Molecular Mechanism Analysis of the Activation of Human Olfactory Receptor OR9Q2 by 4-Methylphenol
Abstract
1. Introduction
- To characterize the binding mode and identify key residues involved in 4-methylphenol recognition through molecular docking.
- To evaluate the binding stability and quantify binding affinity using molecular dynamics simulations and MM-PBSA analysis.
- To evaluate the activation of 4-methylphenol on hOR9Q2 by constructing hOR9Q2-expressing HEK293 cells.
- To confirm the functional significance of predicted residues through site-directed mutagenesis.
2. Materials and Methods
2.1. Chemicals
2.2. Determination of Odor Threshold
2.3. Structure Model of hOR9Q2 and 4-Methylphenol
2.4. Molecular Docking
2.5. Molecular Dynamics (MD) Simulations
2.6. Calculation of Binding Free Energy Change
2.7. Molecular Cloning of hOR9Q2
2.8. PCR-Based Site-Directed Mutagenesis
2.9. Cell Culture and Transfection
2.10. Luminescence Assay
2.11. Data Analysis
3. Results and Discussion
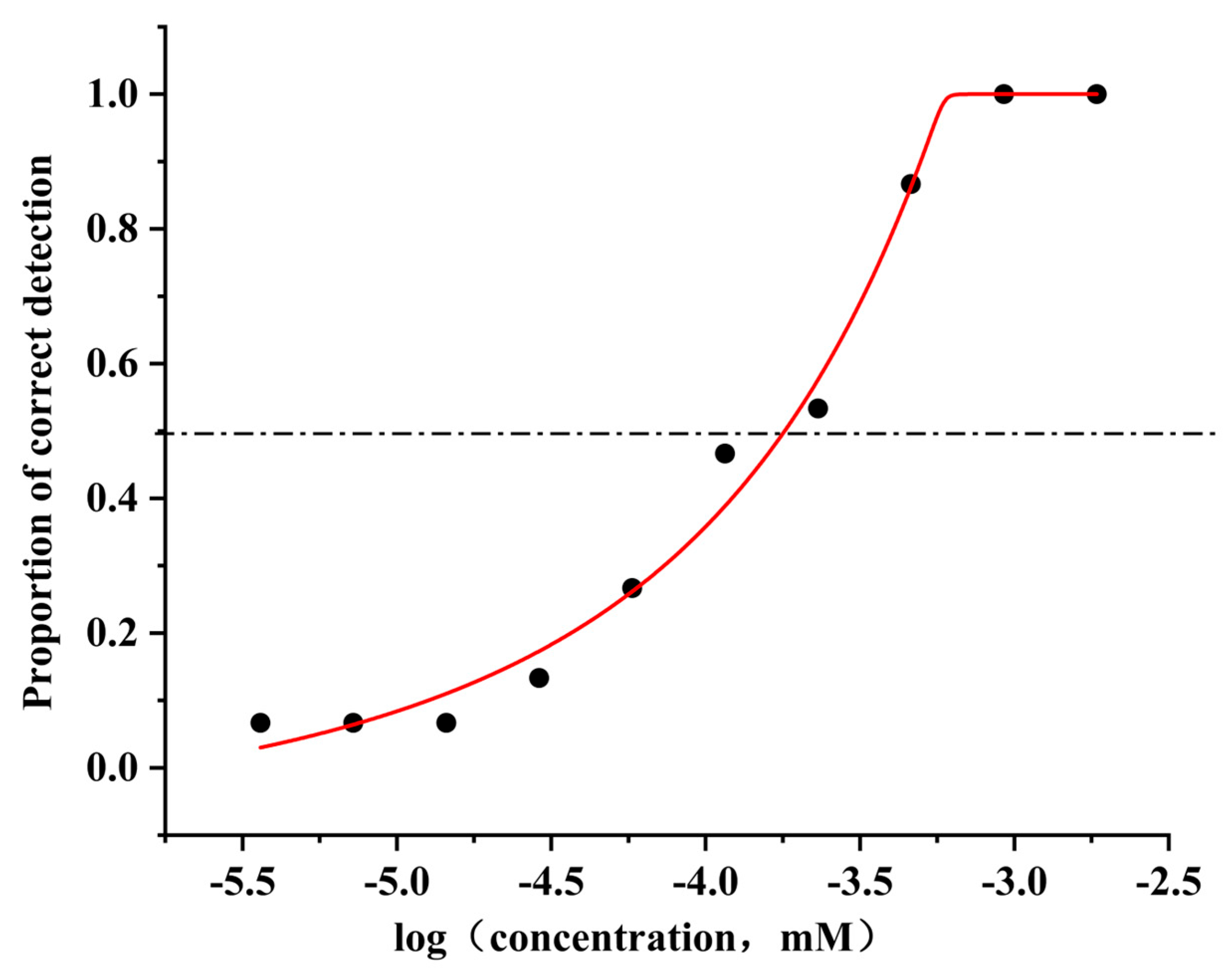
3.1. Odor Characteristics and Thresholds of 4-Methylphenol
3.2. Analysis of Molecular Docking Results
3.3. Stability Analysis of the hOR9Q2-4-Methylphenol Complex
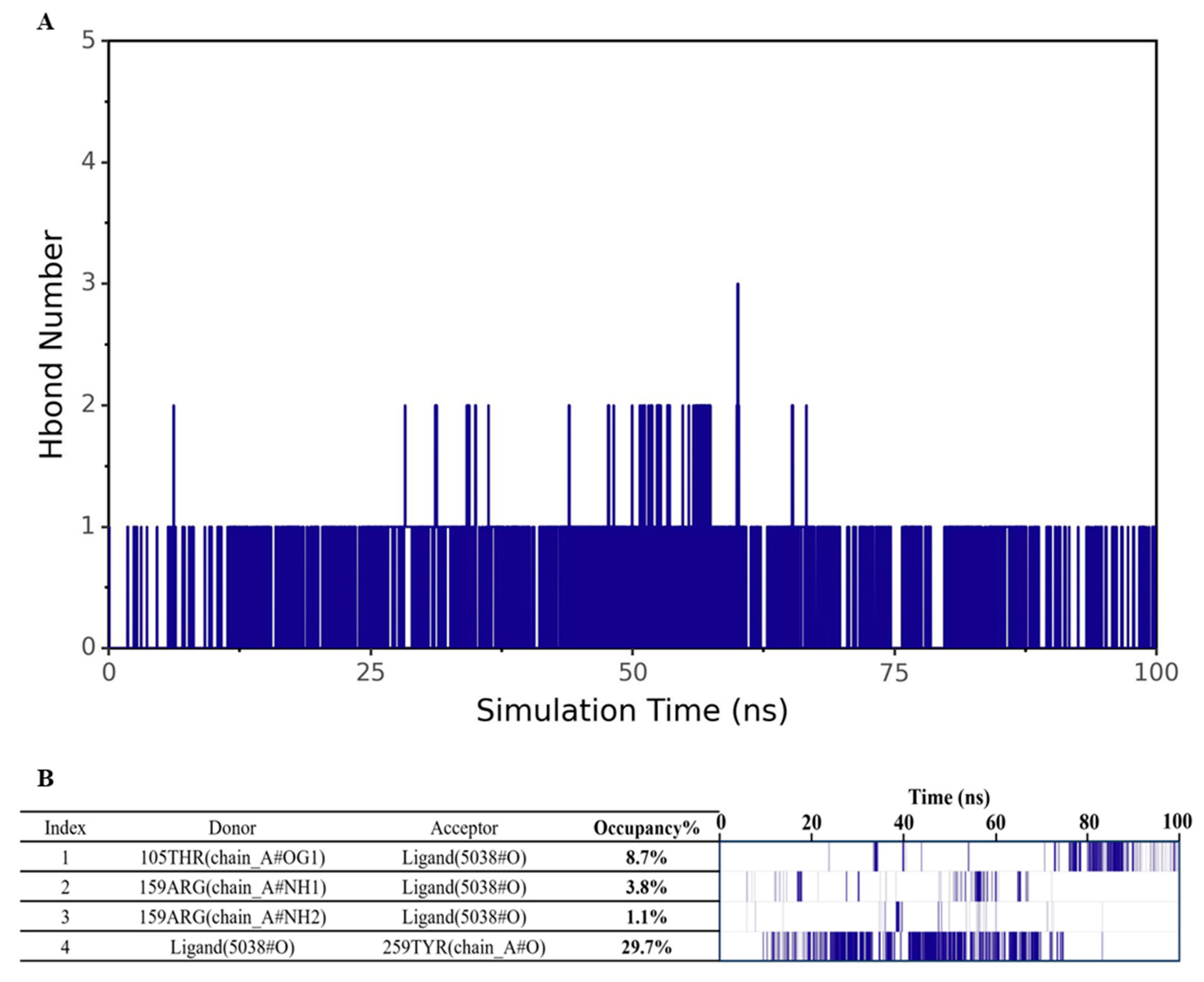
3.4. Analysis of Hydrogen Bond Interaction Between 4-Methylphenol and hOR9Q2
3.5. Thermodynamic Study of 4-Methylphenol and hOR9Q2
3.6. Site-Directed Mutagenesis of Active Site Amino Acids
3.7. Concentration–Response Relationships of hOR9Q2 and Its Mutants to 4-Methylphenol
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thompson, D.C.; Perera, K.; London, R. Studies on the mechanism of hepatotoxicity of 4-methylphenol (p-cresol): Effects of deuterium labeling and ring substitution. Chem. Interact. 1996, 101, 1–11. [Google Scholar] [CrossRef]
- Cicha-Wojciechowicz, D.; Frank, S.; Steinhaus, M.; Majcher, M.A. Key Odorants Forming Aroma of Polish Mead: Influence of the Raw Material and Manufacturing Processes. J. Agric. Food Chem. 2024, 72, 10548–10557. [Google Scholar] [CrossRef] [PubMed]
- Sollai, G.; Barbarossa, I.T.; Usai, P.; Hummel, T.; Crnjar, R. Association between human olfactory performance and ability to detect single compounds in complex chemical mixtures. Physiol. Behav. 2020, 217, 112820. [Google Scholar] [CrossRef] [PubMed]
