To conduct interpretability analysis of diabetic retinopathy (DR) classification models, this study established a comprehensive experimental framework encompassing dataset construction, model selection, interpretability analysis, and evaluation. For the dataset, a total of 2060 fundus images were collected from six hospitals. All images were deidentified prior to data collection. Each image was independently annotated by at least two professionally trained ophthalmologists. In addition to disease grading, polygonal bounding boxes were used to outline lesion areas, including hard exudates, hemorrhages, microaneurysms, and soft exudates, providing pixel-level annotations for model interpretability evaluation. In terms of model selection, four representative architectures and their variants—VGG, DenseNet, ResNet, and EfficientNet—were chosen as the classification models for analysis. These models cover a spectrum from shallow networks with simple linear stacking, to deep networks with dense or cross-layer connections, and further to highly optimized lightweight networks. This selection comprehensively reflects the potential impact of different model depths, connection mechanisms, and feature fusion strategies on interpretability. For interpretability analysis, multiple interpretability methods were applied, including gradient-based techniques (Gradient, SmoothGrad, Integrated Gradients), Shapley value-based methods (SHAP), backpropagation-based attribution methods (DeepLIFT), and activation mapping methods (Grad-CAM++, ScoreCAM). The combination of these analytical approaches enables a multi-faceted analysis of model decisions from complementary dimensions such as input sensitivity, feature contribution distribution, attribution propagation mechanisms, and visual region saliency. Finally, for the qualitative and quantitative assessment of the generated saliency maps, this study employed perturbation-based curve analysis and trend correlation analysis to evaluate whether the highlighted regions contribute meaningfully to model decisions and to examine the consistency between model reasoning and clinical logic. Additionally, the entropy, recall, and Dice coefficient of the saliency maps were calculated to measure the concentration of explanatory signals and their spatial overlap accuracy with ground-truth lesion regions.
3.2. Preparation of the Models to Be Tested
In order to analyze the interpretability of DR classification model, this study selects four deep learning network architectures—VGG [
25], DenseNet [
26], ResNet [
27] and EfficientNet [
28], and trains a total of 16 DR classification models, which are then tested on experimental dataset.
In recent years, network architectures such as VGG, DenseNet, ResNet, and EfficientNet have demonstrated remarkable performance in image recognition tasks, each featuring unique structural designs and advantages that represent different developmental stages of neural networks. The VGG network, renowned for its simplicity and high modularity, primarily extracts image features through stacking multiple 3 × 3 convolutional kernels and 2 × 2 max pooling layers. Although it requires substantial parameters and computational complexity, its unified convolutional architecture facilitates transferability and scalability, making it a widely adopted foundational template for subsequent network architectures across various visual tasks. DenseNet introduces dense connection mechanisms, enabling feature-level connections between all preceding layers, which significantly enhances feature reuse efficiency and gradient flow performance. This connection approach not only effectively mitigates the common gradient vanishing problem in deep networks but also drastically reduces parameter redundancy, allowing models to maintain strong expressive capabilities with fewer parameters. ResNet addresses the performance degradation issue during deep network training by introducing residual connections. Its basic unit employs identity mappings to directly skip-connect inputs to outputs, ensuring stable information flow within the network. This design enables more layers to be stacked without causing performance degradation, leading to groundbreaking advancements in multiple image recognition tasks. EfficientNet is a lightweight convolutional neural network architecture optimized through composite scaling strategies. By holistically considering network depth, width, and image resolution, it automatically determines the optimal scaling ratio to maximize performance under computational constraints. Combining low model complexity with excellent classification accuracy, EfficientNet proves ideal for efficient deployment scenarios. These models span from shallow networks with simple linear stacking, to deep networks employing dense or cross-layer connections, and further extend to highly optimized lightweight networks. This selection comprehensively captures the potential impacts of varying model depths, connection mechanisms, and feature fusion strategies on interpretability.
In order to evaluate the interpretability of different networks and structures in DR classification tasks, as shown in
Table 2, this study selected a number of representative network variants as test models based on four mainstream deep learning network architectures.
Variants within different architectures differ in network depth, structural complexity, and regularization strategies. The primary distinction between VGG16 and VGG19 lies in the number of stacked convolutional layers. VGG16 contains 13 convolutional layers and 3 fully connected layers, while VGG19 introduces additional convolutional layers within each convolution block, resulting in a deeper overall structure that theoretically enhances feature extraction capabilities. Additionally, VGG16-BN and VGG19-BN incorporate Batch Normalization (BN) layers into their original architectures, which helps accelerate model convergence and improve training stability. Variants of the DenseNet architecture primarily adjust the number of network layers and the depth of dense blocks to create models like DenseNet121, 161, 169, and 201. These variants maintain the dense connection mechanism while progressively increasing the number of convolutional layers to enhance network expressiveness. Larger numbers indicate deeper networks, with corresponding increases in model parameters and computational costs, though they also improve the modeling capability for fine-grained features. In the ResNet series, ResNet18 and ResNet34 are shallow variants constructed using basic residual units, suitable for small to medium-scale data tasks. In contrast, ResNet50 and ResNet152 belong to deep networks that incorporate bottleneck blocks to control parameter scale while enhancing nonlinear modeling capabilities. As network depth increases, model expressiveness improves significantly, but training strategies and data requirements also rise accordingly. The EfficientNet series employs composite scaling strategies, systematically adjusting network depth, width, and input image resolution to achieve efficient performance scalability. Variants B0 through B3 represent progressively expanded network architectures. With increasing numbering, these variants demonstrate enhanced classification accuracy and improved perception of complex patterns.
