Role of Virulence Determinants in Candida albicans’ Resistance to Novel 2-bromo-2-chloro-2-(4-chlorophenylsulfonyl)-1-phenylethanone
Abstract
:1. Introduction
2. Materials and Methods
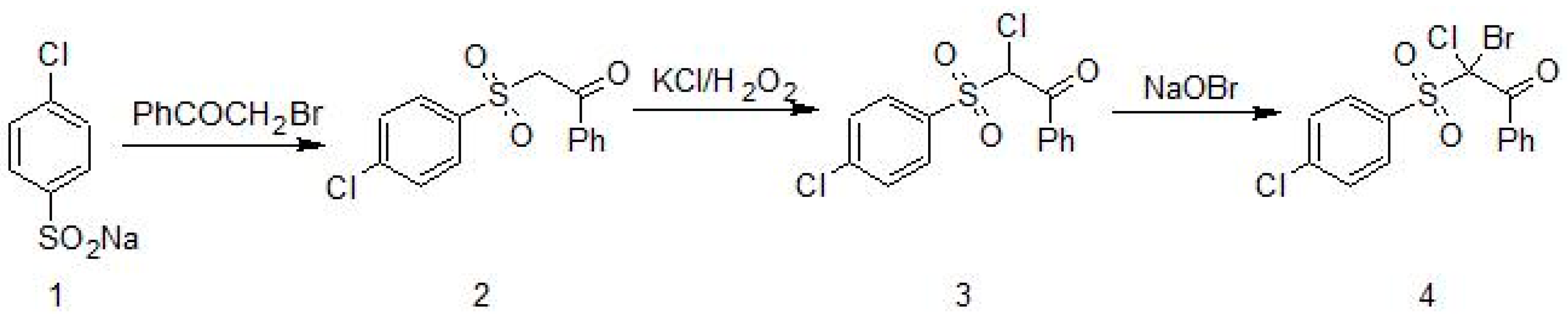
2.1. Synthesis of 2-Bromo-2-Chloro-2-[(4-Chlorophenyl)Sulfonyl]-1-Phenylethanone (Compound 4)
2.2. Strains, Media, and Growth Conditions
2.3. Compound 4 Susceptibility Testing
2.4. Yeast Cell Growth Inhibition under Compound 4 in Time Intervals
2.5. Mononuclear Cell PBMCs. In Vitro Candida Stimulation
2.6. Fluorescence Microscopy
2.7. Cytokine Analysis
2.8. Assay of Candida Adherence to Human Line Caco-2 (ATCC HTB27, LGC, Poland). Inhibition of Adhesion: Microwell-Based Assay
2.9. Analysis of Gene Expression
2.10. Statistical Analysis
3. Results
3.1. In Vitro Anti-Candida Activity of Compound 4
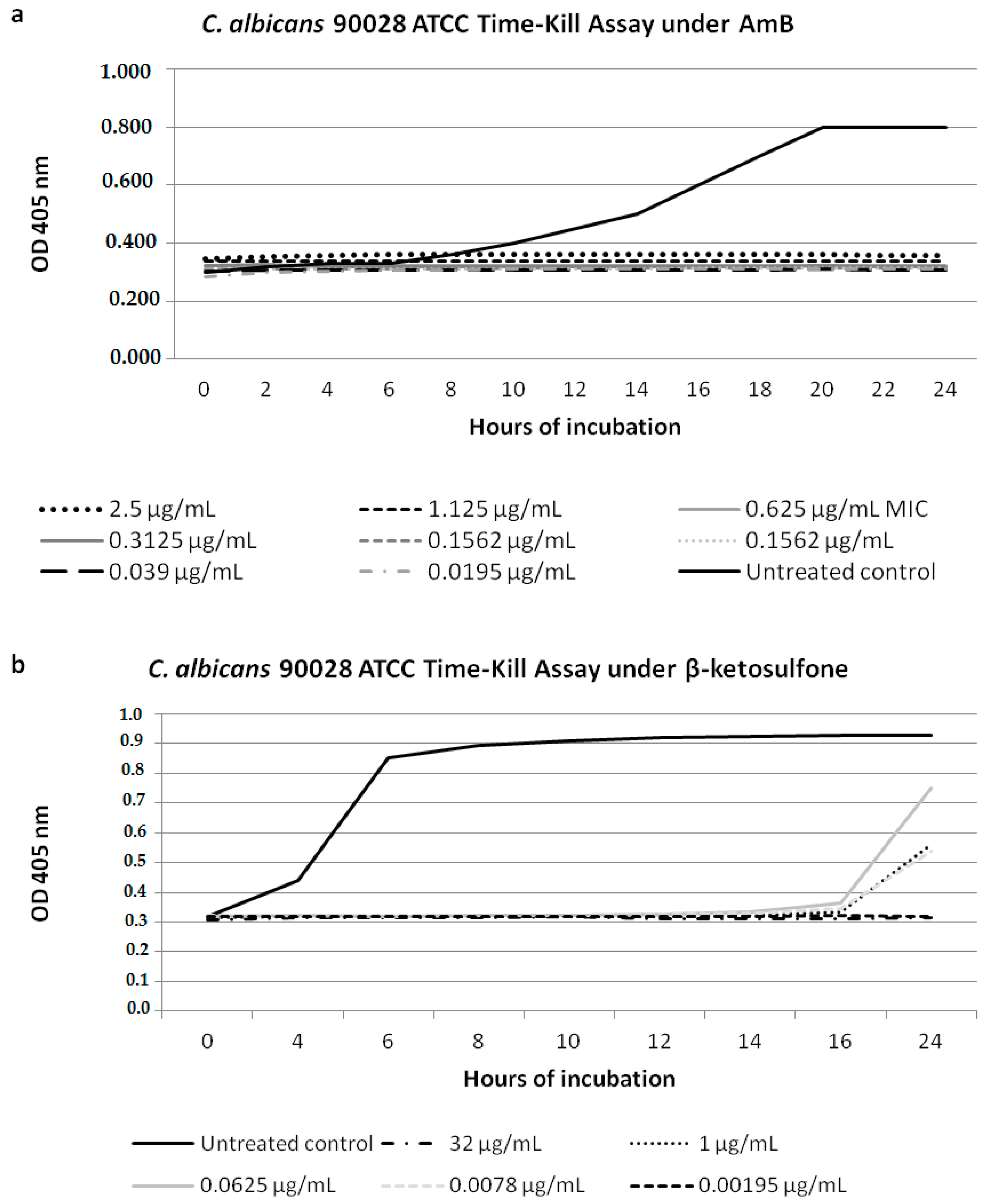
3.2. Candida Cell Growth Inhibition under Compound 4
3.3. Compound 4 Treatment Results in Inhibition of Adhesive Properties of Candida Strains
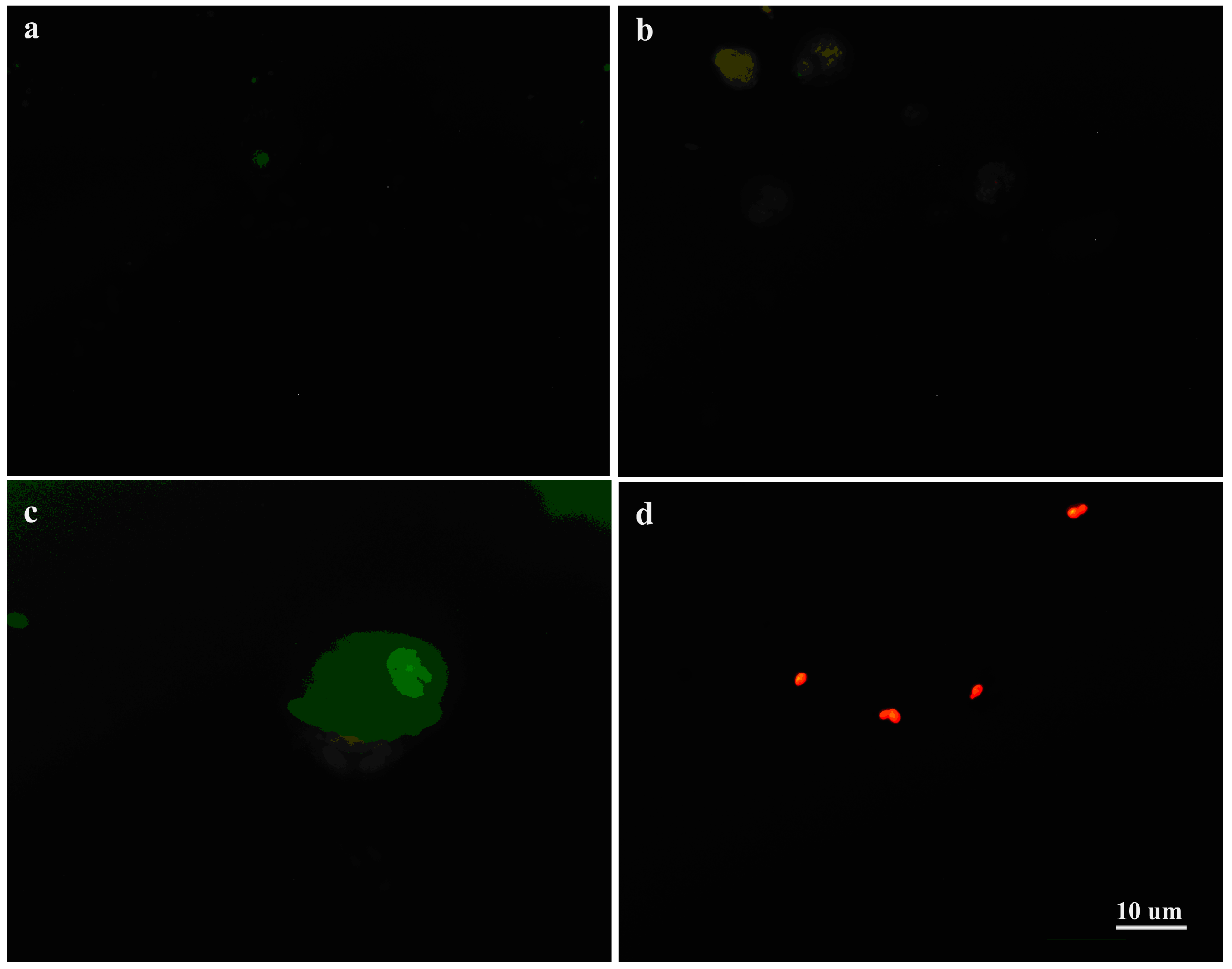
3.4. Interaction of C. albicans Cells with Macrophages and Fungal Morphology under Compound 4
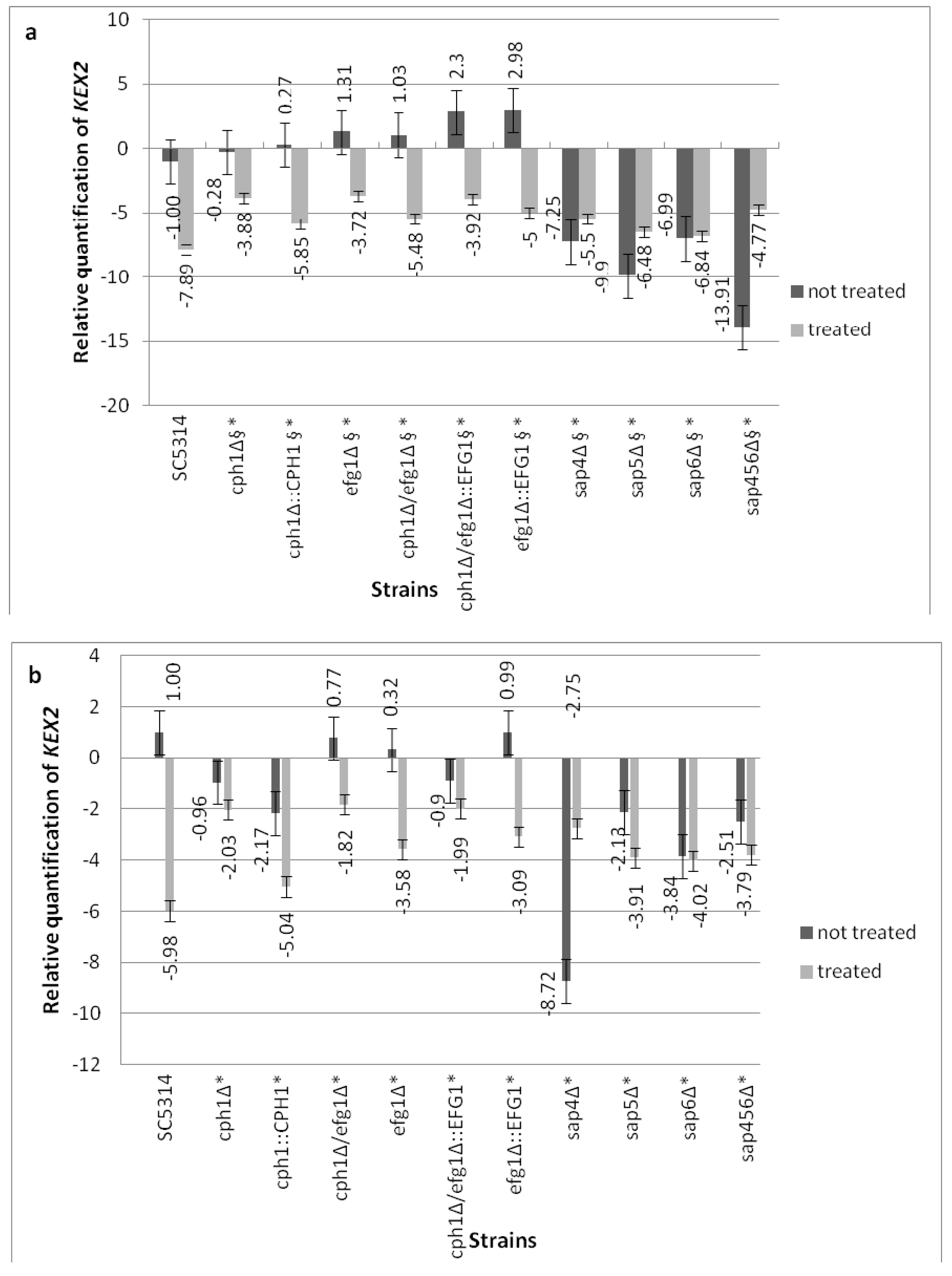
3.5. Compound 4 Modulates the KEX2 mRNA Expression
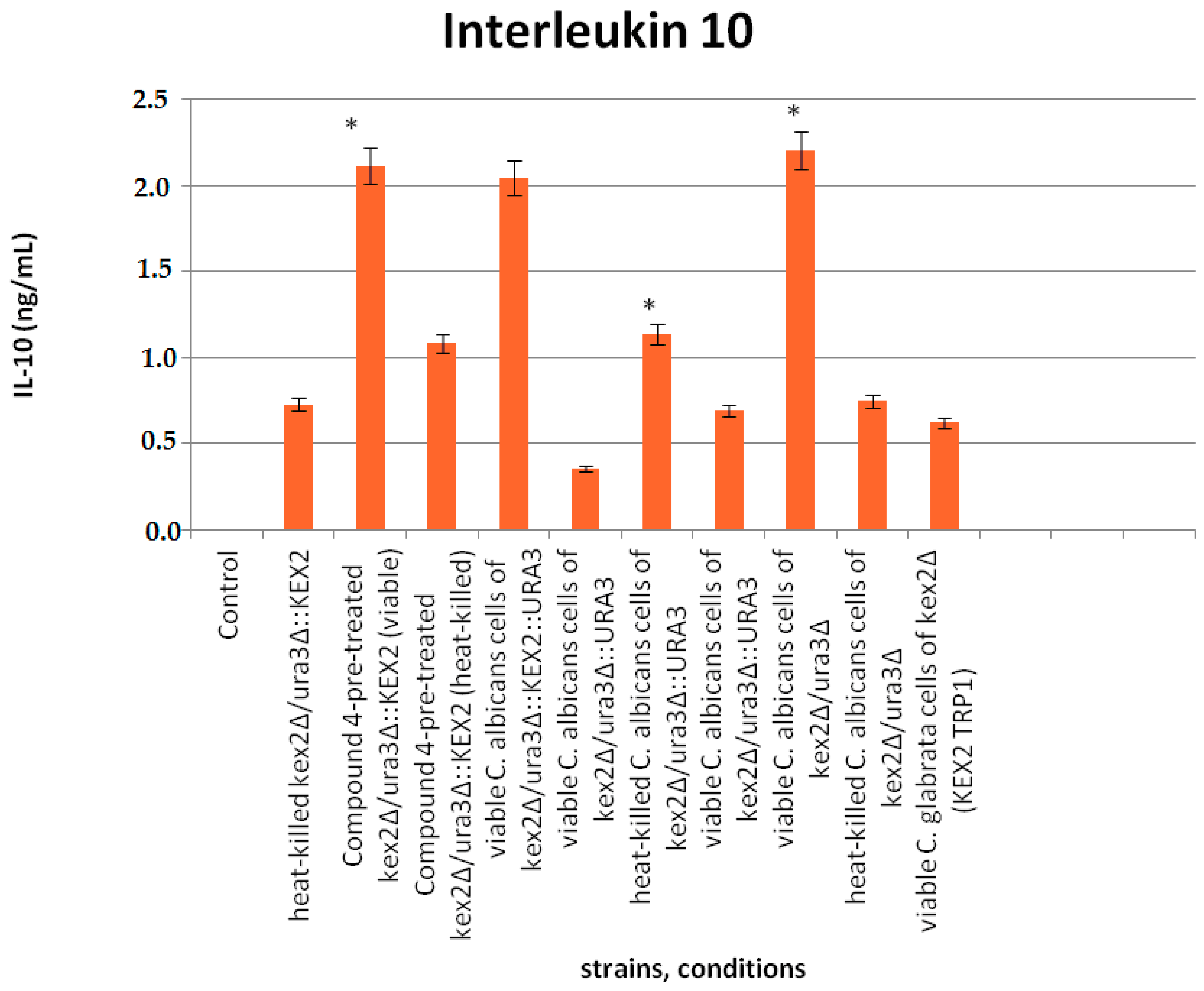
3.6. Interaction of Compound 4 with Interleukine 10
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Staniszewska, M.; Bondaryk, M.; Kazek, M.; Gliniewicz, A.; Braunsdorf, C.; Schaller, M.; Mora-Montes, HM.; Ochal, Z. Effect of the serine protease KEX2 on Candida albicans virulence under halogenated methyl sulfones. Future Microbiol. 2017, 12, 285–306. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M.; Bondaryk, M.; Wieczorek, M.; Estrada-Mata, E.; Mora-Montes, H.M.; Ochal, Z. Antifungal effect of novel 2-bromo-2-chloro-2(4-chlorophenylsulfonyl)-1-phenylethanone against Candida strains. Front. Microbiol. 2016, 7, 1309. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, D.; Gao, S.; Lin, J.; Chen, Z.; Zhao, W. Role of Candida albicans-secreted aspartyl proteinases (Saps) in severe early childhood caries. Int. J. Mol. Sci. 2014, 15, 10766–10779. [Google Scholar] [CrossRef] [PubMed]
