The Eminence of Co-Expressed Ties in Schizophrenia Network Communities
Abstract
:1. Introduction
2. Related Work
2.1. Network Approach for Disease Modeling
2.2. Prominence of Community Detection in Biological Networks
2.3. Tie Structure Analysis
3. Methods
3.1. Collecting Gene Data
3.2. Identifying Functional Modules and Creating the Gene Network
3.3. Categorizing the Gene Components
3.4. Modularity Based Community Detection
3.5. Optimizing Modularity Using Nonlinear Embedding
3.6. Implementing LTSACom for Community Detection
3.7. Validation of Gene Communities
3.8. Discovering the Tie Structure from Communities
3.9. Multiple Correspondence Analysis
4. Results
4.1. Description of the Gene Dataset
4.2. Supervised LDA for Topic Modeling
4.3. Modularity-Based Community Detection
| Algorithm 1 LTSACom for community detection | |
| Input | Input the modularity matrix M derived from schizophrenia gene dataset for detection of gene communities |
| Step 1 | Compute the nearest neighbors using the local information among genes in tangent space |
| Step 2 | Construct the unweighted alignment matrix A based on the embedded vectors in the matrix M |
| Step 3 | Global optimization of A based on local tangents using eigenvector decomposition |
| Output | Compute the modularity index for the dataset to identify gene communities |
4.4. Validating the Community Structure
4.5. Performance Analysis of LTSACom
4.6. Identifying Tie Structure from Gene Communities
4.7. Multiple Correspondence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gilmore, J.H. Understanding what causes schizophrenia: A developmental perspective. Am. J. Psychiatry 2010, 167, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Ebisch, S.J.; Mantini, D.; Northoff, G.; Salone, A.; De Berardis, D.; Ferri, F.; Gallese, V. Altered brain long-range functional interactions underlying the link between aberrant self-experience and self-other relationship in first-episode schizophrenia. Schizophr. Bull. 2014, 40, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.A.; Pettegrew, J.W.; Keshavan, M.S. Magnetic resonance spectroscopy in schizophrenia: Methodological issues and findings—Part I. Biol. Psychiatry 2000, 48, 357–368. [Google Scholar] [CrossRef]
- Monk, C.S.; Peltier, S.J.; Wiggins, J.L.; Weng, S.J.; Carrasco, M.; Risi, S.; Lord, C. Abnormalities of intrinsic functional connectivity in autism spectrum disorders. Neuroimage 2009, 47, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Braff, D.L.; Freedman, R.; Schork, N.J.; Gottesman, I.I. Deconstructing schizophrenia: An overview of the use of endophenotypes in order to understand a complex disorder. Schizophr. Bull. 2007, 33, 21–32. [Google Scholar] [CrossRef]
- Iasevoli, F.; Giordano, S.; Balletta, R.; Latte, G.; Formato, M.V.; Prinzivalli, E.; de Bartolomeis, A. Treatment resistant schizophrenia is associated with the worst community functioning among severely-ill highly-disabling psychiatric conditions and is the most relevant predictor of poorer achievements in functional milestones. Prog. Neuro Psychopharmacol. Biol. Psychiatry 2016, 65, 34–48. [Google Scholar] [CrossRef]
- Orsolini, L.; Tomasetti, C.; Valchera, A.; Vecchiotti, R.; Matarazzo, I.; Vellante, F.; Martinotti, G. An update of safety of clinically used atypical antipsychotics. Expert Opin. Drug Saf. 2016, 15, 1329–1347. [Google Scholar] [CrossRef]
- De Berardis, D.; Rapini, G.; Olivieri, L.; Di Nicola, D.; Tomasetti, C.; Valchera, A.; Serafini, G. Safety of antipsychotics for the treatment of schizophrenia: A focus on the adverse effects of clozapine. Ther. Adv. Drug Saf. 2018, 9, 237–256. [Google Scholar] [CrossRef]
- Rolls, E.T.; Loh, M.; Deco, G.; Winterer, G. Computational models of schizophrenia and dopamine modulation in the prefrontal cortex. Nat. Rev. Neurosci. 2008, 9, 696–709. [Google Scholar] [CrossRef]
- Blom, J.D.; van Praag, H.M. Schizophrenia: It’s broken and it can’t be fixed. A conceptual analysis at the centenary of Bleuler’s Dementia praecox oder Gruppe der Schizophrenien. Isr. J. Psychiatry Relat. Sci. 2011, 48, 240–248. [Google Scholar]
- Maatz, A.; Hoff, P. The birth of schizophrenia or a very modern Bleuler: A close reading of Eugen Bleuler’s “Die Prognose der Dementia praecox” and a re-consideration of his contribution to psychiatry. Hist. Psychiatry 2014, 25, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Rish, I.; Cecchi, G.; Thyreau, B.; Thirion, B.; Plaze, M.; Paillere-Martinot, M.L.; Martelli, C.; Martinot, J.L.; Poline, J.B. Schizophrenia as a Network Disease: Disruption of Emergent Brain Function in Patients with Auditory Hallucinations. PLoS ONE 2013, 8, e50625. [Google Scholar] [CrossRef] [PubMed]
- Anticevic, A.; Murray, J.D.; Barch, D.M. Bridging Levels of Understanding in Schizophrenia through Computational Modeling. Clin. Psychol. Sci. 2015, 3, 433–459. [Google Scholar] [CrossRef] [PubMed]
- Looijestijn, J.; Blom, J.D.; Aleman, A.; Hoek, H.W.; Goekoop, R. An integrated network model of psychotic symptoms. Neurosci. Biobehav. Rev. 2015, 59, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Galderisi, S.; Rucci, P.; Kirkpatrick, B.; Mucci, A.; Gibertoni, D.; Rocca, P.; Bellomo, A. Interplay Among Psychopathologic Variables, Personal Resources, Context-Related Factors, and Real-Life Functioning in Individuals with Schizophrenia: A Network Analysis. JAMA Psychiatry 2018, 75, 396–404. [Google Scholar] [CrossRef]
