p from Candida glabrata: Generation of adhesin mutants with different carbohydrate specificity. Glycobiology 2014, 24, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, M.G.; Bauer, F.F.; Marmur, J.; Pretorius, I.S. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 8419–8424. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.-S.; Dranginis, A.M. The Cell Surface Flocculin Flo11 Is Required for Pseudohyphae Formation and Invasion by Saccharomyces cerevisiae. Mol. Biol. Cell. 1998, 9, 161–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupp, S. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999, 18, 1257–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, R.L.; Fink, G.R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 1994, 8, 2974–2985. [Google Scholar] [CrossRef] [PubMed]
- Van Mulders, S.E.; Christianen, E.; Saerens, S.M.G.; Daenen, L.; Verbelen, P.J.; Willaert, R.; Verstrepen, K.J.; Delvaux, F.R. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 178–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffery-Smith, A.; Taori, S.K.; Schelenz, S.; Jeffery, K.; Johnson, E.M.; Borman, A.; Manuel, R.; Brown, C.S. Candida auris: A Review of the Literature. Clin. Microbiol. Rev. 2018, 31, e00029-17. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Oh, S.-H.; Jones, R.; Garnett, J.A.; Salgado, P.S.; Rusnakova, S.; Matthews, S.J.; Hoyer, L.L.; Cota, E. The Peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J. Biol. Chem. 2014, 289, 18401–18412. [Google Scholar] [CrossRef] [PubMed]
- Salgado, P.S.; Yan, R.; Taylor, J.D.; Burchell, L.; Jones, R.; Hoyer, L.L.; Matthews, S.J.; Simpson, P.J.; Cota, E. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 2011, 108, 15775–15779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraushaar, T.; Brückner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mösch, H.-U.; Essen, L.-O. Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-like Adhesin Domain Girdled by Aromatic Bands. Structure 2015, 23, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 is essential for flor formation caused by the C-terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2004, 237, 425–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kron, S.J.; Styles, C.A.; Fink, G.R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1994, 5, 1003–1022. [Google Scholar] [CrossRef] [PubMed]
- Mösch, H.U. Pseudohyphal Development of Saccharomyces cerevisiae. Contrib. Microbiol. 2000, 5, 185–200. [Google Scholar]
- Reynolds, T.B.; Fink, G.R. Bakers’ Yeast, a Model for Fungal Biofilm Formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.B.; Jansen, A.; Peng, X.; Fink, G.R. Mat Formation in Saccharomyces cerevisiae Requires Nutrient and pH Gradients. Eukaryot. Cell 2008, 7, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.A.; Budroni, M. FLO11-Based Model for Air-Liquid Interfacial Biofilm Formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 Is the Primary Factor in Flor Formation Caused by Cell Surface Hydrophobicity in Wild-Type Flor Yeast. Biosci. Biotechnol. Biochem. 2014, 70, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, H.D.; Dupont, K.; Jespersen, L.; Willats, W.G.T.; Arneborg, N. Identification of amino acids involved in the Flo11p-mediated adhesion of Saccharomyces cerevisiaeto a polystyrene surface using phage display with competitive elution. J. Appl. Microbiol. 