- Newport, G.; Agabian, N. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J. Biol. Chem. 1997, 272, 28954–28961. [Google Scholar] [CrossRef] [PubMed]
- Bader, O.; Schaller, M.; Klein, S.; Kukula, J.; Haack, K.; Mühlschlegel, F.; Korting, H.C.; Schäfer, W.; Hube, B. The KEX2 gene Candida glabrata is required for cell surface integrity. Mol. Microbiol. 2001, 41, 1431–1444. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Budge, S.; Kaloriti, D.; Tillmann, A.; Jacobsen, M.D.; Yin, Z.; Ene, J.V.; Bohovych, I.; Sandai, D.; Kastora, S.; et al. Stress adaptation in a pathogenic fungus. J. Exp. Biol. 2014, 217, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Sudbery, I.; Ramsdale, M. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA 2003, 100, 14327–14332. [Google Scholar] [CrossRef] [PubMed]
- Bondaryk, M.; Grabowska-Jadach, I.; Ochal, Z.; Sygitowicz, G.; Staniszewska, M. Possible role of hydrolytic enzymes (Sap, Kex2) in Candida albicans response to aromatic compounds bearing a sulfone moiety. Chem. Papers 2016, 70, 1336–1350. [Google Scholar] [CrossRef]
- Tripathi, H.S.; Luqman, S.; Meena, A.; Khan, F. Genomic identification of potential targets unique to Candida albicans for the discovery of antifungal agents. Curr. Drug Targets 2014, 15, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.J.; Wang, J.S.; Lin, C.Y.; Chen, C.G.; Hsiao, T.Y.; Hsu, C.T.; Su, C.L.; Fann, M.J.; Ching, Y.T.; Yang, Y.L. Efg1 involved in drug resistance by regulating the expression of ERG3 in Candida albicans. Antimicrob. Agents Chemother. 2005, 49, 1213–1215. [Google Scholar] [CrossRef] [PubMed]
- Prasad, T.; Hameed, S.; Manoharlal, R.; Biswas, S.; Mukhopadhyay, C.K.; Goswami, S.K.; Prasad, R. Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS Yeast Res. 2010, 10, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Bale, M.; Buschelman, B.; Lancaster, M.; Espinel-Ingroff, A.; Rex, J.H.; Rinaldi, M.G. Selection of candidate quality control isolates and tentative quality control ranges for in vitro susceptibility testing of yeast isolates by National Committee for Clinical Laboratory Standards proposed standard methods. J. Clin. Microbiol. 1994, 32, 1650–1653. [Google Scholar] [PubMed]