All models in
Table 2 were trained using the PyTorch 2.1.1 deep learning framework in this experiment. As a widely adopted deep learning platform, PyTorch provides robust tools and modules that enable flexible network design and efficient training processes. To ensure models effectively capture data features and achieve good generalization capabilities, all models were initialized with parameters pre-trained on the ImageNet large-scale image dataset [
29]. Each model was fine-tuned on the dataset constructed in this study, which was randomly split into 80% for training and 20% for testing. After 50 training epochs on the training set, model performance was evaluated on the validation set every two epochs. To ensure the comparability and fairness of the experimental results, all models adopted a unified set of hyperparameters in the cross-comparative analysis. Specifically, the configurations included an initial learning rate of 0.0001, a batch size of 32, and the Adam optimization algorithm.
3.3. Method
The clinical grading of diabetic retinopathy (DR) is typically determined based on the type of lesions and their distribution. The International Clinical Diabetic Retinopathy (ICDR) severity scale [
30], one of the most widely adopted standards, categorizes DR into five grades: Grade 1 (no apparent retinopathy); Grade 2 (mild non-proliferative DR, characterized only by microaneurysms); Grade 3 (moderate non-proliferative DR, with more lesions than microaneurysms alone but insufficient to meet the criteria for severe non-proliferative DR); Grade 4 (severe non-proliferative DR, characterized by: (i) ≥20 intraretinal hemorrhages in each of the four quadrants; (ii) venous beading in two or more quadrants; (iii) prominent intraretinal microvascular abnormalities [IRMA] in at least one quadrant without neovascularization); and Grade 5 (proliferative DR, defined by the presence of neovascularization or vitreous/preretinal hemorrhage [
31]). These criteria indicate that the essence of DR grading lies in identifying typical lesions as well as evaluating their quantity and spatial distribution patterns.