2007, 103, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Bayly, J.; Douglas, L.; Pretorius, I.; Bauer, F.; Dranginis, A. Characteristics of Flo11-dependent flocculation in. FEMS Yeast Res. 2005, 5, 1151–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purevdorj-Gage, B.; Orr, M.E.; Stoodley, P.; Sheehan, K.B.; Hyman, L.E. The role of FLO11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow-cell system. FEMS Yeast Res. 2007, 7, 372–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, L.M.; Li, L.; Yang, Y.; Dranginis, A.M. Expression and Characterization of the Flocculin Flo11/Muc1, a Saccharomyces cerevisiae Mannoprotein with Homotypic Properties of Adhesion. Eukaryot. Cell 2007, 6, 2214–2221. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.; Willaert, R.G. The N-terminal domain of the Flo11 protein from Saccharomyces cerevisiae is an adhesin without mannose-binding activity. FEMS Yeast Res. 2011, 12, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smit, G.; Straver, M.H.; Lugtenberg, B.J.; Kijne, J.W. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl. Environ. Microbiol. 1992, 58, 3709–3714. [Google Scholar] [PubMed]
- Straver, M.H.; Aar, P.C.V.D.; Smit, G.; Kijne, J.W. Determinants of flocculence of brewer’s yeast during fermentation in wort. Yeast 1993, 9, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Wilcocks, K. The importance of surface charge and hydrophobicity for the flocculation of chain-forming brewing yeast strains and resistance of these parameters to acid washing. FEMS Microbiol. Lett. 1995, 134, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Li, L.; Lipke, P.N.; Dranginis, A.M. Molecular Basis for Strain Variation in the Saccharomyces cerevisiaeAdhesin Flo11p. mSphere 2016, 1, e00129-16. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Bailey, C.W.; Huang, X.; Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998, 284, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Simpson, P.J.; Matthews, S.J.; Cota, E. Backbone 1H, 15N, 13C and Ile, Leu, Val methyl chemical shift assignments for the 33.5 kDa N-terminal domain of Candida albicans ALS1. Biomol. NMR Assign. 2010, 4, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Salgado, P.S.; Yan, R.; Rowan, F.; Cota, E. Expression, crystallization and preliminary X-ray data analysis of NT-Als9-2, a fungal adhesin from Candida albicans. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2011, 67, 467–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnuraj, K.; Bowden, M.G.; Davis, S.; Gurusiddappa, S.; Moore, D.; Choe, D.; Xu, Y.; Höök, M.; Narayana, S.V.L. A “dock, lock, and latch” Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen. Cell 2003, 115, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Bowden, M.G.; Heuck, A.P.; Ponnuraj, K.; Kolosova, E.; Choe, D.; Gurusiddappa, S.; Narayana, S.V.L.; Johnson, A.E.; Höök, M. Evidence for the “Dock, Lock, and Latch” Ligand Binding Mechanism of the Staphylococcal Microbial Surface Component Recognizing Adhesive Matrix Molecules (MSCRAMM) SdrG. J. Biol. Chem. 2008, 283, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, V.K.; Rivera, J.J.; Smeds, E.; Ko, Y.