- Gillum, A.M.; Tsai, E.Y.; Kirsch, D.R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984, 198, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Fonzi, W.A.; Irwin, M.Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics 1993, 134, 717–728. [Google Scholar] [PubMed]
- Liu, H.; Köhler, J.; Fink, G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.J.; Köhler, J.R.; DiDomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R. Nonfilamentous C. albicans mutants are avirulent. Cell 1997, 90, 939–949. [Google Scholar] [CrossRef]
- Lermann, U.; Morschhäuser, J. Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiolology 2008, 154, 3281–3295. [Google Scholar] [CrossRef] [PubMed]
- Newport, G.; Kuo, A.; Flattery, A.; Gill, C.; Blake, J.J.; Kurtz, M.B.; Abruzzo, G.K.; Agabian, N. Inactivation of Kex2p diminishes the virulence of Candida albicans. J. Biol. Chem. 2003, 278, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M.; Bondaryk, M.; Ochal, Z. Susceptibility of Candida albicans to new synthetic sulfone derivatives. Arch. Pharm. 2015, 348, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. CLSI Document M27-A3, Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, Approved Standard, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Shields, R.K.; Nguyen, M.H.; Du, C.; Press, E.; Cheng, S.; Clancy, C.J. Paradoxical effect of caspofungin against Candida bloodstream isolates is mediated by multiple pathways but eliminated in human serum. Antimicrob. Agents Chemother. 2011, 55, 2641–2647. [Google Scholar] [CrossRef] [PubMed]
- Bink, A.; Govaert, G.; Vandenbosh, D.; Kuchariková, S.; Coenye, T.; Nelis, H.; Van Dijck, P.; Cammue, B.P.; Thevissen, K. Transcription factor Efg1 contributes to the tolerance of Candida albicans biofilms against antifungal agents in vitro and in vivo. J. Med. Microbiol. 2012, 61, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Temple, S.E.; Pham, K.; Glendenning, P.; Philips, M.; Waterer, G.W. Endotoxin induced TNF and IL-10 mRNA production is higher in male than female donors, correlation with elevated expression of TLR4. Cell. Immunol. 2008, 251, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M.; Bondaryk, M.; Ochal, Z. Polish Patent Application PL-P.408765. 4 July 2014. [Google Scholar]