- Ruan, J.; Zhang, W. Identification and Evaluation of Functional Modules in Gene Co-expression Networks. In RCP 2006: Systems Biology and Computational Proteomics; Springer: Berlin/Heidelberg, Germany, 2006; pp. 57–76. [Google Scholar]
- Wang, J.; Li, M.; Chen, J.; Pan, Y.A. Fast Hierarchical Clustering Algorithm for Functional Modules Discovery in Protein Interaction Networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 607–620. [Google Scholar] [CrossRef]
- Spirin, V.; Mirny, L.A. Protein complexes and functional modules in molecular networks. Proc. Natl. Acad. Sci. USA 2003, 100, 12123–12128. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E.J. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef]
- Easley, D.; Kleinberg, J. Strong and Weak Ties. In Networks, Crowds, and Markets: Reasoning about a Highly Connected World; Cambridge University Press: Cambridge, UK, 2010; pp. 47–84. [Google Scholar]
- Granovetter, M.S. The Strength of Weak Ties. Am. J. Sociol. 1973, 78, 1360–1380. [Google Scholar] [CrossRef]
- Chung, F.; Lu, L.; Dewey, T.G.; Galas, D.J. Duplication Models for Biological Networks. J. Comput. Biol. 2003, 10, 677–687. [Google Scholar] [CrossRef]
- Almaas, E. Biological impacts and context of network theory. J. Exp. Biol. 2007, 210, 1548–1558. [Google Scholar] [CrossRef] [PubMed]
- Rish, I.; Thyreau, B.; Thirion, B.; Plaze, M.; Paillere-martinot, M.L.; Martelli, C.; Cecchi, G.A. Discriminative Network Models of Schizophrenia. Adv. Neural Inf. Process. Syst. 2009, 22, 252–260. [Google Scholar]
- Lynall, M.E.; Bassett, D.S.; Kerwin, R.; McKenna, P.J.; Kitzbichler, M.; Muller, U.; Bullmore, E. Functional Connectivity and Brain Networks in Schizophrenia. J. Neurosci. 2010, 30, 9477–9487. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, J.L.; Giedd, J.N.; Gogtay, N. Neurodevelopmental model of schizophrenia: Update 2012. Mol. Psychiatry 2012, 17, 1228–1238. [Google Scholar] [CrossRef]
- Nekovarova, T.; Fajnerova, I.; Horacek, J.; Spaniel, F. Bridging disparate symptoms of schizophrenia: A triple network dysfunction theory. Front. Behav. Neurosci. 2014, 8, 171. [Google Scholar] [CrossRef]
- Steeds, H.; Carhart-Harris, R.L.; Stone, J.M. Drug models of schizophrenia. Ther. Adv. Psychopharmacol. 2015, 5, 43–58. [Google Scholar] [CrossRef]
- Gheiratmand, M.; Rish, I.; Cecchi, G.A.; Brown, M.R.; Greiner, R.; Polosecki, P.I.; Dursun, S.M. Learning stable and predictive network-based patterns of schizophrenia and its clinical symptoms. NPJ Schizophr. 2017, 3, 22. [Google Scholar] [CrossRef]
- Wolfers, T.; Doan, N.T.; Kaufmann, T.; Alnæs, D.; Moberget, T.; Agartz, I.; Andreassen, O.A. Mapping the Heterogeneous Phenotype of Schizophrenia and Bipolar Disorder Using Normative Models. JAMA Psychiatry 2018, 75, 1146–1155. [Google Scholar] [CrossRef]
- Trossbach, S.V.; Hecher, L.; Schafflick, D.; Deenen, R.; Popa, O.; Lautwein, T.; Malchow, B. Dysregulation of a specific immune-related network of genes biologically defines a subset of schizophrenia. Transl. Psychiatry 2019, 9, 156. [Google Scholar] [CrossRef]
- Palla, G.; Derényi, I.; Farkas, I.; Vicsek, T. Uncovering the overlapping community structure of complex networks in nature and society. Nature 2005, 435, 814–818. [Google Scholar] [CrossRef]
- Clauset, A.; Moore, C.; Newman, M.E. Hierarchical structure and the prediction of missing links in networks. Nature 2008, 453, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Kuchinsky, A.; Morris, J.H.; States, D.J.; Meng, F. GLay: Community structure analysis of biological networks. Bioinformatics 2010, 26, 3135–3137. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Cai, Z.; Musolesi, M.; Wang, Y.; Tennant, D.A.; Weber, R.J.; He, S. Community Detection in Social and Biological Networks Using Differential Evolution. Lect. Notes Comput. Sci. 2012, 7219, 71–85. [Google Scholar]
- Sah, P.; Singh, L.O.; Clauset, A.; Bansal, S. Exploring community structure in biological networks with random graphs. BMC Bioinform. 2014, 15, 220. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, S.; Moutari, S.; Dehmer, M.; Emmert-Streib, F. Comparison of module detection algorithms in protein networks and investigation of the biological meaning of predicted modules. BMC Bioinform. 2016, 17, 129. [Google Scholar] [CrossRef] [Green Version]
- Didier, G.; Brun, C.; Baudot, A. Identifying communities from multiplex biological networks by randomized optimization of modularity. F1000Research 2018, 7, 1–34. [Google Scholar] [CrossRef]
- Tripathi, B.; Parthasarathy, S.; Sinha, H.; Raman, K.; Ravindran, B. Adapting Community Detection Algorithms for Disease Module Identification in Heterogeneous Biological Networks. Bioinform. Comput. Biol. 2019, 10, 164. [Google Scholar] [CrossRef]