- Czerny, M.; Christlbauer, M.; Fischer, A.; Granvogl, M.; Hammer, M.; Hartl, C.; Hernandez, N.M.; Schieberle, P. Re-investigation on odour thresholds of key food aroma compounds and development of an aroma language based on odour qualities of defined aqueous odorant solutions. Eur. Food Res. Technol. 2008, 228, 265–273. [Google Scholar] [CrossRef]
- Baldacchino, F.; Manon, S.; Puech, L.; Buatois, B.; Dormont, L.; Jay-Robert, P. Olfactory and behavioural responses of tabanid horseflies to octenol, phenols and aged horse urine. Med. Vet. Entomol. 2013, 28, 201–209. [Google Scholar] [CrossRef]
- Díaz-Santiz, E.; Rojas, J.C.; Casas-Martínez, M.; Cruz-López, L.; Malo, E.A. Rat volatiles as an attractant source for the Asian tiger mosquito, Aedes albopictus. Sci. Rep. 2020, 10, 5170. [Google Scholar] [CrossRef]
- Li, G.; Wang, L.; Ye, F.; Li, S.; Yu, H. Molecular determinants of olfactory receptor activation: Comparative analysis of Olfr205 and Olfr740 family member responses to indole. Arch. Biochem. Biophys. 2024, 758, 110061. [Google Scholar] [CrossRef]
- Watkins, P.J.; Jaborek, J.R.; Teng, F.; Day, L.; Castada, H.Z.; Baringer, S.; Wick, M. Branched chain fatty acids in the flavour of sheep and goat milk and meat: A review. Small Rumin. Res. 2021, 200, 106398. [Google Scholar] [CrossRef]
- Schroepfer, M.; Czerny, M.; Schulz, H.; Schieberle, P. The odor of leather. J. Am. Leather Chem. Assoc. 2013, 108, 94–107. [Google Scholar]
- Du, L.; Wu, C.; Liu, Q.; Huang, L.; Wang, P. Recent advances in olfactory receptor-based biosensors. Biosens. Bioelectron. 2013, 42, 570–580. [Google Scholar] [CrossRef]
- Lalis, M.; Hladiš, M.; Khalil, S.A.; Deroo, C.; Marin, C.; Bensafi, M.; Baldovini, N.; Briand, L.; Fiorucci, S.; Topin, J. A status report on human odorant receptors and their allocated agonists. Chem. Senses 2024, 49, bjae037. [Google Scholar] [CrossRef]
- Verbeurgt, C.; Wilkin, F.; Tarabichi, M.; Gregoire, F.; Dumont, J.E.; Chatelain, P. Profiling of olfactory receptor gene expression in whole human olfactory mucosa. PLoS ONE 2014, 9, e96333. [Google Scholar] [CrossRef]
- Geithe, C.; Noe, F.; Kreissl, J.; Krautwurst, D. The broadly tuned odorant receptor OR1A1 is highly selective for 3-methyl-2,4-nonanedione, a key food odorant in aged wines, tea, and other foods. Chem. Senses 2016, 42, 181–193. [Google Scholar] [CrossRef]
- Haag, F.; Di Pizio, A.; Krautwurst, D. The key food odorant receptive range of broadly tuned receptor OR2W1. Food Chem. 2022, 375, 131680. [Google Scholar] [CrossRef] [PubMed]