- Amberg, D.; Burke, D.; Strathern, J. (Eds.) Yeast RNA isolations, techniques and protocols. In Methods in Yeast Genetics; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2007; pp. 127–131. [Google Scholar]
- Naglik, J.R.; Moyes, D.; Makwana, J.; Kanzaria, P.; Tsichlaki, E.; Weindl, G.; Tappuni, A.R.; Rogers, C.A.; Woodman, A.J.; Challacombe, S.J.; et al. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidasis. Microbiology 2008, 154, 3266–3280. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Shukla, P.K. Amphotericin B resistance leads to enhanced proteinase and phospholipase activity and reduced germ tube formation in Candida albicans. Fungal Biol. 2010, 114, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.P.; Gursky, L.C.; Rosa, R.T.; Rymovicz, A.U.; Campelo, P.M.; Grégio, A.M.; Koga-Ito, C.Y.; Samaranayake, L.P.; Rosa, E.A. Non-steroidal anti-inflammatory drugs may modulate the protease activity of Candida albicans. Microb. Pathog. 2010, 49, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Samaranayake, Y.H.; Cheung, B.P.; Yau, J.Y.; Yeung, S.K.; Samaranayake, L.P. Human serum promotes Candida biofilm growth and virulence gene expression on silicone biomaterial. PLoS ONE 2013, 8, e6292. [Google Scholar] [CrossRef] [PubMed]
- Bondaryk, M.; Ochal, Z.; Staniszewska, M. Sulfone derivatives reduce growth, adhesion and aspartic protease SAP2 gene expresion. World J. Microbiol. Biotechnol. 2014, 30, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Ripeau, J.S.; Aumont, F.; Belhumeur, P.; Ostrosky-Zeichner, L.; Rex, J.H.; de Repentigny, L. Effect of the echinocandin caspofungin on expression of Candida albicans secretory aspartyl proteinases and phospholipase in vitro. Antimicrob. Agents Chemother. 2002, 46, 3096–3100. [Google Scholar] [CrossRef] [PubMed]