The saliency map derived from interpretable analysis methods reveals how different models measure and calculate the contribution of input features. Models with strong interpretability are expected to demonstrate that their interpretation methods highlight regions consistent with critical lesion areas, reflecting alignment between the model’s decision-making process and clinical logic. Furthermore, if changes in lesions observed in fundus images correspond to trends in highlighted regions of the saliency map, it would demonstrate the model’s interpretability and clinical credibility. Evaluating the interpretability of DR classification models involves assessing whether their decision-making rationale aligns with clinical grading logic, as well as evaluating the quality of saliency maps generated through interpretability methods.
To achieve the aforementioned objectives, this paper designs an experimental protocol. As shown in
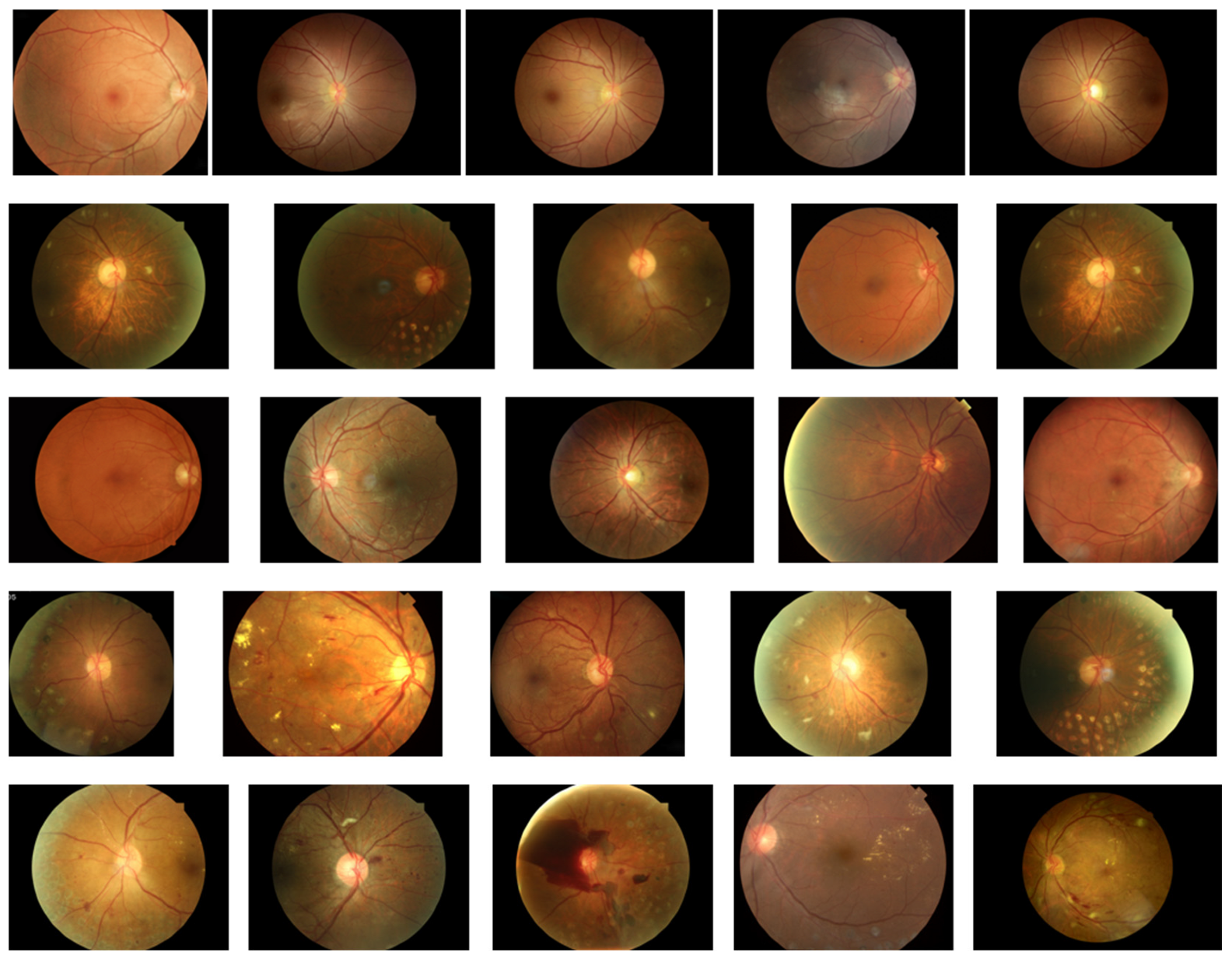
Figure 2, the first phase constitutes the preparation stage, which includes the test model, test images, image masks, and corresponding label information. All test models are stored in.pth format and loaded via the PyTorch framework for inference operations. Test images and their corresponding mask images are read as.jpg files and uniformly resized to 224 × 224 resolution to meet model input requirements. The labels (denoted as DR) consist of five graded categories: normal, mild NPDR, moderate NPDR, severe NPDR, and PDR. To accommodate model evaluation formats, these labels are converted into One-Hot encoding with values ranging from 0 to 4. Test images are selected from the dataset constructed in this study, with five representative images chosen for each of the five lesion grades to ensure diagnostic feature representation and minimal noise interference.
Figure 3 displays the selected image samples arranged from top to bottom: normal, mild NPDR, moderate NPDR, severe NPDR, and PDR. Each pathological grade contains five fundus images.
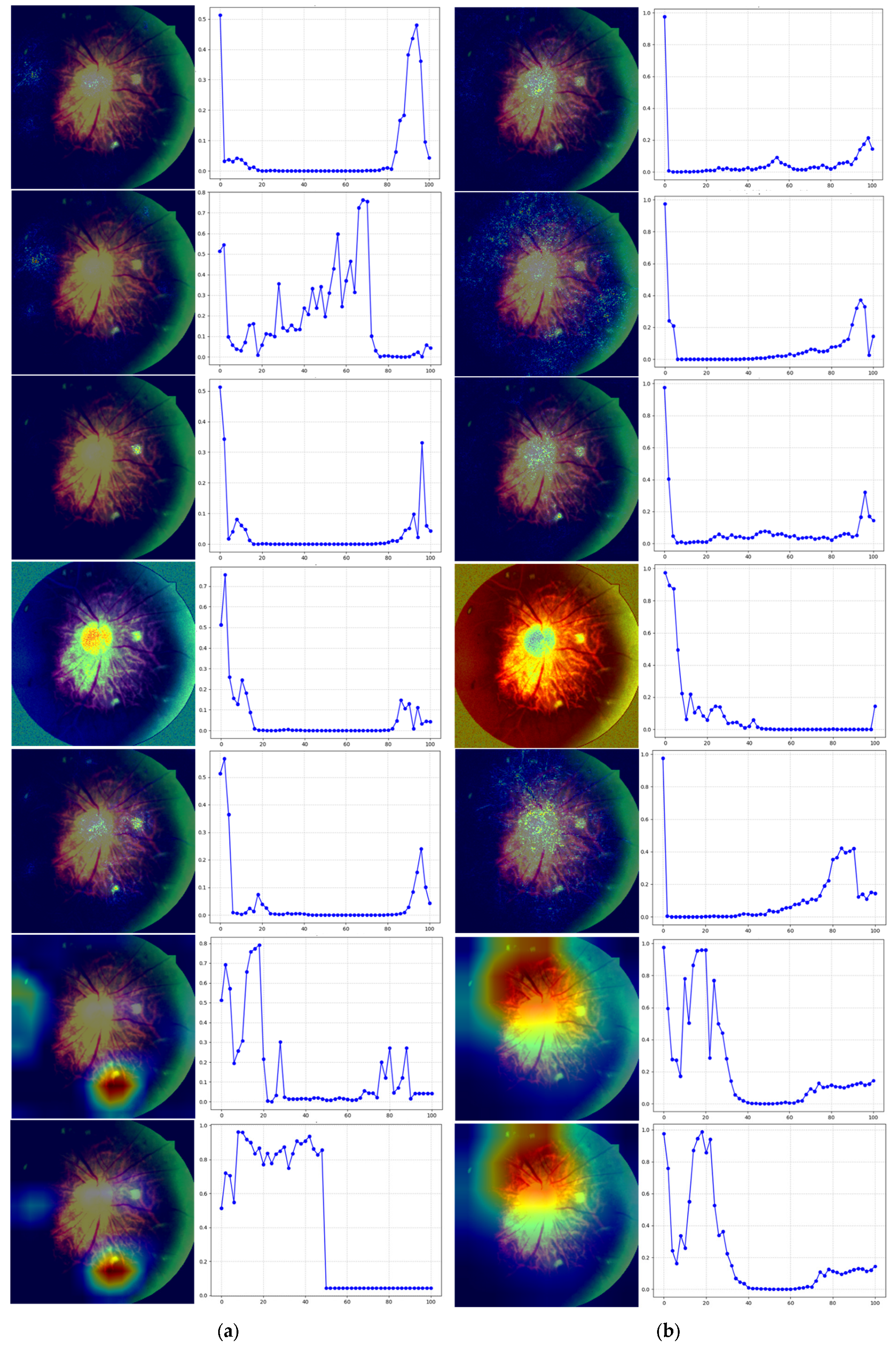
In the second step, we utilized seven mainstream interpretable analysis methods (Gradient, SmoothGrad, Integrated Gradients, SHAP, DeepLIFT, Grad-CAM++, and ScoreCAM) to generate saliency maps for each model, enabling visualization of model focus areas. To ensure accuracy and stability of interpretation results, this study did not adopt the LIME method. While LIME typically relies on superpixel segmentation and region masking for local perturbations in image tasks, followed by linear model fitting to approximate the original model’s local behavior, its segmentation strategy struggles with small, poorly defined lesion areas in diabetic retinopathy images. This makes it difficult to precisely locate critical pathological features, potentially leading to information loss or incorrect segmentation. Furthermore, the method’s reliance on high randomness in perturbation sample generation results in poor stability and reproducibility of interpretation outcomes, failing to meet medical requirements for consistency and reliability in explainable results.