- Onnela, J.P.; Saramäki, J.; Hyvönen, Y.; Szabó, G.; Lazer, D.; Kaski, K.; Kertész, J.; Barabási, A.L. Structure and tie strengths in mobile communication networks. Proc. Natl. Acad. Sci. USA 2007, 104, 7332–7336. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Hoi, S.C.; Ester, M.; Bu, J.; Chen, C. Learning Personalized Preference of Strong and Weak Ties for Social Recommendation. In Proceedings of the 26th International Conference on World Wide Web, Perth, Australia, 3–7 April 2017; pp. 1601–1610. [Google Scholar]
- Hu, H.H.; Wang, L.; Jiang, L.; Yang, W. Strong ties versus weak ties in word-of-mouth marketing. BRQ Bus. Res. Q. 2018, 22, 245–256. [Google Scholar] [CrossRef]
- Fujita, Y.; Kichikawa, Y.; Fujiwara, Y.; Souma, W.; Iyetomi, H. Local bow-tie structure of the web. Appl. Netw. Sci. 2019, 4, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yao, Y.G.; Luo, X.J. SZDB: A Database for Schizophrenia Genetic Research. Schizophr. Bull. 2017, 43, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, P.; Han, G.; Zhao, J.; Lu, P.; Zhao, Z. SZGR 2.0: A one-stop shop of schizophrenia candidate genes. Nucleic Acids Res. 2017, 45, D915–D924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkpatrick, B.; Miller, B. Inflammation and Schizophrenia. Schizophr. Bull. 2013, 39, 1174–1179. [Google Scholar] [CrossRef] [Green Version]
- Müller, N.; Schwarz, M.J. Immune System and Schizophrenia. Curr. Immunol. Rev. 2010, 6, 213–220. [Google Scholar] [CrossRef]
- Carlsson, A. Antipsychotic drugs, neurotransmitters, and schizophrenia. Am. J. Psychiatry 1978, 135, 165–173. [Google Scholar] [CrossRef]
- Corcoran, C.; Walker, E.; Huot, R.; Mittal, V.; Tessner, K.; Kestler, L.; Aalaspina, D. The Stress Cascade and Schizophrenia: Etiology and Onset. Schizophr. Bull. 2003, 29, 671–692. [Google Scholar] [CrossRef] [Green Version]
- De Hert, M.; Schreurs, V.; Vancampfort, D.; Van Winkel, R. Metabolic syndrome in people with schizophrenia: A review. World Psychiatry 2013, 8, 15–22. [Google Scholar] [CrossRef]
- Perkovic, M.N.; Erjavec, G.N.; Strac, D.S.; Uzun, S.; Kozumplik, O.; Pivac, N. Theranostic Biomarkers for Schizophrenia. Int. J. Mol. Sci. 2017, 18, 733. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, J.D.; Blei, D.M. Supervised Topic Models. In Proceedings of the NIPS’07 20th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 3–6 December 2007; pp. 121–128. [Google Scholar]
- Litvin, O.; Causton, H.C.; Chen, B.J.; Pe’er, D. Modularity and interactions in the genetics of gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 6441–6446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, D.M.; Jeng, A.; Deem, M.W. The emergence of modularity in biological systems. Phys. Life Rev. 2011, 8, 129–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roweis, S.T.; Saul, L.K. Nonlinear Dimensionality Reduction by Locally Linear Embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Sun, S. Local Tangent Space Discriminant Analysis. Neural Process. Lett. 2016, 43, 727–744. [Google Scholar] [CrossRef]
- Li, W. Modularity Embedding. In Proceedings of the International Conference on Neural Information Processing, Daegu, Korea, 3–7 November 2013; pp. 92–99. [Google Scholar]
- Zhang, Z.; Zha, H. Principal Manifolds and Nonlinear Dimension Reduction via Local Tangent Space Alignment. SIAM J. Sci. Comput. 2002, 26, 313–338. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Algesheimer, R.; Tessone, C.J. A Comparative Analysis of Community Detection Algorithms on Artificial Networks. Sci. Rep. 2018, 6, 30750. [Google Scholar] [CrossRef] [Green Version]
- Abdi, H.; Valentin, D. Multiple Correspondence Analysis. In Encyclopedia of Measurement and Statistics; Sage: Sauzend Oakes, CA, USA, 2007; pp. 1–11. [Google Scholar]
- Chang, J. Lda: Collapsed Gibbs Sampling Methods for Topic Models. 2015. Available online: https://cran.r-project.org/web/packages/lda/lda.pdf (accessed on 23 May 2019).
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International Conference on Weblogs and Social Media, ICWSM 2009, San Jose, CA, USA, 17–20 May 2009; pp. 361–362. [Google Scholar]
- Newman, M.E.J. Fast algorithm for detecting community structure in networks. Phys. Rev. E 2004, 69, 066133. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Cheung, W.; Liu, J. Community Mining from Signed Social Networks. IEEE Trans. Knowl. Data Eng. 2007, 19, 1333–1348. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 10, P10008. [Google Scholar] [CrossRef] [Green Version]
- He, D.; Liu, J.; Yang, B.; Huang, Y.; Liu, D.; Jin, D. An ant-based algorithm with local optimization for community detection in large-scale networks. Adv. Complex Syst. 2012, 15, 1250036. [Google Scholar] [CrossRef] [Green Version]
- Rosvall, M.; Bergstrom, C.T. Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA 2008, 105, 1118–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosvall, M.; Bergstrom, C.T. Multilevel Compression of Random Walks on Networks Reveals Hierarchical Organization in Large Integrated Systems. PLoS ONE 2011, 6, e18209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husson, F.; Josse, J.; Le, S.; Mazet, J. FactoMineR: Multivariate Exploratory Data Analysis and Data Mining. 2017. Available online: https://cran.r-project.org/web/packages/FactoMineR/FactoMineR.pdf (accessed on 28 May 2019).