- Copping, V.M.; Berelle, C.J.; Hube, B.; Gow, N.A.; Brown, A.J.; Odds, F.C. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted aspartyl proteinase genes. J. Antimicrob. Chemother. 2005, 55, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, E.; Schneider, A.E.; Sándor, N.; Lermann, U.; Staib, P.; Kremlitzka, M.; Bajtay, Z.; Barz, D.; Erdei, A.; Józsi, M. Secreted aspartic protease 2 of Candida albicans inactivates factor H and the macrophage factor H-receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18). Immunol. Lett. 2015, 168, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Zavrel, M.; Majer, O.; Kuchler, K.; Rupp, S. Transcription factor Efg1 shows a haploinsufficiency phenotype in modulating the cell wall architecture and immunogenicity of Candida albicans. Eukaryot. Cell 2012, 129, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Rueda, C.; Cuenca-Estrella, M.; Zaragoza, O. Paradoxical growth of Candida albicans in the presence of caspofungin is associated with multiple cell well rearrangements and decreased virulence. Antimicrob. Agents Chemother. 2014, 58, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Juvvadi, P.R.; Muñoz, A.; Lamoth, F.; Soderblom, E.J.; Moseley, M.A.; Read, N.D.; Steinbach, W.J. Calcium mediated induction of paradoxical growth following caspofungin treatment is associated with calcineurin activation and phosphorylation in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2015, 59, 4946–4955. [Google Scholar] [CrossRef] [PubMed]
| Species | Strains | Parental | Genotype/Characteristic | Reference |
|---|---|---|---|---|
| Candida albicans | 90028 | None | Prototrophic wild-type strain | [12] |
| SC5314 | None | Prototrophic wild-type strain | [13] | |
| CAI4 | SC5314 | ura3Δ::1imm434/ura3Δ::1imm434 | [14] | |
| cph1∆ | CAI4 | ura3Δ::1imm434/ura3Δ::1imm434 cph1Δ::hisGΔ/cph1::hisG-URA3-hisG | [15] | |
| cph1∆ (CPH1) | CAI4 | ura3Δ::1imm434/ura3Δ::1imm434 cph1Δ::hisG/cph1Δ::hisG (CPH1) | [16] | |
| efg1∆ | CAI4 | ura3Δ::1imm434/ura3Δ::1imm434 efg1Δ::hisG/efg1Δ::hisG-URA3-hisG | [16] | |