-P.; Bowden, M.G.; Wann, E.R.; Gurusiddappa, S.; Fitzgerald, J.R.; Höök, M. A Structural Model of the Staphylococcus aureus ClfA–Fibrinogen Interaction Opens New Avenues for the Design of Anti-Staphylococcal Therapeutics. PLoS Pathog. 2008, 4, e1000226. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Green, C.B.; Oh, S.-H.; Zhao, X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 2008, 46, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Phan, Q.T.; Fu, Y.; Ibrahim, A.S.; Filler, S.G.; Zhang, M.; Waring, A.J.; Edwards, J.E., Jr. Functional and Structural Diversity in the Als Protein Family of Candida albicans. J. Biol. Chem. 2004, 279, 30480–30489. [Google Scholar] [CrossRef] [PubMed]
- Loza, L.; Fu, Y.; Ibrahim, A.S.; Sheppard, D.C.; Filler, S.G.; Edwards, J.E. Functional analysis of theCandida albicans ALS1 gene product. Yeast 2004, 21, 473–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 2004, 150, 2415–2428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otoo, H.N.; Lee, K.G.; Qiu, W.; Lipke, P.N. Candida albicans Als Adhesins Have Conserved Amyloid-Forming Sequences. Eukaryot. Cell 2008, 7, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Garcia, M.C.; Lipke, P.N.; Dufrêne, Y.F. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20744–20749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipke, P.N.; Garcia, M.C.; Alsteens, D.; Ramsook, C.B.; Klotz, S.A.; Dufrêne, Y.F. Strengthening relationships: Amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 2012, 20, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Staab, J.F.; Bradway, S.D.; Fidel, P.L.; Sundstrom, P. Adhesive and Mammalian Transglutaminase Substrate Properties of Candida albicans Hwp1. Science 1999, 283, 1535–1538. [Google Scholar] [CrossRef] [PubMed]
- Tsuchimori, N.; Sharkey, L.L.; Fonzi, W.A.; French, S.W.; Edwards, J.E.; Filler, S.G. Reduced Virulence of HWP1-Deficient Mutants of Candida albicans and Their Interactions with Host Cells. Infect. Immun. 2000, 68, 1997–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staab, J.F.; Bahn, Y.-S.; Tai, C.-H.; Cook, P.F.; Sundstrom, P. Expression of Transglutaminase Substrate Activity on Candida albicans Germ Tubes through a Coiled, Disulfide-bonded N-terminal Domain of Hwp1 Requires C-terminal Glycosylphosphatidylinositol Modification. J. Biol. Chem. 2004, 279, 40737–40747. [Google Scholar] [CrossRef] [PubMed]
- Dalle, F.; Wächtler, B.; L’Ollivier, C.; Holland, G.; Bannert, N.; Wilson, D.; Labruère, C.; Bonnin, A.; Hube, B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 2010, 12, 248–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naglik, J.R.; Moyes, D.L.; Wächtler, B.; Hube, B. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011, 13, 963–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; He, H.; Dong, Y.; Pan, H. Hyphae-Specific Genes HGC1, ALS3, HWP1, and ECE1 and Relevant Signaling Pathways in Candida albicans. Mycopathologia 2013, 176, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Nguyen, Q.B.; Wolyniak, M.J.; Frechette, G.; Lehman, C.R.; Fox, B.K.; Sundstrom, P. Release of transcriptional repression through the HCR promoter region confers uniform expression of HWP1 on surfaces of Candida albicans germ tubes. PLoS ONE 2018, 13, e0192260-24. [Google Scholar] [CrossRef] [PubMed]