- Monaco, A.; Monda, A.; Amoroso, N.; Bertolino, A.; Blasi, G.; Di Carlo, P.; Bellotti, R. A complex network approach reveals a pivotal substructure of genes linked to schizophrenia. PLoS ONE 2018, 13, e0190110. [Google Scholar] [CrossRef] [Green Version]
- Kos, M.Z.; Duan, J.; Sanders, A.R.; Blondell, L.; Drigalenko, E.I.; Carless, M.A.; Göring, H.H. Dopamine perturbation of gene co-expression networks reveals differential response in schizophrenia for translational machinery. Transl. Psychiatry 2018, 8, 278. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Z.Z.; Tang, L.; Tang, Y.N.; Gao, Y.Y.; Li, H.J. Global vs local modularity for network community detection. PLoS ONE 2018, 13, e0205284. [Google Scholar] [CrossRef] [Green Version]





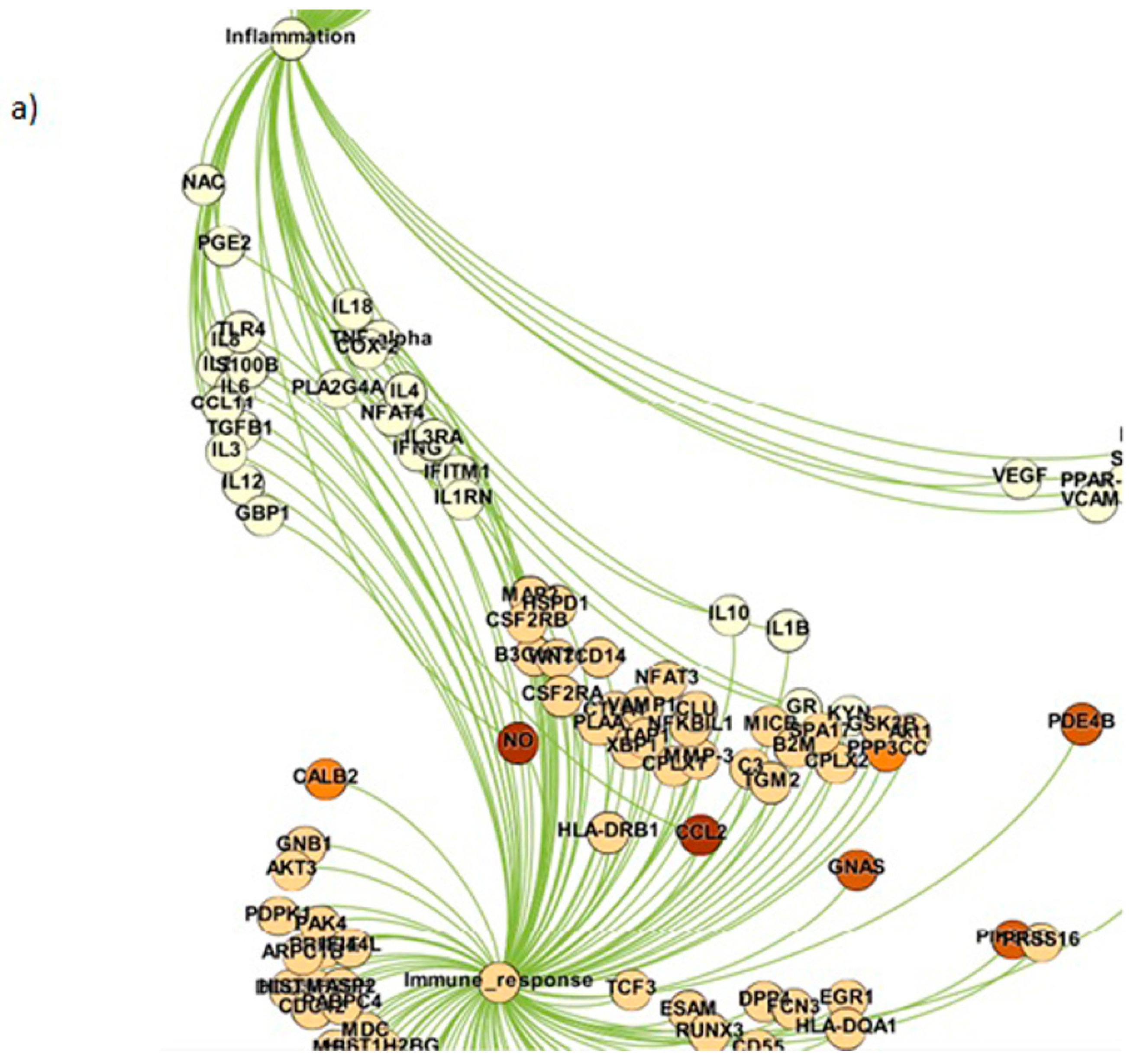
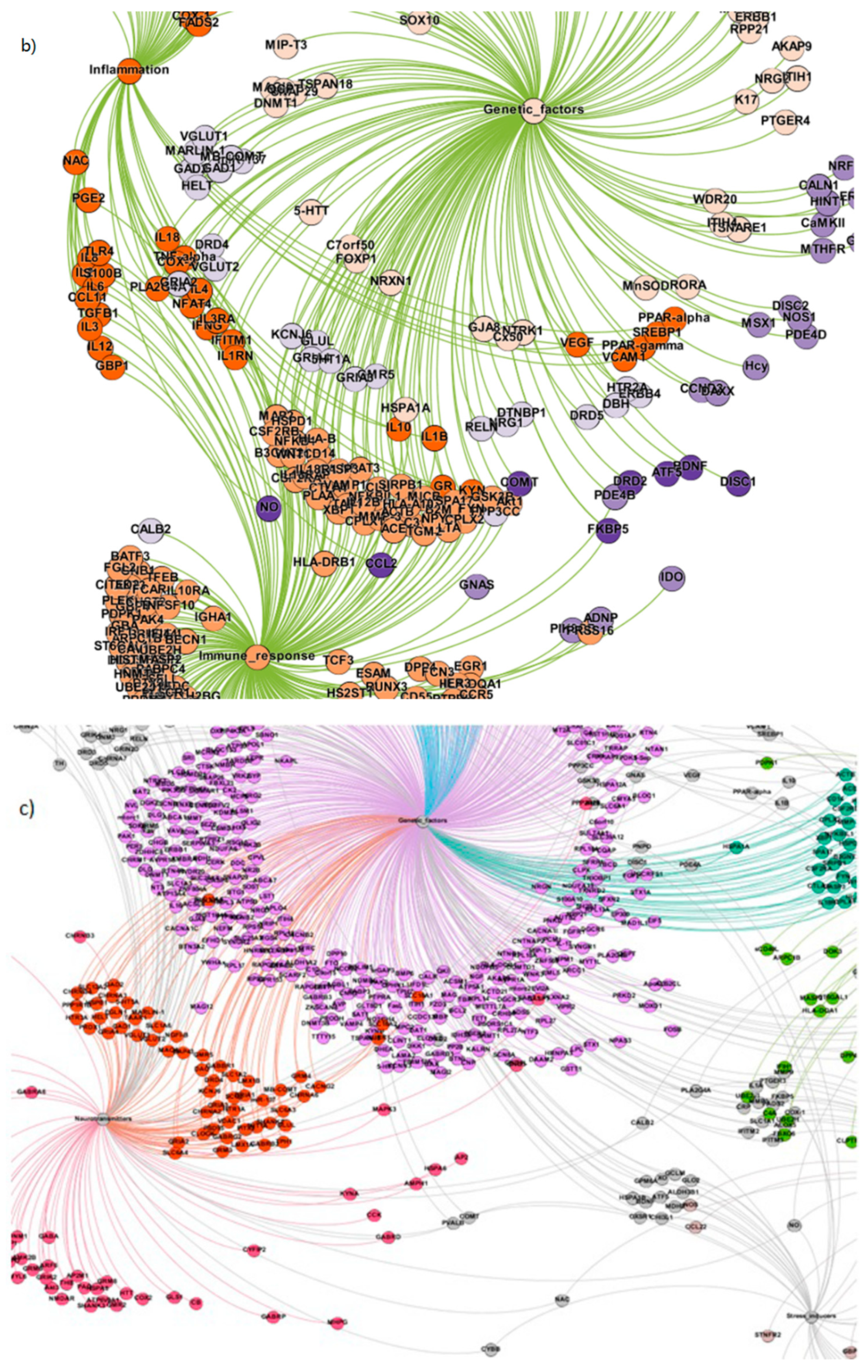
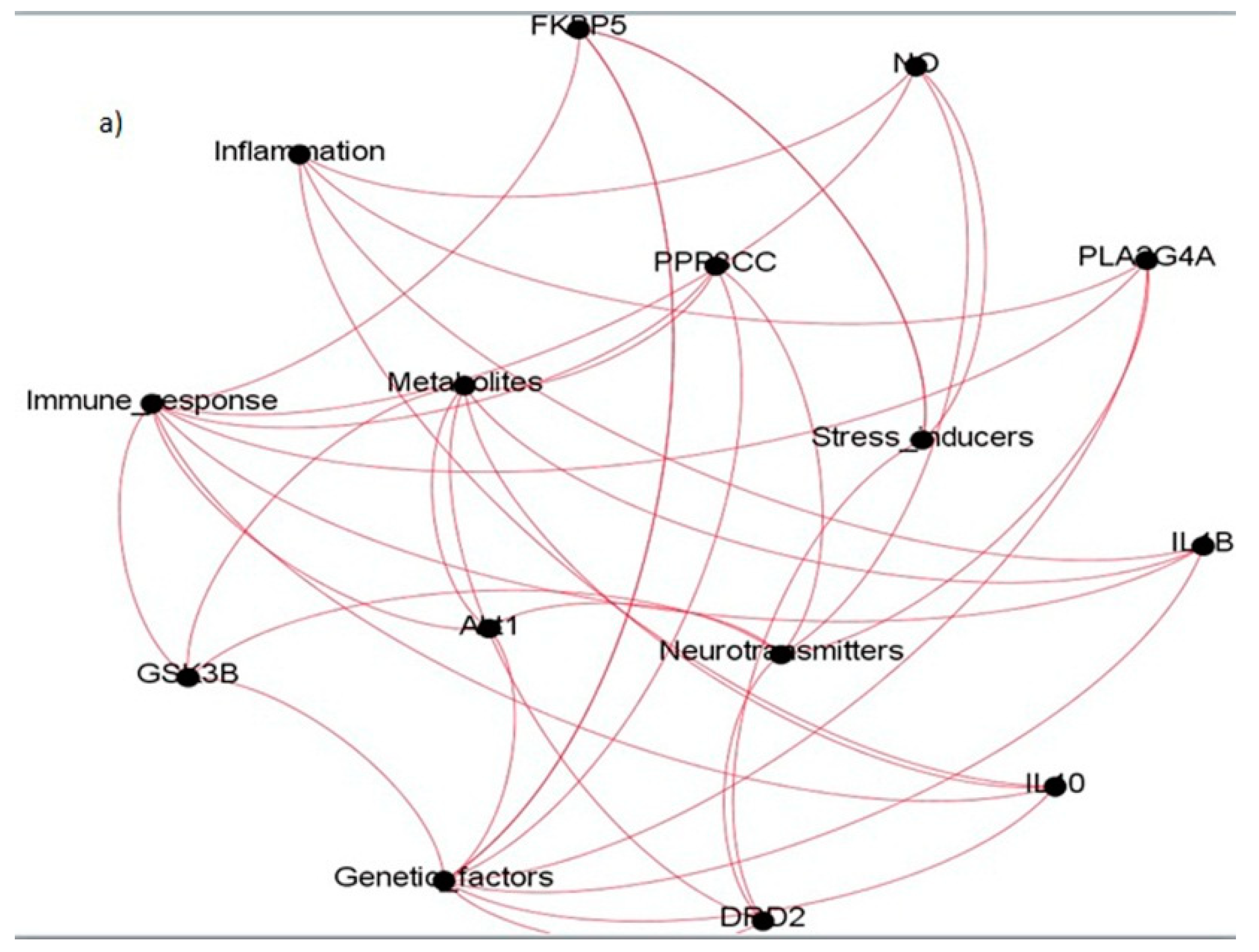
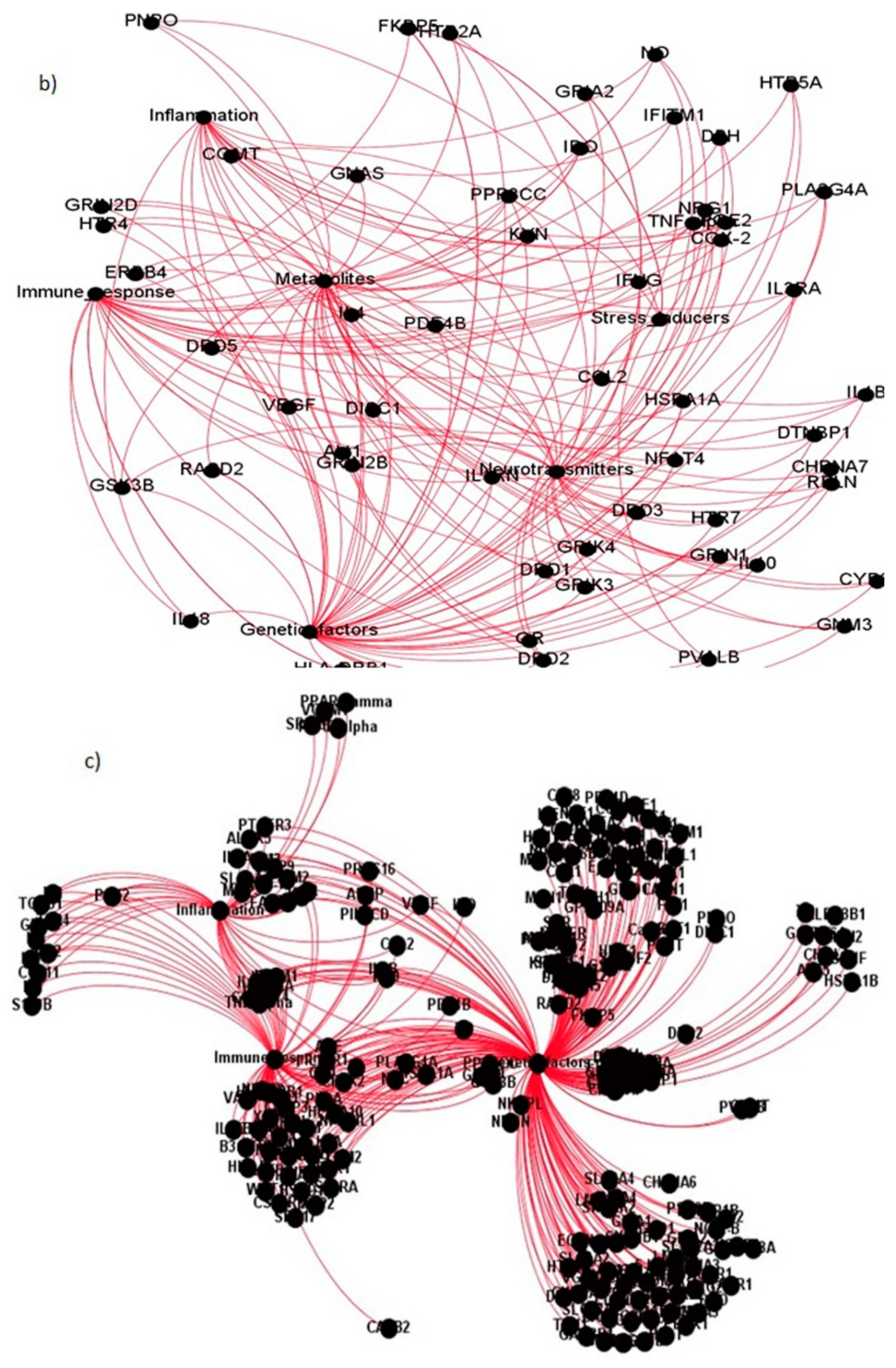


| Sl. No. | Study | Methodology Adopted | Significant Contributions |
|---|---|---|---|
| 1 | [24] | Disrupted brain function is modeled using non-linear models like gaussian naïve bayes and SVM (Support Vector Machine) | Non-linear models resulted in better functional brain mapping with 86% accuracy |
| 2 | [25] | Functional connectivity between brain components are investigated | Network topology derived from fMRI (functional Magnetic Resonance Imaging) indicated robustness of schizophrenia brain connectivity with minimal hub regions |
| 3 | [26] | Neurodevelopmental model is developed for detecting schizophrenia using longitudinal population studies | Postmortem gene expression and brain imaging denotes phenotypic characterization with developmental risk |
| 4 | [14] | Disruption in brain patterns are detected from functional networks using fMRI experiment | Disruptions are found to be global in nature owing to long distance correlations with 93% classification accuracy differentiating schizophrenia and control samples |
| 5 | [27] | Triple brain network model is proposed based on central executive network (CEN) and default mode to detect aberrant behavior in schizophrenia | The unitary mechanism of the disease is identified in cognitive, negative and positive domains |