| cph1∆/efg1∆ | CAI4 | ura3Δ::1imm434/ura3Δ::1imm434Δ cph1::hisG/cph1Δ::hisG Δefg1::hisG/efg1Δ::hisG-URA3-hisG | [16] | |
| efg1∆ (EFG1) | CAI4 | ura3Δ::1 imm434/ura3Δ::1 imm434Δ efg1::hisG/efg1Δ::hisG (EFG1) | [16] | |
| cph1∆/efg1∆ (EFG1) | CAI4 | ura3Δ::1 imm434/ura3Δ::1 imm434 cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG (EFG1) | [16] | |
| sap4∆ | SC5314 | sap4-1Δ::FRT/Δsap4-2::FRT | [17] | |
| sap5∆ | SC5314 | sap5-1Δ::FRT/Δsap5-2::FRT | [17] | |
| sap6∆ | SC5314 | sap6-1Δ::FRT/sap6-2 Δ::FRT | [17] | |
| sap456∆ | SC5314 | sap4-1Δ::FRT/sap4-2Δ::FRT; Δsap5-1::FRT/sap5Δ-2::FRT; sap6-1Δ::FRT/sap6-2Δ::FRT | [17] | |
| kex2∆CNA1 | CAI4 | ura3Δ::imm434/ura3Δ::imm434KEX2kex2Δ::hisGURA3hisG | [18] | |
| kex2∆CNA2 | CAI4 | ura3Δ::imm434/ura3Δ::imm434KEX2kex2Δ::hisG | [18] | |
| kex2∆CNA3 | CAI4 | ura3Δ::imm434/ura3Δ::imm434kex2Δ::hisGkex2Δ::hisGURA3hisG | [18] | |
| kex2∆CNA4 | CAI4 | ura3Δ::imm434/ura3Δ::imm434kex2Δ::hisGkex2Δ::hisG | [18] | |
| Candida glabrata | kex2∆1006 | Cg∆HT6 | his3Δ rp1Δt kex2Δ::HIS3 | [5] |
| kex2∆1008 | 1005 | his3 Δtrp1Δ kex2Δ::HIS3 pKRT1(KEX2 TRP1) | [5] | |
| kex2∆1010 | 1005 | his3Δ trp1Δ kex2Δ::HIS3 pACT-14(TRP1) | [5] |
| Primers | Sequence (5′-3′) |
|---|---|
| ACT1-1 | GACAATTTCTCTTTCAGCACTAGTAGTGA |
| ACT1-2 | GCTGGTAGAGACTTGACCAACCA |
| KEX2-1 | TTTATATTGGGATATTTATTATCA |
| KEX2-2 | TGGGATTTTAATAATAAAGGCAAA |
| Candida Spp. | µg/mL | |||||||
|---|---|---|---|---|---|---|---|---|
| 16 | 8 | 4 | 2 | 1 | 0.5 | 0.5 | 0.5 1 | |
| SC5314 | 99.69 ± 0.01 | 99.55 ± 0.03 | 95.01 ± 0.01 | 92.70 ± 0.21 | 93.14 ± 0.05 | 95.59 ± 0.01 | 93.50 ± 0.01 | 100 |
| Δsap4 3 | 97.97 ± 0.22 | 96.90 ± 0.02 | 95.50 ± 0.01 | 92.85 ± 0.02 | 100 ± 0.04 2 | 99.90 ± 0.03 | 98.89 ± 0.02 | 100 |
| Δsap5 3 | 99.50 ± 0.00 | 99.31 ± 0.01 | 95.10 ± 0.20 | 40.05 ± 0.04 | 100 ± 0.03 2 | 98.32 ± 0.10 | 97.00 ± 0.04 | 100 |
| Δsap6 3 | 99.80 ± 0.05 | 98.90 ± 0.01 | 99.42 ± 0.01 | 99.90 ± 0.01 | 96.95 ± 0.02 | 100 ± 0.01 2 | 97.67 ± 0.01 | 100 |
| Δsap456 3 | 100 ± 0.02 | 98.70 ± 0.02 | 99.13 ± 0.01 | 99.85 ± 0.02 | 100 ± 0.01 | 97.89 ± 0.02 | 98.00 ± 0.04 | 100 |
| Can16 3 | 99.40 ± 0.03 | 99.42 ± 0.04 | 99.94 ± 0.05 | 100 ± 0.216 | 100 ± 0.01 | 100 ± 0.04 2 | 99.00 ± 0.02 | 100 |