- Haag, F.; Hoffmann, S.; Krautwurst, D. Key Food furanones furaneol and sotolone specifically activate distinct odorant receptors. J. Agric. Food Chem. 2021, 69, 10999–11005. [Google Scholar] [CrossRef] [PubMed]
- Noe, F.; Polster, J.; Geithe, C.; Kotthoff, M.; Schieberle, P.; Krautwurst, D. OR2M3: A highly specific and narrowly tuned human odorant receptor for the sensitive detection of onion key food odorant 3-mercapto-2-methylpentan-1-ol. Chem. Senses 2016, 42, 195–210. [Google Scholar] [CrossRef]
- Haag, F.; Frey, T.; Hoffmann, S.; Kreissl, J.; Stein, J.; Kobal, G.; Hauner, H.; Krautwurst, D. The multi-faceted food odorant 4-methylphenol selectively activates evolutionary conserved receptor OR9Q2. Food Chem. 2023, 426, 136492. [Google Scholar] [CrossRef]
- Ollitrault, G.; Achebouche, R.; Dreux, A.; Murail, S.; Audouze, K.; Tromelin, A.; Taboureau, O. Pred-O3, a web server to predict molecules, olfactory receptors and odor relationships. Nucleic Acids Res. 2024, 52, W507–W512. [Google Scholar] [CrossRef] [PubMed]
- El Rhabori, S.; Alaqarbeh, M.; El Allouche, Y.; Naanaai, L.; El Aissouq, A.; Bouachrine, M.; Chtita, S.; Khalil, F. Exploring innovative strategies for identifying anti-breast cancer compounds by integrating 2D/3D-QSAR, molecular docking analyses, ADMET predictions, molecular dynamics simulations, and MM-PBSA approaches. J. Mol. Struct. 2024, 1320, 139500. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Q.; Fan, W.; Shi, Q.; Mao, J.; Xie, J.; Chai, G.; Zhang, C. Deciphering olfactory receptor binding mechanisms: A structural and dynamic perspective on olfactory receptors. Front. Mol. Biosci. 2025, 11, 1498796. [Google Scholar] [CrossRef]
- Kaneshiro, H.; Sato, M.; Tanaka, A.; Nakata, S.; Aihara, Y.; Kitoh-Nishioka, H.; Mori, Y.; Tanaka, S. A Structure-Based Approach for Predicting Odor Similarity of Molecules via Docking Simulations with Human Olfactory Receptors. ACS Omega 2025, 10, 39933–39945. [Google Scholar] [CrossRef]
- Ben Khemis, I.; Aouaini, F.; Knani, S.; Graba, B.; Lefi, N.; Ben Lamine, A. New insights into the docking mechanism of vanillin on three mammalian olfactory receptors via a new statistical physics model. Sci. Rep. 2025, 15, 32271. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, X.; Sun, Z.; Shen, T.; Kou, X.; Niu, Y.; Xiao, Z. Unraveling the interaction mechanism between enantiomers of lactone compounds (γ-octalactone and γ-undecalactone) in Longjing tea and OR1A1 olfactory receptor using molecular docking and molecular dynamics simulation. Food Biosci. 2025, 66, 106282. [Google Scholar] [CrossRef]