| 6 | [28] | Several drugs including dopaminergic, cholinergic, glutamatergic, GABA (Gamma-Aminobutyric Acid), kappa opioid, cannabinoid and serotonergic are evaluated to understand their interaction patterns in schizophrenia | The stimulants impacting progression of schizophrenia are identified from the drug models |
| 7 | [12] | Multiple alterations in brain disorders are identified using a network model | The network model detected the positive symptoms of diseases using integrated approach from social, biological and psychological factors. |
| 8 | [29] | Predictive model is developed based on functional network patterns to detect schizophrenia | Sparse multivariate regression model applied on whole-brain functionality resulted in 74% accuracy for predicting schizophrenia |
| 9 | [30] | Magnetic resonance imaging data is utilized for mapping differences in brain structure | Overlapping regions of 2% is observed in cerebral, frontal and temporal regions. |
| 10 | [31] | Differentially expressed schizophrenia transcripts are identified using dysregulated genes | Two markers RGS1 and CCL4 are identified with 97% accuracy from 27% of patient subset |
| Sl. No. | Study | Algorithm/Methodology Adopted for Community Detection | Contributions/Future Scope |
|---|---|---|---|
| 1 | [32] | Overlapping communties are detected based on modularity metric in complex networks | Non-trivial interactions are detected across protein and word graphs |
| 2 | [33] | Hierarchy structure is inferred from biochemical networks to identify topological properties with enhanced accuracy | Several network phenomenas are identified by considering hierarchy as the central dogma for network orientation |
| 3 | [34] | Several community detection algorithms are designed for network visualization and clustering | GLay platform is found to be suitable for analysis of large biological networks using community detection |
| 4 | [35] | Differential evolution algorithm is designed for detecting network communities using modularity as fitness function | The algorithm does not require prior knowledge of the network structure, hence outperforms other community detection algorithms on real world datasets |
| 5 | [36] | Generative model is developed from undirected graphs to detect network communities | Community structure helps in detecting functional connections and dynamics for uncovering biological mechanisms |