Figure 2’s Stage Two summarizes the applicable conditions for each method. All methods require specifying labels for test images to guide models in generating decision features corresponding to categories. Notably, Integrated Gradients, SHAP, and DeepLIFT also require a baseline input. Differences in theoretical foundations and computational approaches result in variations in implementation. As shown in
Table 3, Gradient calculates gradients between input features and model outputs to reveal feature influence on decision-making. This method has low computational complexity, requires no image category designation, and doesn’t depend on baseline inputs. SmoothGrad applies random noise to input samples and calculates gradient averages to reduce high-frequency noise in gradient computation, thereby enhancing feature attribution stability. While not requiring baseline inputs, this method demands image category information and higher computational costs. Integrated Gradients employs integral gradient calculation, measuring cumulative feature contributions through gradient accumulation from baseline to actual inputs. This approach necessitates selecting appropriate baseline inputs and specifying image categories, leading to high computational requirements. Both SHAP and DeepLIFT compute feature importance based on baseline inputs: SHAP uses Shapley values to allocate feature contributions, while DeepLIFT performs attribution analysis by referencing activation values. Both methods require specifying image category information and reference inputs, making them suitable for various tasks with high and medium computational complexity, respectively. Grad-CAM++ and ScoreCAM are both visualization approaches based on convolutional feature maps, primarily applicable to models with convolutional architectures. Grad-CAM++ employs gradient weighting to calculate the influence of feature maps on prediction results, which is ideal for handling complex target objects or multi-instance scenarios. ScoreCAM enhances stability by treating feature maps as masks that repeatedly interact with original images, observing model predictions to assess feature map importance without requiring gradient calculations.