- Hayek, P.; Dib, L.; Yazbeck, P.; Beyrouthy, B.; Khalaf, R.A. Characterization of Hwp2, a Candida albicans putative GPI-anchored cell wall protein necessary for invasive growth. Microbiol. Res. 2010, 165, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.S.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Schneider, H.A.; Nett, J.E.; Sheppard, D.C.; Filler, S.G.; Andes, D.R.; Mitchell, A.P. Complementary Adhesin Function in C. albicans Biofilm Formation. Curr. Biol. 2008, 18, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Andes, D.R.; Nett, J.E.; Smith, F.J.; Yue, F.; Phan, Q.-T.; Edwards, J.E.; Filler, S.G.; Mitchell, A.P. Critical Role of Bcr1-Dependent Adhesins in C. albicans Biofilm Formation In Vitro and In Vivo. PLoS Pathog. 2006, 2, e63. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Bennett, R.J. Hwp1 and Related Adhesins Contribute to both Mating and Biofilm Formation in Candida albicans. Eukaryot. Cell 2009, 8, 1909–1913. [Google Scholar] [CrossRef] [PubMed]
- Nobbs, A.H.; Vickerman, M.M.; Jenkinson, H.F. Heterologous Expression of Candida albicans Cell Wall-Associated Adhesins in Saccharomyces cerevisiae Reveals Differential Specificities in Adherence and Biofilm Formation and in Binding Oral Streptococcus gordonii. Eukaryot. Cell 2010, 9, 1622–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwivedi, P.; Thompson, A.; Xie, Z.; Kashleva, H.; Ganguly, S.; Mitchell, A.P.; Dongari-Bagtzoglou, A. Role of Bcr1-Activated Genes Hwp1 and Hyr1 in Candida Albicans Oral Mucosal Biofilms and Neutrophil Evasion. PLoS ONE 2011, 6, e16218. [Google Scholar] [CrossRef] [PubMed]
- Orsi, C.F.; Borghi, E.; Colombari, B.; Neglia, R.G.; Quaglino, D.; Ardizzoni, A.; Morace, G.; Blasi, E. Impact of Candida albicans hyphal wall protein 1 (HWP1) genotype on biofilm production and fungal susceptibility to microglial cells. Microb. Pathog. 2014, 69–70, 20–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponniah, G.; Rollenhagen, C.; Bahn, Y.-S.; Staab, J.F.; Sundstrom, P. State of differentiation defines buccal epithelial cell affinity for cross-linking to Candida albicans Hwp1. J. Oral Patho.l Med. 2007, 36, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.; Stassen, C.; Stals, I.; Donohue, D.S.; Devreese, B.; De Greve, H.; Willaert, R.G. The N-Terminal Domain of the Flo1 Flocculation Protein from Saccharomyces cerevisiae Binds Specifically to Mannose Carbohydrates. Eukaryot. Cell 2011, 10, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Donohue, D.S.; Ielasi, F.S.; Goossens, K.V.Y.; Willaert, R.G. The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans. Mol. Microbiol. 2011, 80, 1667–1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-Glycan Interaction Network-Based Identification of Host Receptors of Microbial Pathogenic Adhesins. MBio 2016, 7, e00584-16. [Google Scholar] [CrossRef] [PubMed]
- Bony, M.; Thines-Sempoux, D.; Barre, P.; Blondin, B. Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein Flo1p. J. Bacteriol. 1997, 179, 4929–4936. [Google Scholar] [CrossRef] [PubMed]
- Spillmann, D.; Burger, M.M. Carbohydrate-carbohydrate interactions in adhesion. J. Cell. Biochem. 1996, 61, 562–568. [Google Scholar] [CrossRef]