| 6 | [37] | Modularity-based community detection agorithms are compared on benchmark networks | Protein interaction networks are investigated to identify multiple biological modules and hierarchical organization |
| 7 | [38] | Community detection is performed for detecting modularity in multiplex networks based on recursive clustering | Randomization improves the quality of community detection based on threshold p-value for disease dataset |
| 8 | [39] | Biologically relevant modules are deteted as non-overlapping communities using modularity and conductance metrics | It is noted that overlapping community detection algorithms are preferred for identification of diseases modules |
| Sl. No. | Study | Methodology Adopted | Significant Conclusion |
|---|---|---|---|
| 1 | [21] | The cohesive power of weak ties helps in analyzing connected components | Emphasis on weak ties reveals the connectivity and topological features in the network |
| 2 | [40] | Communication patterns are observed in mobile networks based on strength of interaction across the ties | Information diffusion revealed neither strong nor weak ties are effective |
| 3 | [41] | Importance of ties is explored and a recommender system is developed using probability-based matrix factorization algorithm | Different types of social network ties are classified using the recommendation system with enhanced accuracy and specificity |
| 4 | [42] | The word of mouth implication is demonstrated using agent-based modeling approach | Strong ties are found to disseminate information accurately compared to weak ties |
| 5 | [43] | Strongly connected components are analyzed based on the local bow-tie structures in world wide web graphs | Differences were observed between WWW graphs and other graphs due to global bow-tie associations |
| Sl. No | Gene Modules | Number of Genes |
|---|---|---|
| 1. | Inflammation | 73 |
| 2. | Immune response | 213 |
| 3. | Genetic factors | 674 |
| 4. | Neurotransmitters | 149 |
| 5. | Metabolites | 168 |
| 6. | Stress Inducers | 36 |
| Trials | Modularity | SP | FUA | MABA | FN | FEC | InfoMap | LTSACom |
|---|---|---|---|---|---|---|---|---|
| 20 | 0.1 | 0.6784 | 0.7622 | 0.7981 | 0.8021 | 0.7123 | 0.7892 | 0.9194 |
| 40 | 0.2 | 0.6755 | 0.7544 | 0.8083 | 0.8166 | 0.7851 | 0.7985 | 0.8996 |
| 60 | 0.3 | 0.7081 | 0.765 | 0.8348 | 0.7933 | 0.8037 | 0.7831 | 0.8631 |