| YLO323 | 99.71 ± 0.02 | 99.48 ± 0.03 | 95.67 ± 0.02 | 86.22 ± 0.02 | 92.04 ± 0.02 | 95.53 ± 0.22 | 95.67 ± 004 | 100 |
| HLC52 | 99.63 ± 0.03 | 98.94 ± 0.04 | 99.48 ± 0.025 | 99.74 ± 0.06 | 99.77 ± 0.02 | 98.99 ± 0.04 | 98.90 ± 0.01 | 100 |
| HLC54 3 | 99.75 ± 0.01 | 99.16 ± 0.09 | 99.33 ± 0.08 | 99.83 ± 0.05 | 100 ± 0.06 2 | 99.43 ± 0.09 | 99.00 ± 0.01 | 100 |
| HLC74 3 | 99.76 ± 0.01 | 98.93 ± 0.04 | 98.96 ± 0.04 | 100 ± 0.02 | 99.38 ± 0.01 | 98.85 ± 0.03 | 98.50 ± 0.03 | 100 |
| HLC84 | 99.71 ± 0.02 | 99.31 ± 0.01 | 99.24 ± 0.02 | 98.86 ± 0.04 | 99.15 ± 0.02 | 99.16 ± 0.03 | 99.00 ± 0.01 | 100 |
| CNA1 | 99.06 ± 0.00 | 99.04 ± 0.00 | 99.00 ± 0.01 | 98.98 ± 0.01 | 98.97 ± 0.01 | 98.96 ± 0.01 | 98.00 ± 0.02 | 100 |
| CNA2 | 99.05 ± 0.00 | 99.00 ± 0.01 | 98.99 ± 0.01 | 98.98 ± 0.00 | 98.970 ± 0.00 | 98.95 ± 0.00 | 98.77 ± 0.03 | 100 |
| CNA3 | 99.44 ± 0.00 | 99.19 ± 0.00 | 99.17 ± 0.00 | 98.98 ± 0.00 | 98.77 ± 0.01 | 96.50 ± 0.00 | 96.00 ± 0.01 | 100 |
| CNA4 | 99.96 ± 0.01 | 99.48 ± 0.00 | 99.12 ± 0.01 | 99.11 ± 0.01 | 98.67 ± 0.01 | 95.64 ± 0.01 | 94.78 ± 0.01 | 100 |
| 1006 | 100 ± 0.00 | 100 ± 0.21 2 | 99.47 ± 0.00 | 99.35 ± 0.01 | 99.27 ± 0.00 | 99.01 ± 0.01 | 99.00 ± 0.03 | 100 |
| 1008 | 99.08 ± 0.00 | 99.04 ± 0.01 | 98.99 ± 0.00 | 98.99 ± 0.01 | 98.97 ± 0.01 | 98.97 ± 0.00 | 97.90 ± 0.02 | 100 |
| 1010 | 100 ± 0.00 2 | 99.27 ± 0.01 | 99.21 ± 0.00 | 99.16 ± 0.00 | 98.94 ± 0.01 | 98.94 ± 0.00 | 98.94 ± 0.03 | 100 |
| Species | Strains | Genotypes | Cells Treated at 0.25 µg/mL | Non Treated Cells |
|---|---|---|---|---|
| Candida albicans | SC5314 (wt) | Prototrophic wild-type strain | 29.99 ± 0.34 | 59.34 ± 0.06 |
| sap4Δ | sap4-1Δ::FRT/Δsap4-2::FRT | 76.67 ± 0.15 | 46.32 ± 0.16 | |
| sap5Δ | sap5-1Δ::FRT/Δsap5-2::FRT | 48.88 ± 0.17 | 35.55 ± 0.21 | |
| sap6Δ | sap6-1Δ::FRT/sap6-2 Δ::FRT | 25.29 ± 0.16 | 22.99 ± 0.04 | |
| sap4Δ/sap5Δ/sap6 | sap4-1Δ::FRT/sap4-2Δ::FRT; Δsap5-1::FRT/sap5Δ-2::FRT; sap6-1Δ::FRT/sap6-2Δ::FRT | 52.91 ± 0.33 | 15.88 ± 0.03 | |
| cph1Δ | ura3Δ::1imm434/ura3Δ::1imm434 cph1Δ::hisG/cph1Δ::hisG | 63.89 ± 0.25 | 35.13 ± 0.24 | |