- Mei, S.; Ding, J.; Chen, X. Identification of differential volatile and non-volatile compounds in coffee leaves prepared from different tea processing steps using HS-SPME/GC–MS and HPLC-Orbitrap-MS/MS and investigation of the binding mechanism of key phytochemicals with olfactory and taste receptors using molecular docking. Food Res. Int. 2023, 168, 112760. [Google Scholar] [CrossRef] [PubMed]
- Chaohui, Y.; Xuehai, G.; Changrong, G.; Ping, Z.; Shuangmin, L.; Zhichao, X. Taste characterization and molecular docking study of novel umami flavor peptides in Yanjin black bone Chicken meat. Food Chem. 2024, 464, 141695. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Andrews, C.W.; Capelli, A.-M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2005, 49, 5912–5931. [Google Scholar] [CrossRef]
- Fan, J.; Fu, A.; Zhang, L. Progress in molecular docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- O’DOwd, B.F.; Hnatowich, M.; Regan, J.W.; Leader, W.M.; Caron, M.G.; Lefkowitz, R.J. Site-directed mutagenesis of the cytoplasmic domains of the human beta 2-adrenergic receptor. Localization of regions involved in G protein-receptor coupling. J. Biol. Chem. 1988, 263, 15985–15992. [Google Scholar] [CrossRef]
- Stocker, W.A.; Howard, J.A.; Maskey, S.; Luan, H.; Harrison, S.G.; Hart, K.N.; Hok, L.; Thompson, T.B.; Walton, K.L.; Harrison, C.A. Characterization of the molecular mechanisms that govern anti-Müllerian hormone synthesis and activity. FASEB J. 2024, 38, e23377. [Google Scholar] [CrossRef]
- Wu, C.; Xu, M.; Dong, J.; Cui, W.; Yuan, S. The structure and function of olfactory receptors. Trends Pharmacol. Sci. 2024, 45, 268–280. [Google Scholar] [CrossRef]
- Sekharan, S.; Ertem, M.Z.; Zhuang, H.; Block, E.; Matsunami, H.; Zhang, R.; Wei, J.N.; Pan, Y.; Batista, V.S. QM/MM model of the mouse olfactory receptor MOR244-3 validated by site-directed mutagenesis experiments. Biophys. J. 2014, 107, L5–L8. [Google Scholar] [CrossRef]
- Mitamura, T.; Umata, T.; Nakano, F.; Shishido, Y.; Toyoda, T.; Itai, A.; Kimura, H.; Mekada, E. Structure-function analysis of the diphtheria toxin receptor toxin binding site by site-directed mutagenesis. J. Biol. Chem. 1997, 272, 27084–27090. [Google Scholar] [CrossRef]