- Haseley, S.R.; Vermeer, H.J.; Kamerling, J.P.; Vliegenthart, J.F.G. Carbohydrate self-recognition mediates marine sponge cellular adhesion. Proc. Natl. Acad. Sci. USA 2001, 98, 9419–9424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucior, I.; Burger, M.M. Carbohydrate–carbohydrate interactions in cell recognition. Curr. Opin. Struct. Biol. 2004, 14, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Busquets, X.; Kornig, A.; Bucior, I.; Burger, M.M.; Anselmetti, D. Self-Recognition and Ca2+-Dependent Carbohydrate-Carbohydrate Cell Adhesion Provide Clues to the Cambrian Explosion. Mol. Biol. Evol. 2009, 26, 2551–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mill, P.J. The Nature of the Interactions between Flocculent Cells in the Flocculation of Saccharomyces cerevisiae. J. Gen. Microbiol. 1964, 35, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- JAYATISSA, P.M.; ROSE, A.H. Role of Wall Phosphomannan in Flocculation of Saccharomyces cerevisiae. J. Gen. Microbiol. 1976, 96, 165–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, B.L.A.; Poon, N.H.; James, A.P.; Seligy, V.L. Possible Mechanism for Flocculation Interactions Governed by Gene FLO1 in Saccharomyces cerevisiae. J. Bacteriol. 1982, 150, 878–889. [Google Scholar] [PubMed]
- Willaert, R.; Kasas, S.; Devreese, B.; Dietler, G. Yeast Nanobiotechnology. Fermentation 2016, 2, 18. [Google Scholar] [CrossRef]
- Alsteens, D.; Beaussart, A.; Derclaye, S.; El-Kirat-Chatel, S.; Park, H.R.; Lipke, P.N.; Dufrêne, Y.F. Single-cell force spectroscopy of Als-mediated fungal adhesion. Anal. Methods 2013, 5, 3657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Kirat-Chatel, S.; Beaussart, A.; Derclaye, S.; Alsteens, D.; Kucharíková, S.; Van Dijck, P.; Dufrêne, Y.F. Force Nanoscopy of Hydrophobic Interactions in the Fungal Pathogen Candida glabrata. ACS Nano 2015, 9, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Touhami, A. Aggregation of yeast cells: Direct measurement of discrete lectin-carbohydrate interactions. Microbiology 2003, 149, 2873–2878. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Šťovíček, V.; Hlaváček, O.; Chernyavskiy, O.; Štěpánek, L.; Kubínová, L.; Palková, Z. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J. Cell. Biol. 2011, 194, 679–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klotz, S.A.; Gaur, N.K.; Lake, D.F.; Chan, V.; Rauceo, J.; Lipke, P.N. Degenerate Peptide Recognition by Candida albicans Adhesins Als5p and Als1p. Infect. Immun. 2004, 72, 2029–2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argimón, S.; Wishart, J.A.; Leng, R.; Macaskill, S.; Mavor, A.; Alexandris, T.; Nicholls, S.; Knight, A.W.; Enjalbert, B.; Walmsley, R.; et al. Developmental Regulation of an Adhesin Gene during Cellular Morphogenesis in the Fungal Pathogen Candida albicans. Eukaryot. Cell 2007, 6, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a Multifunctional Adhesin and Invasin. Eukaryot. Cell 2011, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Issa, S.; Moran, A.P.; Ustinov, S.N.; Lin, J.H.-H.; Ligtenberg, A.J.; Karlsson, N.G. O-linked oligosaccharides from salivary agglutinin: Helicobacter pylori binding sialyl-Lewis x and Lewis b are terminating moieties on hyperfucosylated oligo-N-acetyllactosamine. Glycobiology 2010, 20, 1046–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, M.P.; Haidaris, C.G. Analysis of Candida albicans Adhesion to Salivary Mucin. Infect. Immun. 1993, 61, 1940–1949. [Google Scholar] [PubMed]