The third phase involves preliminary qualitative evaluation of the saliency map using perturbation curve analysis to assess whether it accurately reflects the model’s critical regions. This method simulates gradual loss of image area information, observing how model predictions evolve to determine the relative importance of saliency map-identified regions in decision-making. The perturbation curve generation process is as follows: For a test image, first rank all pixels by saliency values from highest to lowest to identify the model’s most focused areas. Then, at each perturbation ratio (0–100%, with 2% increments), sequentially mask high-salience regions ranked higher in the saliency list. At each ratio, record the model’s confidence in predicting original categories after masking corresponding high-salience areas. Finally, plot the model’s output probabilities against perturbation ratios on the perturbation curve. If the saliency map accurately identifies the model’s primary focus areas, increased masking should lead to a sharp decline in target category confidence, showing a monotonically decreasing trend with rapid rate of decrease. Conversely, if the masked high-salience regions have minimal impact on predictions, indicating poor representation of actual key areas, the perturbation curve will show no significant change.
The fourth step involves calculating the entropy value of the saliency map and conducting trend correlation analysis to verify whether the model’s decision-making basis aligns with clinical classification logic. The saliency map entropy value measures the concentration of regions of interest within the model, while entropy reflects the randomness or complexity of the saliency map. Higher entropy values indicate scattered distribution of highlighted areas, suggesting unfocused model attention and poor interpretability. Conversely, lower entropy values demonstrate concentrated saliency regions with clear boundaries, indicating more focused model attention and enhanced interpretability and consistency. For saliency maps of images within the same category, similar pattern recognition and regional distribution should be observed—meaning higher spatial consistency in both location and intensity, with relatively stable entropy variations. When comparing saliency maps across different categories, significant trend changes in model attention should emerge as DR severity increases. For instance, in mild lesions, the model may focus on fewer concentrated areas, whereas in severe or proliferative lesions, multiple lesion regions may attract attention, resulting in more complex saliency map structures and correspondingly higher entropy values.
The calculation formula of entropy is shown in Equation (1), where is the normalized pixel intensity of the significant figure.
Normalization of pixel intensity is typically achieved by scaling the range of pixel values to between 0 and 1. If an image’s pixel intensity falls within a specific range (e.g., 0 to 255), the normalized intensity can be calculated using Formula (2). Here, Ii represents the original intensity value of the i-th pixel in the saliency map, while min(I) and max(I) denote the minimum and maximum values of all pixels in the saliency map, respectively.
Trend correlation analysis aims to verify whether the focus areas of the model’s explainable methods change synchronously with the increase in lesions in fundus images. In lesion type selection, hemorrhagic spots and microaneurysms typically appear as small, round, dark-red spots in fundus images with clear boundaries and high contrast. These lesions exhibit stable morphology and distinct optical characteristics, allowing their extraction from real images through edge smoothing and color matching techniques without compromising structural consistency when embedded into other images. Compared to soft exudates (with blurred morphology) and neovascularization (with disorganized structures), adding hemorrhagic spots and microaneurysms proves more feasible. Neovascularization lesions often lack clear boundaries and are usually roughly localized using elliptical boxes or bounding circles in fundus images, making pixel-level localization and extraction challenging. Soft exudates typically feature indistinct edges, irregular shapes, and color distributions that closely match background tissues, lacking clear structural boundaries. Even after cropping from real images, maintaining naturalness becomes difficult, often resulting in “poor fusion” phenomena in target images. Although hard exudates possess higher brightness and prominent color features enabling accurate extraction, their clustered distribution in images makes unnatural fusion likely during synthesis due to brightness variations. Therefore, selecting hemorrhagic spots and microaneurysms as lesion additions in this study represents a practical experimental strategy under current conditions. In terms of analytical methodology, the trend correlation evaluation employs qualitative analysis to focus on the model’s response patterns to changes in lesion quantity. This approach aims to reveal how the model prioritizes critical regions under varying lesion counts. To achieve this, we utilized ScoreCAM’s white-box interpretability method to generate category activation maps. By applying mask processing and forward propagation to feature maps, ScoreCAM constructs discriminative saliency maps that effectively reconstruct the image regions the model focuses on during decision-making. With its strong visualization capabilities, this method proves particularly suitable for explanatory analysis tasks involving lesion quantity trends in our research.