- ISO 8586:2023; Sensory Analysis—Selection and Training of Sensory Assessors. International Organisation of Standardisation (ISO): Geneva, Switzerland, 2023.
- ISO 13301:2018; Sensory Analysis—Methodology—General Guidance for Measuring Odour, Flavour and Taste Detection Thresholds by a Three-Alternative Forced-Choice (3-AFC) Procedure. International Organization for Standardization: Geneva, Switzerland, 2018.
- Liu, D.; Qiu, Y.; Li, Q.; Zhang, H. Atomistic Simulation of Lysozyme in Solutions Crowded by Tetraethylene Glycol: Force Field Dependence. Molecules 2022, 27, 2110. [Google Scholar] [CrossRef]
- Wang, J.; Wang, D.; Huang, M.; Sun, B.; Ren, F.; Wu, J.; Zhang, J.; Li, H.; Sun, X. Decoding Molecular Mechanism Underlying Human Olfactory Receptor OR8D1 Activation by Sotolone Enantiomers. J. Agric. Food Chem. 2024, 72, 5403–5415. [Google Scholar] [CrossRef] [PubMed]
- Jade, D.; Alzahrani, A.; Critchley, W.; Ponnambalam, S.; Harrison, M.A. Identification of FDA-approved drugs against SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) through computational virtual screening. Struct. Chem. 2022, 34, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Gao, X.; Tang, J.; Gao, L.; Cui, X.; Liu, K.; Zhang, X.; Jin, M. Exploring the Efficacy and Target Genes of Atractylodes Macrocephala Koidz Against Alzheimer’s Disease Based on Multi-Omics, Computational Chemistry, and Experimental Verification. Curr. Issues Mol. Biol. 2025, 47, 118. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Jarzynka, H.F. Headspace-Solid Phase Micro Extraction–Gas Chromatography–Mass Spectrometry (HS-SPME-GC-MS) Analysis for Oak Barrel Flavor Compounds in Whiskey. Master’s Thesis, University of Texas, Arlington, TX, USA, 2021. [Google Scholar]
- Strube, A.; Buettner, A.; Czerny, M. Influence of chemical structure on absolute odour thresholds and odour characteristics of ortho- and para-halogenated phenols and cresols. Flavour Fragr. J. 2012, 27, 304–312. [Google Scholar] [CrossRef]
- Mall, V.; Schieberle, P. On the importance of phenol derivatives for the peaty aroma attribute of scotch whiskies from Islay. In Sex, Smoke, and Spirits: The Role of Chemistry; ACS Symposium Series 1321; American Chemical Society: Washington, DC, USA, 2019; pp. 107–116. [Google Scholar]