- de Repentigny, L.; Aumont, F.; Bernard, K.; Belhumeur, P. Characterization of Binding of Candida albicans to Small Intestinal Mucin and Its Role in Adherence to Mucosal Epithelial Cells. Infect. Immun. 2000, 68, 3172–3179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.C.; Lee, J.T.; Ramsook, C.B.; Alsteens, D.; Dufrêne, Y.F.; Lipke, P.N. A Role for Amyloid in Cell Aggregation and Biofilm Formation. PLoS ONE 2011, 6, e17632. [Google Scholar] [CrossRef] [PubMed]
- de Nobel, H.; Lipke, P.N. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell. Biol. 1994, 4, 42–45. [Google Scholar] [CrossRef]
- Lipke, P.N.; Klotz, S.A.; Dufrêne, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like β-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microb. Mol. Biol. Rev. 2018, 82, 237. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Cota, E. Candida albicans Agglutinin-Like Sequence (Als) Family Vignettes: A Review of Als Protein Structure and Function. Front. Microbiol. 2016, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.; Nett, J.; Oschel, P.; Albrecht, R.; Marchillo, K.; Pitula, A. Development and Characterization of an In Vivo Central Venous Catheter Candida albicans Biofilm Model. Infect. Immun. 2004, 72, 6023–6031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuford, J.A.; Rouse, M.S.; Piper, K.E.; Steckelberg, J.M.; Patel, R. Evaluation of Caspofungin and Amphotericin B Deoxycholate against Candida albicans Biofilms in an Experimental Intravascular Catheter Infection Model. J. Infect. Dis. 2006, 194, 710–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzell, A.L.; Chaturvedi, A.K.; Pierce, C.G.; Prasad, D.; Uppuluri, P.; Lopez-Ribot, J.L. Treatment and prevention of Candida albicans biofilms with caspofungin in a novel central venous catheter murine model of candidiasis. J. Antimicrob. Chemother. 2009, 64, 567–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Řičicová, M.; Kucharíková, S.; Tournu, H.; Hendrix, J.; Bujdáková, H.; Van Eldere, J.; Lagrou, K.; Van Dijck, P. Candida albicans biofilm formation in a new in vivo rat model. Microbiology 2010, 156, 909–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nett, J.E.; Brooks, E.G.; Cabezas-Olcoz, J.; Sanchez, H.; Zarnowski, R.; Marchillo, K.; Andes, D.R. Rat Indwelling Urinary Catheter Model of Candida albicans Biofilm Infection. Infect. Immun. 2014, 82, 4931–4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, E.P.; Bui, C.K.; Nett, J.E.; Hartooni, N.; Mui, M.C.; Andes, D.R.; Nobile, C.J.; Johnson, A.D. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol. Microbiol. 2015, 96, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2017, 16, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Dranginis, A.M.; Rauceo, J.M.; Coronado, J.E.; Lipke, P.N. A Biochemical Guide to Yeast Adhesins: Glycoproteins for Social and Antisocial Occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Palecek, S.P. Distinct domains of the Candida albicans adhesin Eap1p mediate cell–cell and cell–substrate interactions. Microbiology 2008, 154, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Frade, J.P.; Arthington-Skaggs, B.A. Effect of serum and surface characteristics on Candida albicans biofilm formation. Mycoses 2010, 54, e154–e162. [Google Scholar] [CrossRef] [PubMed]