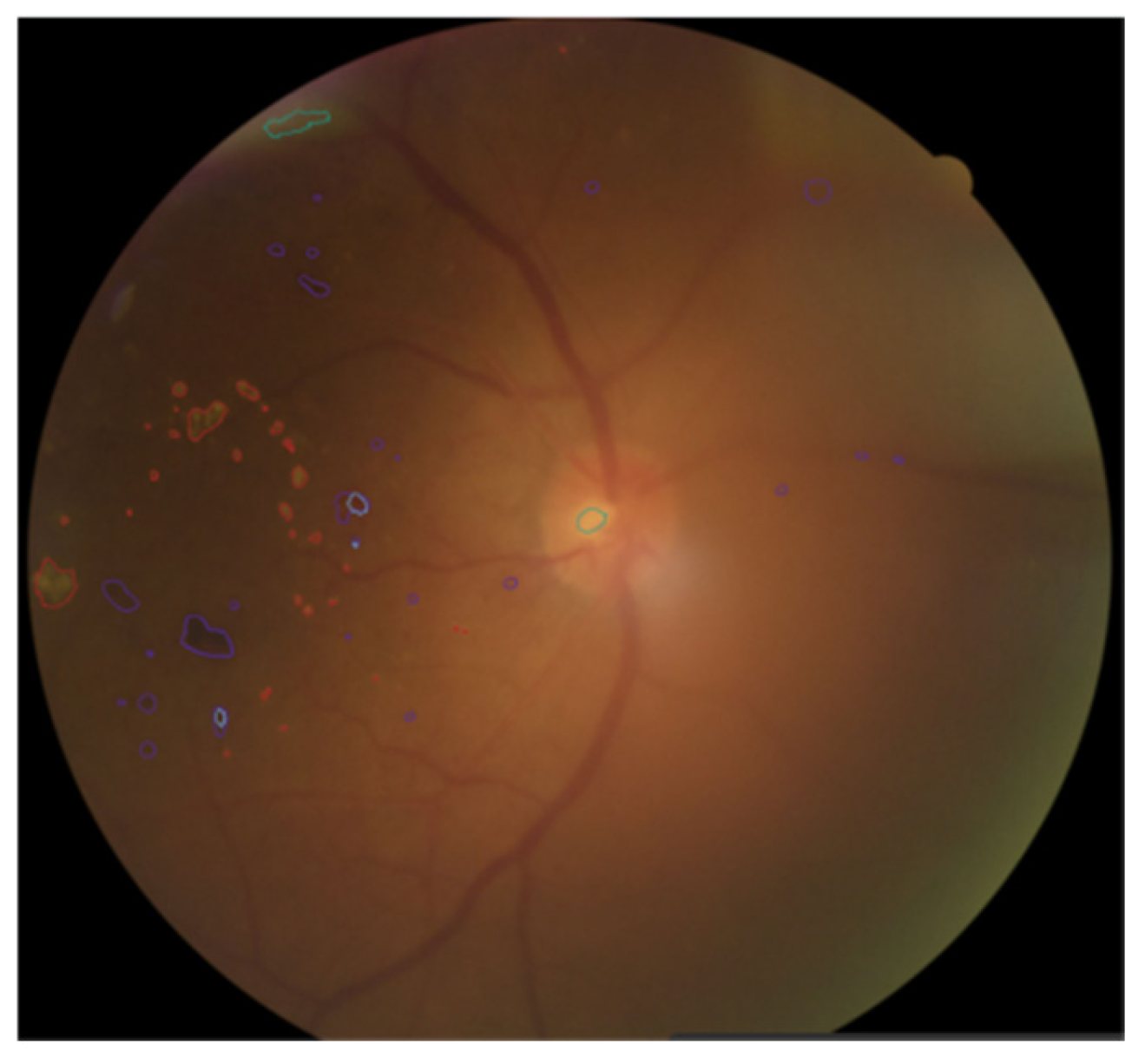
As shown in
Figure 4, when annotating fundus images, doctors use different colors to distinguish various pathological areas. Specifically, the red curve-enclosed area indicates firm exudates, the purple curve-enclosed area represents hemorrhagic points, the blue curve-enclosed area denotes microaneurysms, and the green curve-enclosed area signifies soft exudates.
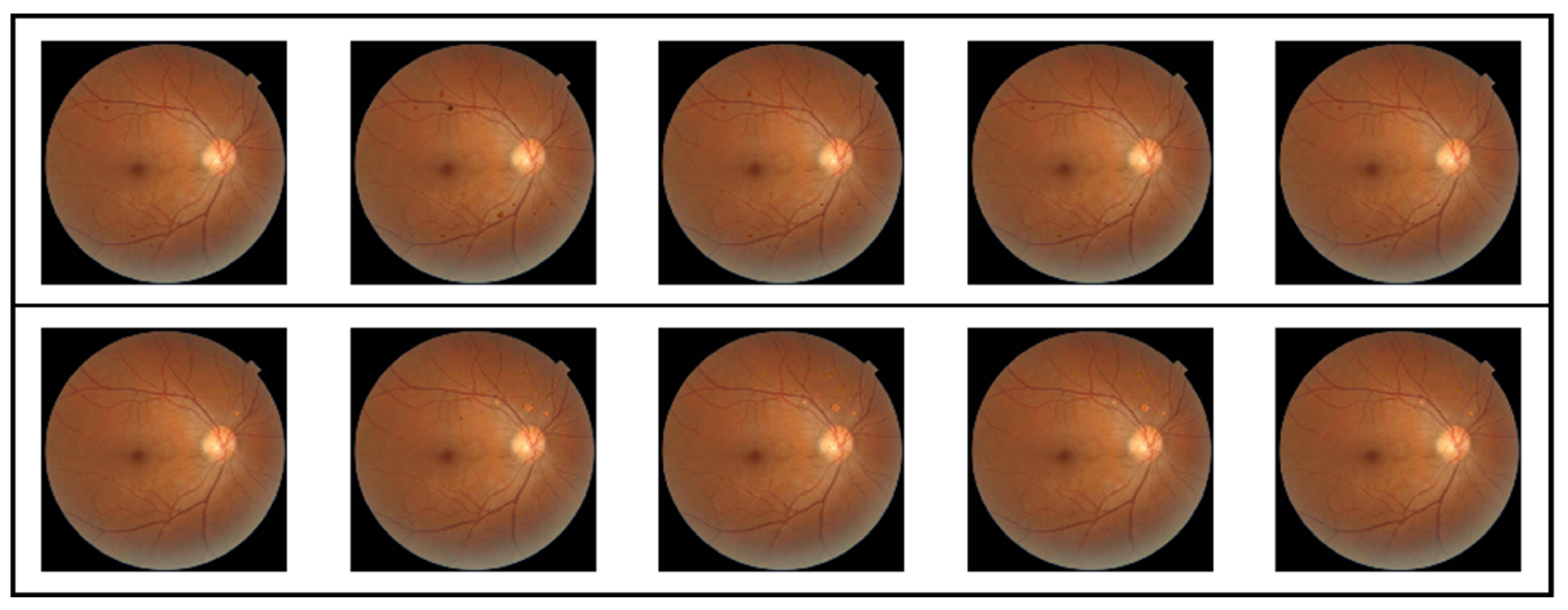
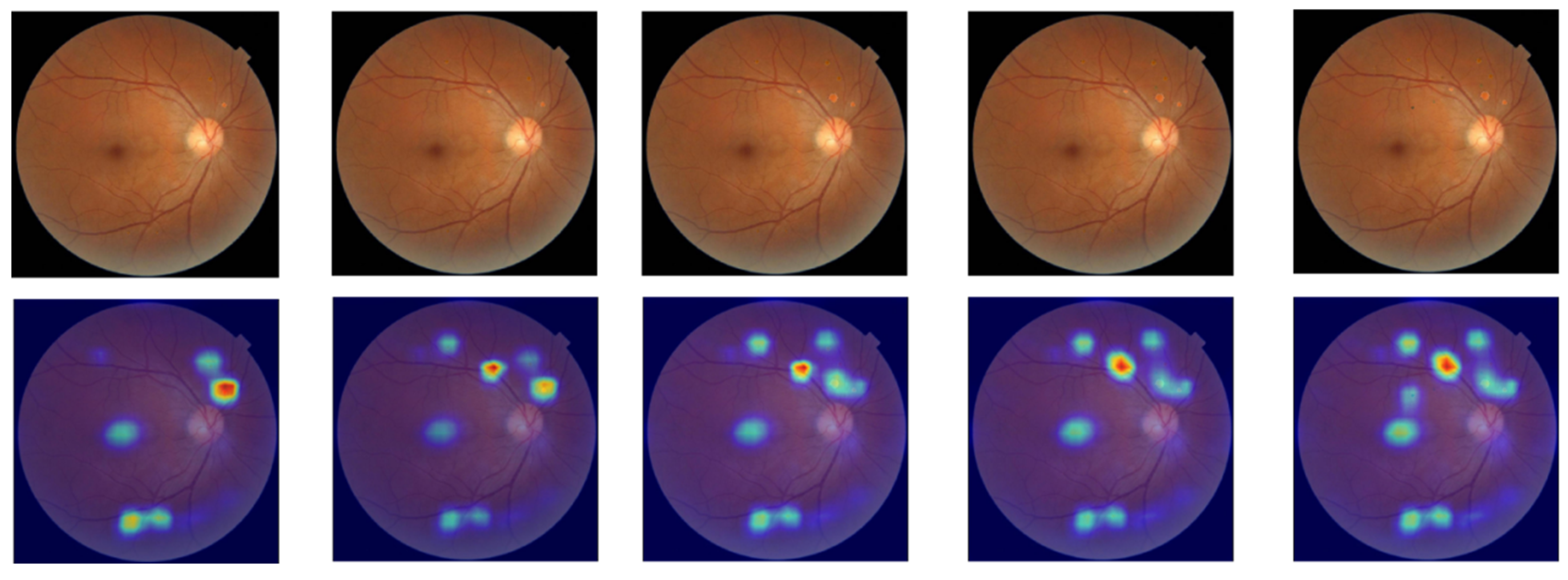
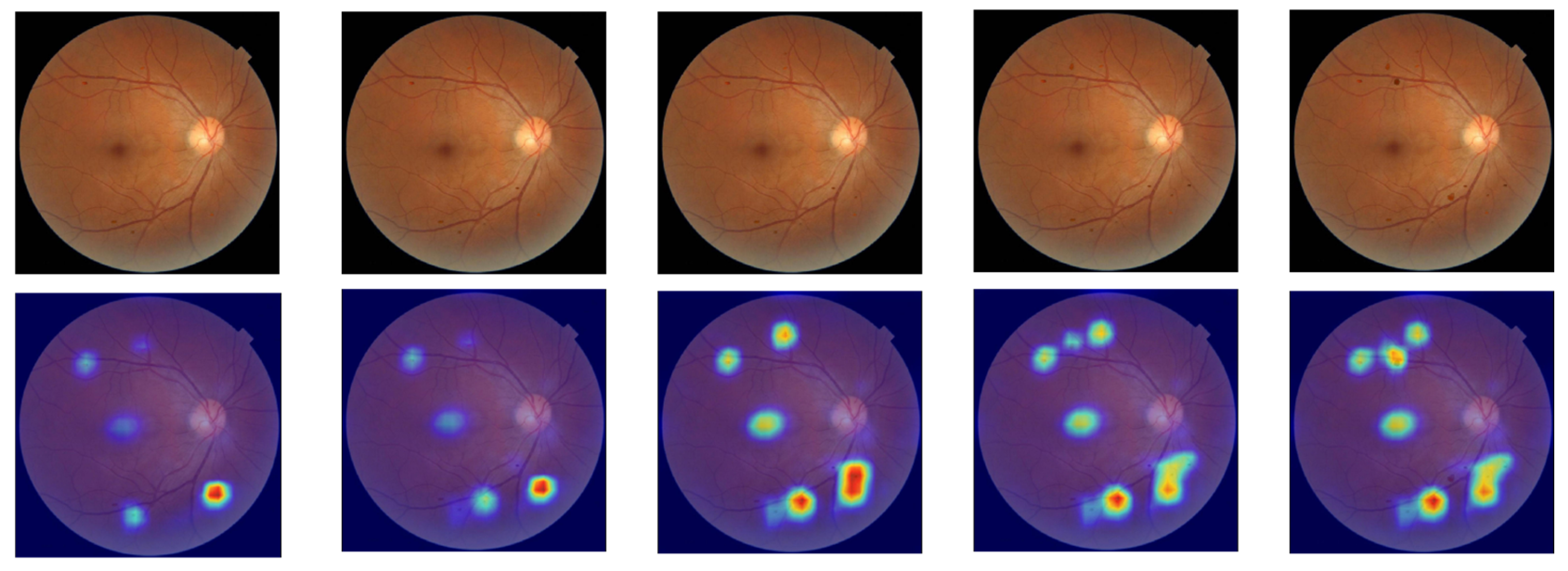
The pathological lesions (bleeding points and microaneurysms) incorporated in this study were all derived from physician-annotated real ophthalmic fundus images. Based on the annotated information, we extracted typical lesion regions and applied image processing techniques including cropping, edge smoothing, and fusion to naturally overlay them onto normal fundus images, thereby generating test samples for trend correlation analysis. In the microaneurysm addition experiment, we selected representative microaneurysm areas from physician-annotated images and overlaid them onto normal fundus images, creating a sequence of images with progressively decreasing numbers of microaneurysms. As shown in the second row of