- Agnieszka, K.B. Thermodynamics of ligand-protein interactions: Implications for molecular design. In Thermodynamics; Juan Carlos, M.-P., Ed.; IntechOpen: Rijeka, Croatia, 2011; Chapter 1. [Google Scholar]
- Bissantz, C.; Kuhn, B.; Stahl, M.A. Medicinal chemist’s guide to molecular interactions. J. Med. Chem. 2010, 14, 5061–5084. [Google Scholar] [CrossRef]
- Mbaye, M.N.; Hou, Q.; Basu, S.; Teheux, F.; Pucci, F.; Rooman, M. A comprehensive computational study of amino acid interactions in membrane proteins. Sci. Rep. 2019, 9, 12043. [Google Scholar] [CrossRef]
- Lanzarotti, E.; Defelipe, L.A.; Marti, M.A.; Turjanski, A.G. Aromatic clusters in protein–protein and protein–drug complexes. J. Cheminformatics 2020, 12, 30. [Google Scholar] [CrossRef]
- McGaughey, G.B.; Gagné, M.; Rappé, A.K. π-Stacking interactions: Alive and Well in Proteins. J. Biol. Chem. 1998, 273, 15458–15463. [Google Scholar] [CrossRef]
- Ringer, A.L.; Senenko, A.; Sherrill, C.D. Models of S/π interactions in protein structures: Comparison of the H2S–benzene complex with PDB data. Protein Sci. 2007, 16, 2216–2223. [Google Scholar] [CrossRef]
- Cunha, K.C.; Rusu, V.H.; Viana, I.F.T.; Marques, E.T.A.; Dhalia, R.; Lins, R.D. Assessing protein conformational sampling and structural stability via de novo design and molecular dynamics simulations. Biopolymers 2015, 103, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Syed Ausaf, A.; Md Imtaiyaz, H.; Asimul, I.; Faizan, A. A Review of Methods Available to Estimate Solvent-Accessible Surface Areas of Soluble Proteins in the Folded and Unfolded States. Curr. Protein Pept. Sci. 2014, 15, 456–476. [Google Scholar] [CrossRef]
- Liu, K.; Watanabe, E.; Kokubo, H. Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations. J. Comput. Mol. Des. 2017, 31, 201–211. [Google Scholar] [CrossRef]
- Ivanov, P. Conformations of some lower-size large-ring cyclodextrins derived from conformational search with molecular dynamics and principal component analysis. J. Mol. Struct. 2012, 1009, 3–10. [Google Scholar] [CrossRef]
- Ben Chorin, A.; Masrati, G.; Kessel, A.; Narunsky, A.; Sprinzak, J.; Lahav, S.; Ashkenazy, H.; Ben-Tal, N. ConSurf-DB: An accessible repository for the evolutionary conservation patterns of the majority of PDB proteins. Protein Sci. 2020, 29, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Man, O.; Gilad, Y.; Lancet, D. Prediction of the odorant binding site of olfactory receptor proteins by human–mouse comparisons. Protein Sci. 2004, 13, 240–254. [Google Scholar] [CrossRef]