- Sandini, S.; Stringaro, A.; Arancia, S.; Colone, M.; Mondello, F.; Murtas, S.; Girolamo, A.; Mastrangelo, N.; De Bernardis, F. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 2011, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.T.; Silva, S.; Pereira, L.; Williams, D.W.; Azeredo, J.; Henriques, M. Effect of progesterone on Candida albicans vaginal pathogenicity. Int J. Med. Microbiol. 2014, 304, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.V.; Mitchell, A.P. Candida albicans Biofilm Development and Its Genetic Control. Microbiol. Spectr. 2015, 3, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Zupancic, M.L.; Frieman, M.; Smith, D.; Alvarez, R.A.; Cummings, R.D.; Cormack, B.P. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 2008, 68, 547–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, K.; Wolf, M. Cryo-electron microscopy for structural analysis of dynamic biological macromolecules. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Hospenthal, M.K.; Zyla, D.; Costa, T.R.D.; Redzej, A.; Giese, C.; Lillington, J.; Glockshuber, R.; Waksman, G. The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod. Structure 2017, 25, 1829–1838. [Google Scholar] [CrossRef] [PubMed]

| Micro-Organism | Adhesin | Ligand in the Structure | Mutations | PDB Code | Interacting Substrate/Function Properties | Refs |
|---|---|---|---|---|---|---|
| C. albicans | NT-Als3p (residues1-313) | Heptathreonine | - | 4LEB | Endothelial and epithelial cells; fibronectin, laminin, type-IV collagen; abiotic surfaces (glass, plastics) | [62] |
| Apo | K59M, A116V, Y301F | 4LEE | [62] | |||
| NT-Als3p (residues 1-299) | Apo | - | 4LE8 | * | ||
| NT-Als9-2p | Apo | - | 2Y7N | [63] | ||
| Apo | G299W | 2Y7O, 2YLH | ||||
| Fg-γ 1 | - | 2Y7L | ||||
| C. glabrata | N-Epa1p | Gal | - | 4A3X | Epithelial cells, fibronectin, mucin | [43] |
| Galβ-1,3-Glc | - | 4AF9 | [44] | |||
| Galβ-1,3-Glc | E227D, Y228N | 4AFC | [44] | |||
| Galβ-1,3-GalNAc (T antigen) | - | 4ASL | [44] | |||
| 4D3W | [52] | |||||
| Galβ-1,4-Glc (lactose) | 4COU | [52] | ||||
| Glycerol | E227D, Y228N, D229N | 4AFA | [44] | |||
| Glycerol | R226I, E227G, Y228K | 4AFB | [44] | |||
| N-Epa6p | Galβ-1,4-Glc (lactose) | 4COU | [52] | |||
| Galβ-1,3-GalNAc (T-antigen) | 4COW | [52] | ||||
| N-acetyl-D-lactosamine | 4COY | [52] | ||||
| Lacto-N-biose | 4COZ | [52] | ||||
| α1-3-galactobiose | 4COV | [52] | ||||
| N-Epa9p | Galβ-1,4-Glc | - | 4CP0 | * | ||
| Galβ-1,3-GlcNAc | - | 4CP1 | * | |||
| Galβ-1,4-GlcNAc | - | 4CP2 | * | |||
| S. cerevisiae | N-Flo1p | Apo | - | 4LHL | Cell-cell interaction via cell surface mannans | [39] |
| Man | - | 4LHK | [39] | |||
| N-Lg-Flo1p | Apo | - | 4GQ7 | Cell-cell interaction via cell surface mannans and phosphomannans | [5] | |
| Manα-1,2-Man | - | 4LHK | [39] | |||
| N-Lg-Flo5p | Apo | - | 2XJQ | Cell-cell interaction via cell surface mannans | [6] | |
| Man | - | 2XJP | [6] | |||
| Man3(D1) | - | 2XJT | [6] | |||
| Man5(D2-3) | - | 2XJR | [6] | |||
| Manα-1,2-Man | - | 2XJS | [6] | |||
| Manα-1,2-Man | S277A | 2XJU | [6] | |||
| Glc | - | 2XJV | [6] | |||
| N-Flo11p | Apo | - | 4UYR | Cell-cell and cell-hydrophobic plastic adhesion via hydrophobic interactions | [64] | |
| - | 4UYS | [64] | ||||
| - | 4UYT | [64] |
| Microorganism | Adhesin | Ligand | Dissociation Constant KD (mM) | Refs |