| 80 | 0.4 | 0.7002 | 0.7598 | 0.8329 | 0.7821 | 0.8136 | 0.8043 | 0.9386 |
| 100 | 0.5 | 0.6923 | 0.7322 | 0.8071 | 0.8031 | 0.8203 | 0.8124 | 0.8931 |
| 120 | 0.6 | 0.7093 | 0.7999 | 0.8199 | 0.7932 | 0.8108 | 0.8342 | 0.9032 |
| 140 | 0.7 | 0.7129 | 0.7132 | 0.8478 | 0.8155 | 0.8093 | 0.8188 | 0.9203 |
| 160 | 0.8 | 0.7361 | 0.6992 | 0.8332 | 0.7973 | 0.8478 | 0.8109 | 0.9201 |
| 180 | 0.9 | 0.7102 | 0.7338 | 0.8554 | 0.8045 | 0.8313 | 0.8013 | 0.9289 |
| 200 | 1.0 | 0.7009 | 0.7221 | 0.8441 | 0.7899 | 0.8396 | 0.8032 | 0.9256 |
| Sl. No. | Coexpressed Gene Ties | Gene Names | Categories They Belong to |
|---|---|---|---|
| 1. | Akt1 | Protein kinase B | Immune response, Genetic factors, Neurotransmitters, Metabolites |
| 2. | DRD2 | Dopamine Receptor 2 | Genetic factors, Neurotransmitters, Metabolites, Stress inducers |
| 3. | IL10 | Interleukin 10 | Inflammatory, Immune response, Genetic factors, Metabolites |
| 4. | PPP3CC | Protein Phosphatase 3 Catalytic Subunit Gamma | Immune response, Genetic factors, Neurotransmitters, Metabolites |
| 5. | PLA2G4A | Phospholipase A2 Group 4A | Inflammatory, Immune response, Genetic factors, Neurotransmitters, |
| 6. | GSK3B | Glycogen synthase kinase-3B | Immune response, Genetic factors, Neurotransmitter, Metabolites |
| 7. | NO | Nitric oxide | Inflammatory, Immune response, Neurotransmitter, Stress inducer |
| 8. | COX2 | Cyclooxygenase 2 | Inflammatory, Immune response, Genetic factor, Neurotransmitter |
| 9. | FKBP5 | FK506 Binding Protein 5 | Immune response, Genetic factors, Stress inducers |
| 10. | IL1B | Interleukin 1 beta | Inflammatory, Immune response, Metabolite |
| 11. | COMT | Catechol-O-methyltransferase | Genetic factors, Neurotransmitter, Stress inducer |
| 12. | DISC1 | Disrupted in Schizophrenia 1 | Genetic factor, Metabolite, Stress inducer |
| 13. | PDE4B | Phosphodiesterase 4B | Immune response, Genetic factor, Metabolite |
| 14. | PNPO | Pyridoxine 5’-phosphatase oxidase | Genetic factor, Metabolite, Stress inducer |
| 15. | NFAT4 | Nuclear Factor of Activated T Cells 4 | Inflammatory, Immune response, Genetic factor |
| 16. | IFNG | Interferon gamma | Inflammatory, Immune response, Genetic factor |
| 17. | TNFA | Tumor Necrosis Factor Alpha | Inflammatory, Immune response, Genetic factor |
| 18. | IL3RA | Interleukin 3 Receptor Subunit Alpha | Inflammatory, Immune response, Genetic factor |