- Katada, S.; Hirokawa, T.; Oka, Y.; Suwa, M.; Touhara, K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: Mapping the odorant-binding site. J. Neurosci. 2005, 25, 1806–1815. [Google Scholar] [CrossRef]
- Serrano, L.; Neira, J.-L.; Sancho, J.; Fersht, A.R. Effect of alanine versus glycine in α-helices on protein stability. Nature 1992, 356, 453–455. [Google Scholar] [CrossRef]
- Scott, K.A.; Alonso, D.O.V.; Sato, S.; Fersht, A.R.; Daggett, V. Conformational entropy of alanine versus glycine in protein denatured states. Proc. Natl. Acad. Sci. USA 2007, 104, 2661–2666. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, C.-R.; Jiang, M.; Zhu, Y.-Y.; Wang, J.; Chen, J.; Shi, J.-H. Binding interaction of atorvastatin with bovine serum albumin: Spectroscopic methods and molecular docking. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 156, 155–163. [Google Scholar] [CrossRef]
- Ben Khemis, I.; Noureddine, O.; Smati, H.; Aouaini, F.; Hassine, S.B.H.; Ben Lamine, A. Advanced investigation of a putative adsorption process of nine non key food odorants (non-KFOs) on the broadly tuned human olfactory receptor OR2W1: Statistical physics modeling and molecular docking study. Int. J. Biol. Macromol. 2023, 233, 123548. [Google Scholar] [CrossRef] [PubMed]
- Smati, H.; Ben Torkia, Y.; Ben Khemis, I.; Aouaini, F.; Ben Lamine, A.; Znaidia, S. Modeling by statistical physics and interpretation of the olfactory process of the two enantiomers 3-mercapto-2-methylbutan-1-ol and 3-mercapto-2-methylpentan-1-ol on the OR2M3 human olfactory receptor. Int. J. Biol. Macromol. 2023, 243, 124896. [Google Scholar] [CrossRef] [PubMed]
- Matsunami, H.; Vaidehi, N.; Ghosh, S.; Yohda, M.; Do, M.; Ikegami, K.; Sharma, R.; Nagai, M.H.; de March, C.A.; Fukutani, Y.; et al. Structural instability and divergence from conserved residues underlie intracellular retention of mammalian odorant receptors. Proc. Natl. Acad. Sci. USA 2020, 177, 2957–2967. [Google Scholar]
- Gavira, J.A.; Gumerov, V.M.; Rico-Jiménez, M.; Petukh, M.; Upadhyay, A.A.; Ortega, A.; Matilla, M.A.; Zhulin, I.B.; Krell, T. How bacterial chemoreceptors evolve novel ligand specificities. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef] [PubMed]

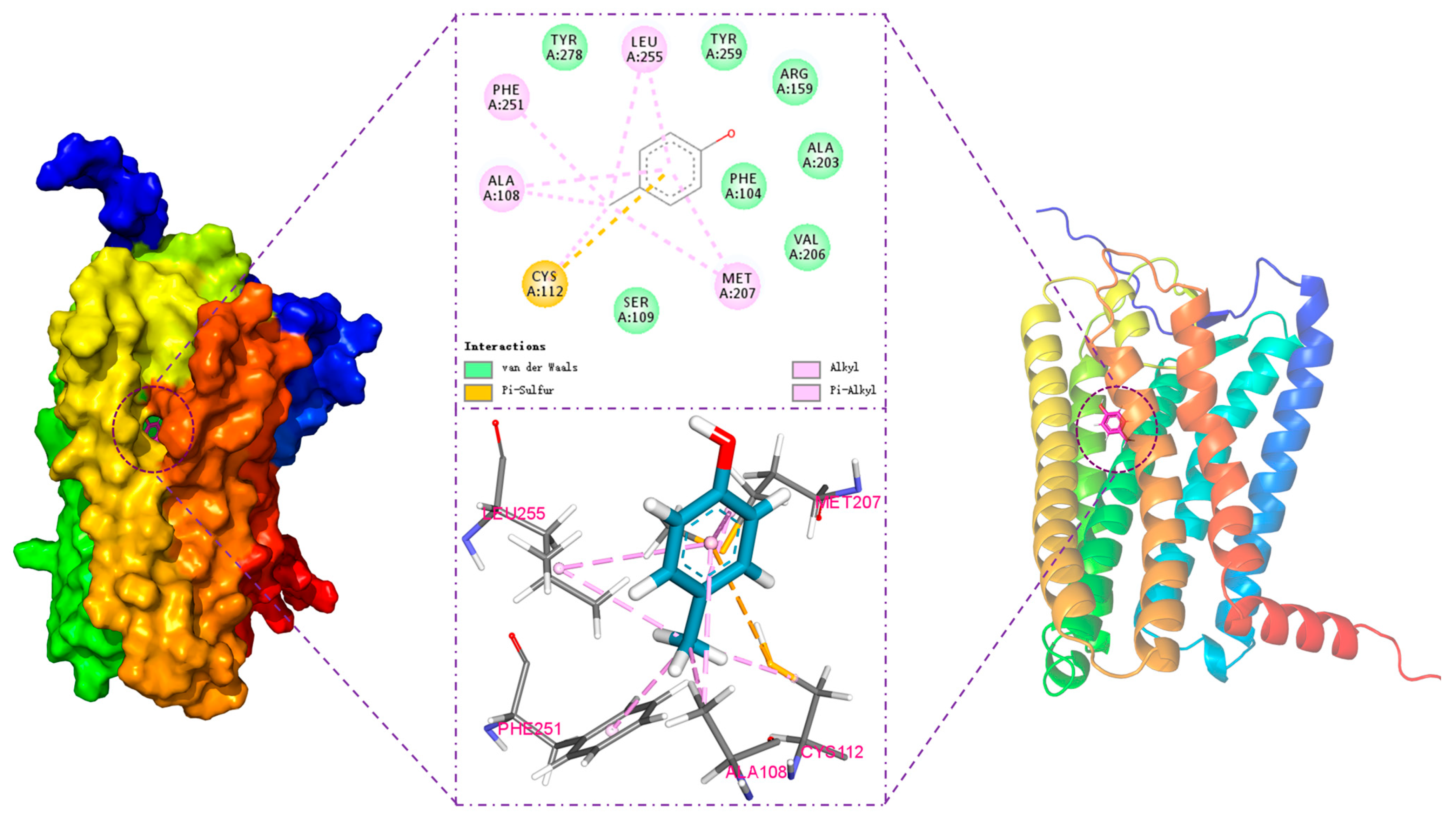