| cph1Δ::CPH1 | ura3Δ::1imm434/ura3Δ::1imm434 cph1Δ::hisG/cph1Δ::hisG (CPH1) | 30.56 ± 0.03 | 12.82 ± 0.09 | |
| efg1Δ | ura3Δ::1imm434/ura3Δ::1imm434 efg1Δ::hisG/efg1Δ::hisG-URA3-hisG | 46.40 ± 0.25 | 29.26 ± 0.16 | |
| cph1Δ/efg1Δ | ura3Δ::1imm434/ura3Δ::1imm434Δ cph1::hisG/cph1Δ::hisG Δefg1::hisG/efg1Δ::hisG-URA3-hisG | 68.00 ± 0.16 | 72.21 ± 0.22 | |
| efg1Δ::EFG1 | ura3Δ::1 imm434/ura3Δ::1 imm434Δ efg1::hisG/efg1Δ::hisG (EFG1) | 48.33 ± 0.12 | 8.34 ± 0.09 | |
| cph1Δ/efg1Δ::EFG1 | ura3Δ::1 imm434/ura3Δ::1 imm434 cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG (EFG1) | 61.87 ± 0.38 | 48.55 ± 0.078 | |
| kex2Δ CNA1 | ura3Δ::imm434/ura3Δ::imm434KEX2kex2Δ::hisGURA3hisG | 37.85 ± 0.04 | 55.8 ± 0.18 | |
| kex2Δ CNA2 | ura3Δ::imm434/ura3Δ::imm434KEX2kex2Δ::hisG | 22.49 ± 0.12 | 62.45 ± 0.27 | |
| kex2Δ CNA3 | ura3Δ::imm434/ura3Δ::imm434kex2Δ::hisGkex2Δ::hisGURA3hisG | 48.17 ± 0.41 | 52.33 ± 0.16 | |
| kex2Δ CNA4 | ura3Δ::imm434/ura3Δ::imm434kex2Δ::hisGkex2Δ::hisG | 7.33 ± 0.06 | 22.20 ± 0.06 | |
| Candida glabrata | Cg kex2Δ 1006 | his3Δ rp1Δt kex2Δ::HIS3 | 45.13 ± 0.22 | 56.99 ± 0.24 |
| Cg kex2Δ 1008 | his3Δtrp1Δ kex2Δ::HIS3 pKRT1 (KEX2 TRP1) | 17.43 ± 0.08 | 82.15 ± 0.19 | |
| Cg kex2Δ 1010 | his3Δ trp1Δ kex2Δ::HIS3 pACT-14 (TRP1) | 18.34 ± 0.07 | 53.85 ± 0.28 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staniszewska, M.; Bondaryk, M.; Ochal, Z. Role of Virulence Determinants in Candida albicans’ Resistance to Novel 2-bromo-2-chloro-2-(4-chlorophenylsulfonyl)-1-phenylethanone. J. Fungi 2017, 3, 32. https://doi.org/10.3390/jof3030032
Staniszewska M, Bondaryk M, Ochal Z. Role of Virulence Determinants in Candida albicans’ Resistance to Novel 2-bromo-2-chloro-2-(4-chlorophenylsulfonyl)-1-phenylethanone. Journal of Fungi. 2017; 3(3):32. https://doi.org/10.3390/jof3030032
Chicago/Turabian StyleStaniszewska, Monika, Małgorzata Bondaryk, and Zbigniew Ochal. 2017. "Role of Virulence Determinants in Candida albicans’ Resistance to Novel 2-bromo-2-chloro-2-(4-chlorophenylsulfonyl)-1-phenylethanone" Journal of Fungi 3, no. 3: 32. https://doi.org/10.3390/jof3030032