| 19. | IL4 | Interleukin 4 | Inflammatory, Immune response, Genetic factor |
| 20. | GRIK3 | Glutamate Ionotropic Receptor Kainate Type Subunit 3 | Genetic factor, Neurotransmitter, Metabolite |
| 21. | VEGF | Vascular endothelial growth factor | Inflammatory, Genetic factor, Metabolite |
| 22. | HTR4 | 5-Hydroxytryptamine Receptor 4 | Genetic factor, Neurotransmitter, Metabolite |
| 23. | GNAS | Guanine nucleotide binding protein, alpha stimulating | Immune response, Genetic factor, Neurotransmitter |
| 24. | CHRNA7 | Cholinergic Receptor Nicotinic Alpha 7 | Genetic factor, Neurotransmitter, Metabolite |
| 25. | DTNBP1 | Dysbindin 1 | Genetic factor, Neurotransmitter, Metabolite |
| 26. | RELN | Reelin | Genetic factor, Neurotransmitter, Metabolite |
| 27. | DRD1 | Dopamine Receptor D1 | Genetic factor, Neurotransmitter, Metabolite |
| 28. | DRD3 | Dopamine Receptor D3 | Genetic factor, Neurotransmitter, Metabolite |
| 29. | DRD5 | Dopamine Receptor D5 | Genetic factor, Neurotransmitter, Metabolite |
| 30. | ERBB4 | Epidermal growth factor receptor family 4 | Genetic factor, Neurotransmitter, Metabolite |
| 31. | GRIA2 | Glutamate Ionotropic Receptor AMPA (Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) Type Subunit 2 | Genetic factor, Neurotransmitter |
| 32. | HSPA1A | Heat Shock Protein Family A Member 1A | Immune response, Genetic factor |
| 33. | RASD2 | RASD Family Member 2 | Genetic factor, Metabolite |
| 34. | IDO | Indoleamine 2,3-dioxygenase | Immune response, Metabolite |
| 35. | PGE2 | Prostaglandin E2 | Inflammatory, Immune response |
| 36. | KYN | Kynurenine | Inflammatory, Immune response |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sridhar, A.; GS, S.; Reddy, A.M.; Bhattacharjee, B.; Nagaraj, K. The Eminence of Co-Expressed Ties in Schizophrenia Network Communities. Data 2019, 4, 149. https://doi.org/10.3390/data4040149
Sridhar A, GS S, Reddy AM, Bhattacharjee B, Nagaraj K. The Eminence of Co-Expressed Ties in Schizophrenia Network Communities. Data. 2019; 4(4):149. https://doi.org/10.3390/data4040149
Chicago/Turabian StyleSridhar, Amulyashree, Sharvani GS, AH Manjunatha Reddy, Biplab Bhattacharjee, and Kalyan Nagaraj. 2019. "The Eminence of Co-Expressed Ties in Schizophrenia Network Communities" Data 4, no. 4: 149. https://doi.org/10.3390/data4040149