Figure 5, the left-to-right images contained 14, 12, 10, 8, and 6 microaneurysm lesions, respectively. For the bleeding point addition experiment, we adopted the same image processing strategy to extract and screen typical hemorrhagic areas from authentic images, then sequentially overlayed them onto normal fundus images. As depicted in the first row of
Figure 5, the left-to-right images contained 13, 11, 9, 7, and 5 hemorrhagic points, respectively.
In the fifth step, AOPC score, Recall and Dice were calculated to analyze the quality of the significant figure.
The AOPC score evaluates the fidelity of saliency maps to model predictions by progressively removing significant regions and observing changes in model outputs, thereby assessing whether the interpretation method accurately identifies key features influencing decision-making. The formula is shown in Equation (3), where represents the model’s prediction for the original input x, denotes the input image after removing the top k% of significant pixels, and K indicates the number of disturbance steps.
The Recall and Dice coefficients serve as key metrics for evaluating the consistency between model interpretation results and actual lesion regions. By quantitatively calculating the overlap between highlighted areas in the saliency map and lesion mask regions, these metrics provide validation of the interpretability of model predictions. In practical implementation, we first normalized the saliency maps generated by different interpretability analysis methods across models. Subsequently, using a predefined percentile threshold (0.9), we performed binary operations to retain only the top 10% high-response regions with significant values as the model’s “highlighted areas”.
The recall rate is used to measure the matching degree between the significant figure highlight area and the real mask area. The calculation formula is shown in Equation (4), where TP represents the number of pixels correctly highlighted, and FN represents the number of pixels not correctly highlighted.
The Dice coefficient is used to measure the similarity between the significant map highlight area and the true mask. The calculation formula is shown in Equation (5), where A is the binary significant map, B is the binary label, and |A ∩ B| represents the overlapping pixels between them.