| Residues of hOR9Q2 | Transmembrane Domains | Distance (Å) | Binding Force | Interaction Category |
|---|---|---|---|---|
| Ala108 | TM3 | 3.9 | Alkyl | Hydrophobic |
| 4.5 | Pi-Alkyl | |||
| Cys112 | TM3 | 4.3 | Alkyl | Hydrophobic |
| 5.0 | Pi-sulfur | Miscellaneous | ||
| Met207 | TM5 | 4.8 | Pi-Alkyl | Hydrophobic |
| 5.2 | Alkyl | |||
| Phe251 | TM6 | 4.6 | Pi-Alkyl | Hydrophobic |
| Leu255 | TM6 | 4.9 | Alkyl | Hydrophobic |
| 5.8 | Pi-Alkyl | Hydrophobic |
| Complex | ΔEvdW (kJ/mol) | ΔEele (kJ/mol) | ΔGPB (kJ/mol) | ΔGSA (kJ/mol) | −TΔS (kJ/mol) | ΔGbind (kJ/mol) |
|---|---|---|---|---|---|---|
| Protein-Ligand | −81.451 ± 0.351 | −4.850 ± 0.091 | 56.924 ± 0.205 | −10.796 ± 0.061 | 5.699 ± 0.675 | −40.173 ± 0.340 |
| No. | Amino Acid Position | Primitive Amino Acid Residues | Original Base Sequence | Mutated Amino Acid Residues | Mutant Base Sequence |
|---|---|---|---|---|---|
| 1 | 71 | Ile | ATC | Val | GTC |
| 2 | 108 | Ala | GCC | Gly | GGC |
| 3 | 112 | Cys | TGC | Ala | GCC |
| 4 | 158 | Val | GTT | Ile | ATT |
| 5 | 204 | Leu | CTT | Val | GTT |
| 6 | 207 | Met | ATG | Ala | GCG |
| 7 | 251 | Phe | TTC | Ala | GCC |
| 8 | 255 | Leu | CTC | Ala | GCC |
| 9 | 259 | Tyr | TAC | Ala | GCC |
| 10 | 277 | Leu | CTC | Phe | TTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, F.; Wang, M.; Zhang, L.; Duan, W.; Sun, B.; Xie, J.; Huang, M.; Sun, S.; Yang, R.; Zhang, Y. Molecular Mechanism Analysis of the Activation of Human Olfactory Receptor OR9Q2 by 4-Methylphenol. Foods 2025, 14, 3738. https://doi.org/10.3390/foods14213738
Wen F, Wang M, Zhang L, Duan W, Sun B, Xie J, Huang M, Sun S, Yang R, Zhang Y. Molecular Mechanism Analysis of the Activation of Human Olfactory Receptor OR9Q2 by 4-Methylphenol. Foods. 2025; 14(21):3738. https://doi.org/10.3390/foods14213738
Chicago/Turabian StyleWen, Fengge, Mengxue Wang, Lili Zhang, Wen Duan, Baoguo Sun, Jianping Xie, Mingquan Huang, Shihao Sun, Rui Yang, and Yuyu Zhang. 2025. "Molecular Mechanism Analysis of the Activation of Human Olfactory Receptor OR9Q2 by 4-Methylphenol" Foods 14, no. 21: 3738. https://doi.org/10.3390/foods14213738
APA StyleWen, F., Wang, M., Zhang, L., Duan, W., Sun, B., Xie, J., Huang, M., Sun, S., Yang, R., & Zhang, Y. (2025). Molecular Mechanism Analysis of the Activation of Human Olfactory Receptor OR9Q2 by 4-Methylphenol. Foods, 14(21), 3738. https://doi.org/10.3390/foods14213738