|---|---|---|---|---|
| C. albicans | N-Als1p | Fibronectin | 0.0016 ± 0.0006 | [111] |
| Laminin | 0.013 ± 0.002 | [111] | ||
| Fucose | 0.21 ± 0.03 | [111] | ||
| N-Als1p | 0.020 ± 0.001 | [111] | ||
| N-Als3p | Fibronectin | 0.41 ± 0.04 | [112] | |
| Laminin | 0.01 ± 0.001 | [112] | ||
| GlcNAc | 0.034 ± 0.004 | [112] | ||
| N-Als3p | 0.014 ± 0.002 | [112] | ||
| C. glabrata | N-Epa1p | Galβ | 0.115 ± 0.011 | [43] |
| Galβ-1,4-Glc 1 | 0.031 ± 0.004 | [43] | ||
| 0.035 ± 0.006 | [3] | |||
| Galα-1,3-Gal 2 | 0.0050 ± 0.0009 | [7] | ||
| Galβ-1,3-Gal 3 | 0.0009 ± 0.00005 | [7] | ||
| Galβ-1,3-GalNAc 4 | 0.0035 ± 0.0004 | [43] | ||
| 0.0021 ± 0.0003 | [3] | |||
| 0.0009 ± 0.0001 | [7] | |||
| Galβ-1,3-GlcNAc 5 | 0.0049 ± 0.003 | [7] | ||
| Galβ-1,4-GlcNAc 6 | 0.0020 ± 0.0002 | [7] | ||
| Mucin | 0.0047 ± 0.0009 | [112] | ||
| Fibronectin | 0.911 10−3 ± 0.122 10−3 | [8] | ||
| N-Epa1p E227A | Fibronectin | 0.317 10−3 ± 0.026 10−3 | [8] | |
| N-Epa1p Y228W | Fibronectin | 0.545 10−3 ± 0.058 10−3 | [8] | |
| N-Epa6p | Galα-1,3-Gal 2 | 0.0004 ± 0.0007 | [7] | |
| Galβ-1,3-Gal 3 | 0.0003 ± 0.0002 | [7] | ||
| Galβ-1,3-GalNAc 4 | 0.0005 ± 0.0011 | [7] | ||
| Galβ-1,3-GlcNAc 5 | 0.0148 ± 0.0001 | [7] | ||
| Galβ-1,4-GlcNAc 6 | 0.0094 ± 0.0013 | [7] | ||
| N-Epa7p | Galα-1,3-Gal 2 | 0.0124 ± 0.0009 | [7] | |
| Galβ-1,3-Gal 3 | 0.0013 ± 0.0001 | [7] | ||
| Galβ-1,3-GalNAc 4 | 0.0015 ± 0.00001 | [7] | ||
| Galβ-1,3-GlcNAc 5 | 0.0147 ± 0.0004 | [7] | ||
| Galβ-1,4-GlcNAc 6 | 0.0048 ± 0.0004 | [7] | ||
| S. cerevisiae | N-Flo1p | Man | 8.7 ± 0.4 | [39] |
| Manα-1,2-Man | 0.6 ± 0.1 | [39] | ||
| Manα-1,3-Man | 3.3 ± 0.3 | [39] | ||
| Manα-1,6-Man | 6.9 ± 0.6 | [39] | ||
| Glc | >100 | [39] | ||
| N-Lg-Flo1p | Man | 0.8 ± 0.03 | [54] | |
| Man | 0.5 ± 0.05 | [39] | ||
| Man1P 7 | 0.06 ± 0.002 | [54] | ||
| Manα-1,2-Man | 4.5 ± 0.38 | [54] | ||
| Manα-1,3-Man | 3.9 ± 0.98 | [54] | ||
| Manα-1,6-Man | 3.0 ± 0.33 | [54] | ||
| Glc | 5.8 ± 0.80 | [54] | ||
| Glc1P 8 | 0.41 ± 0.03 | [54] | ||
| N-Flo5p (S227A) | Man | 29.3 ± 3.6 (9.7 ± 0.6) | [6] | |
| Manα-1,2-Man | 3.5 ± 0.3 (1.6 ± 0.1) | [6] | ||
| Manα-1,3-Man | No binding | [6] | ||
| Manα-1,6-Man | No binding | [6] | ||
| Man3(D1) | 2.8 ± 0.2 | [6] | ||
| Man5(D2-3) | 2.2 ± 0.5 | [6] | ||
| Glc | >1000 | [6] | ||
| N-Flo11p | N-Flo11 | 0.0195 ± 0.0023 | [64] |
| Cell type | Adhesin | Ligand | Rupture force | Comments | Refs |
|---|---|---|---|---|---|
| C. albicans | Als5p | Fibronectin | 2800 ± 600 pN | SMFS 1 in vitro | [122] |
| C. glabrata | Epa6p | Hydrophobic surface (dodecane-thiol) | 31–52 nN | SCFC 2: cell immobilization on the AFM probe using dopamine | [123] |
| S. cerevisiae | Flo1p | Flo1p | 300 (100–600) | SMFS | [39] |
| S. pastorianus | Lg-Flo1p | Glucose | 121 ± 53 pN | Amylose-coated AFM probe interact with cell surface | [124] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willaert, R.G. Adhesins of Yeasts: Protein Structure and Interactions. J. Fungi 2018, 4, 119. https://doi.org/10.3390/jof4040119
Willaert RG. Adhesins of Yeasts: Protein Structure and Interactions. Journal of Fungi. 2018; 4(4):119. https://doi.org/10.3390/jof4040119
Chicago/Turabian StyleWillaert, Ronnie G. 2018. "Adhesins of Yeasts: Protein Structure and Interactions" Journal of Fungi 4, no. 4: 119. https://doi.org/10.3390/jof4040119





