Probiotic Properties, Prebiotic Fermentability, and GABA-Producing Capacity of Microorganisms Isolated from Mexican Milk Kefir Grains: A Clustering Evaluation for Functional Dairy Food Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, Enzymes and Bacterial Strains
2.2. Kefir Grains
2.3. Isolation of Bacteria and Yeast from Kefir Grains
2.4. Microorganism Identification by MALDI-TOF MS
2.5. Aggregation Experiments
2.6. Antibiotic Susceptibility
2.7. Antimicrobial Activity
2.8. Resistance to Simulated In Vitro Gastrointestinal Digestion
2.9. Evaluation of Fermentability with Commercial Prebiotics
2.10. GABA Production
2.11. Statistical Analysis
3. Results
3.1. Microorganism Identification
3.2. Aggregation Experiments
3.3. Antibiotic Susceptibility
3.4. Antimicrobial Activity
3.5. In Vitro Digestion Assay
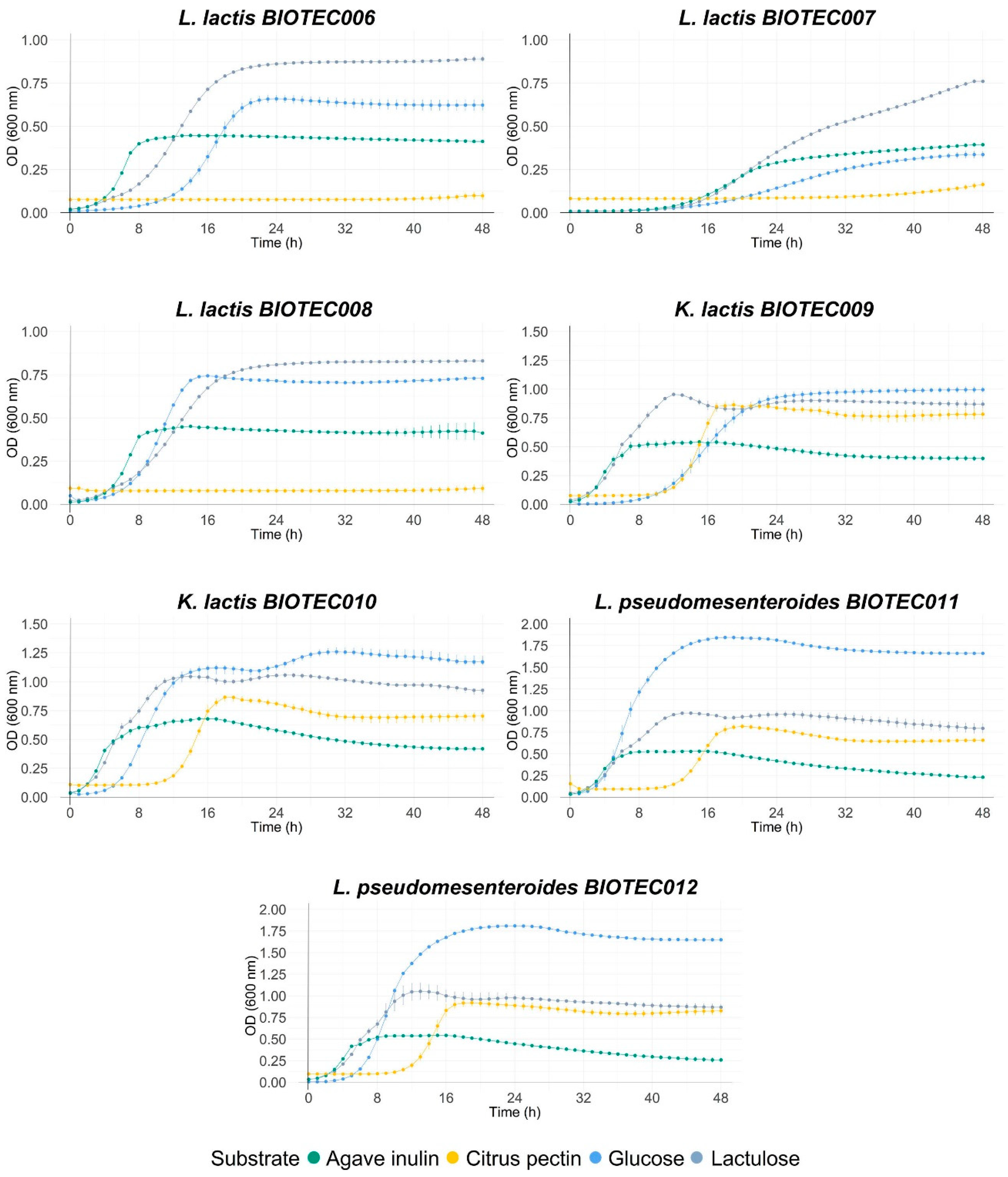
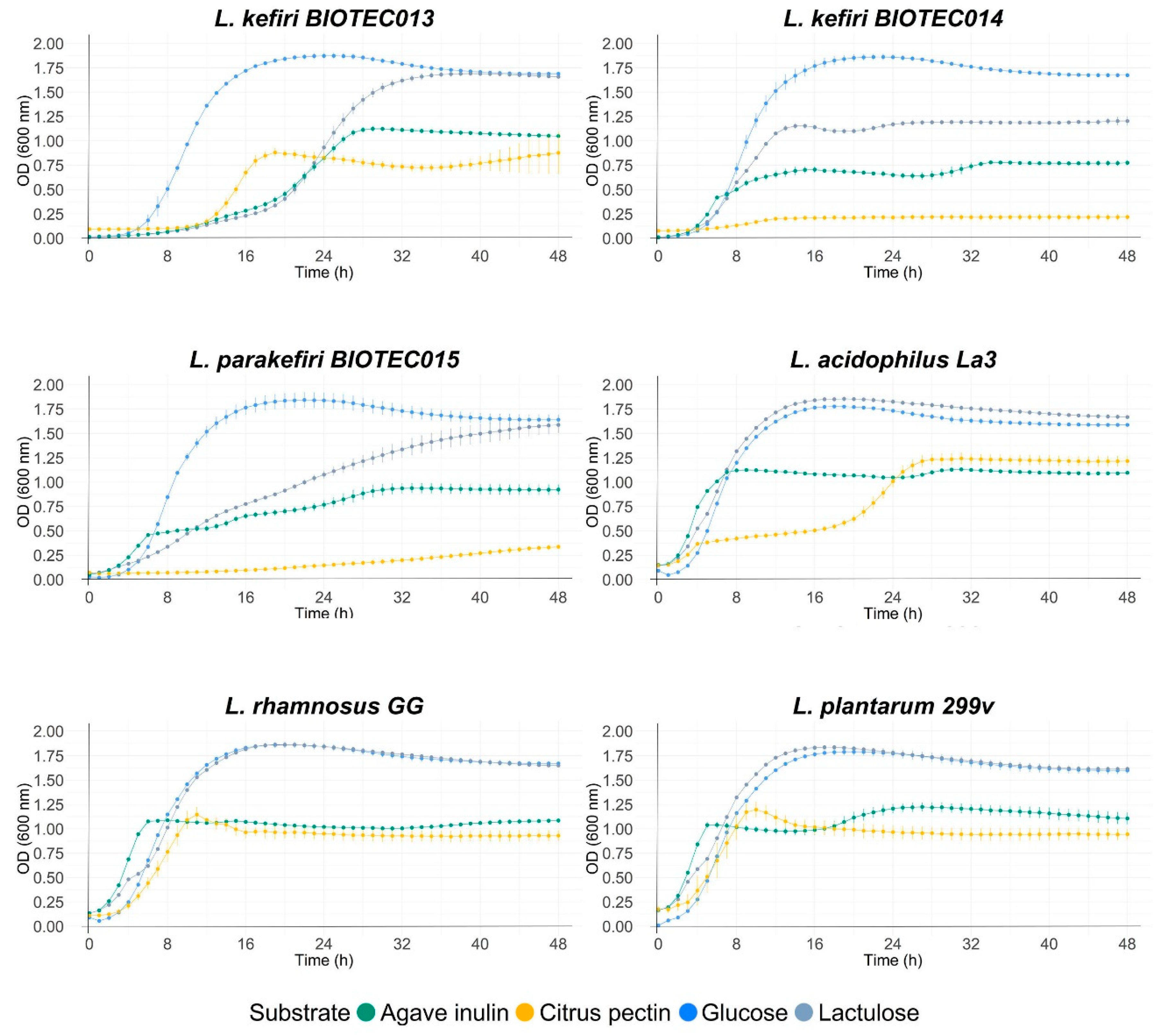
3.6. Evaluation of Fermentability with Commercial Prebiotics
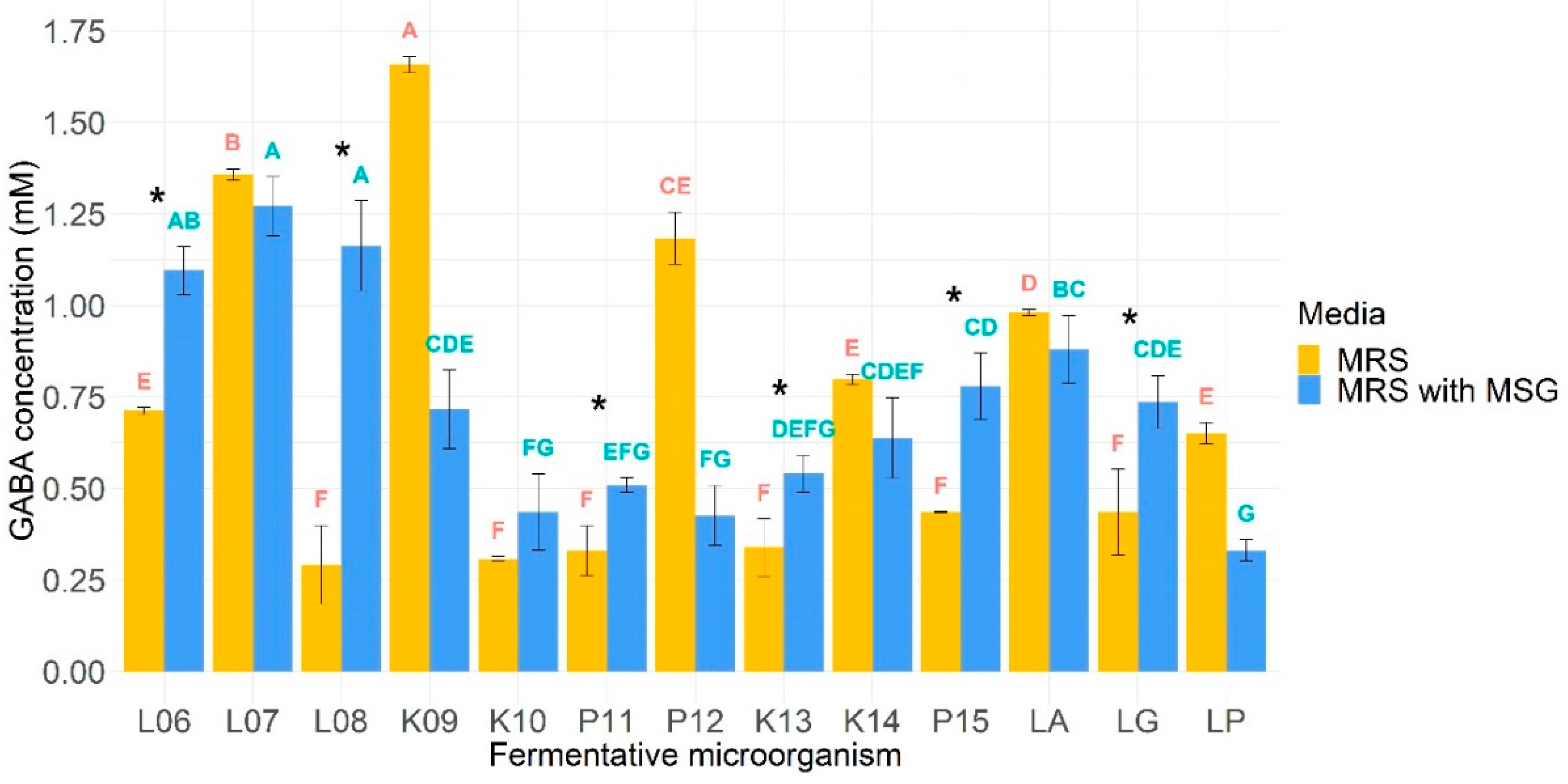
3.7. GABA Production
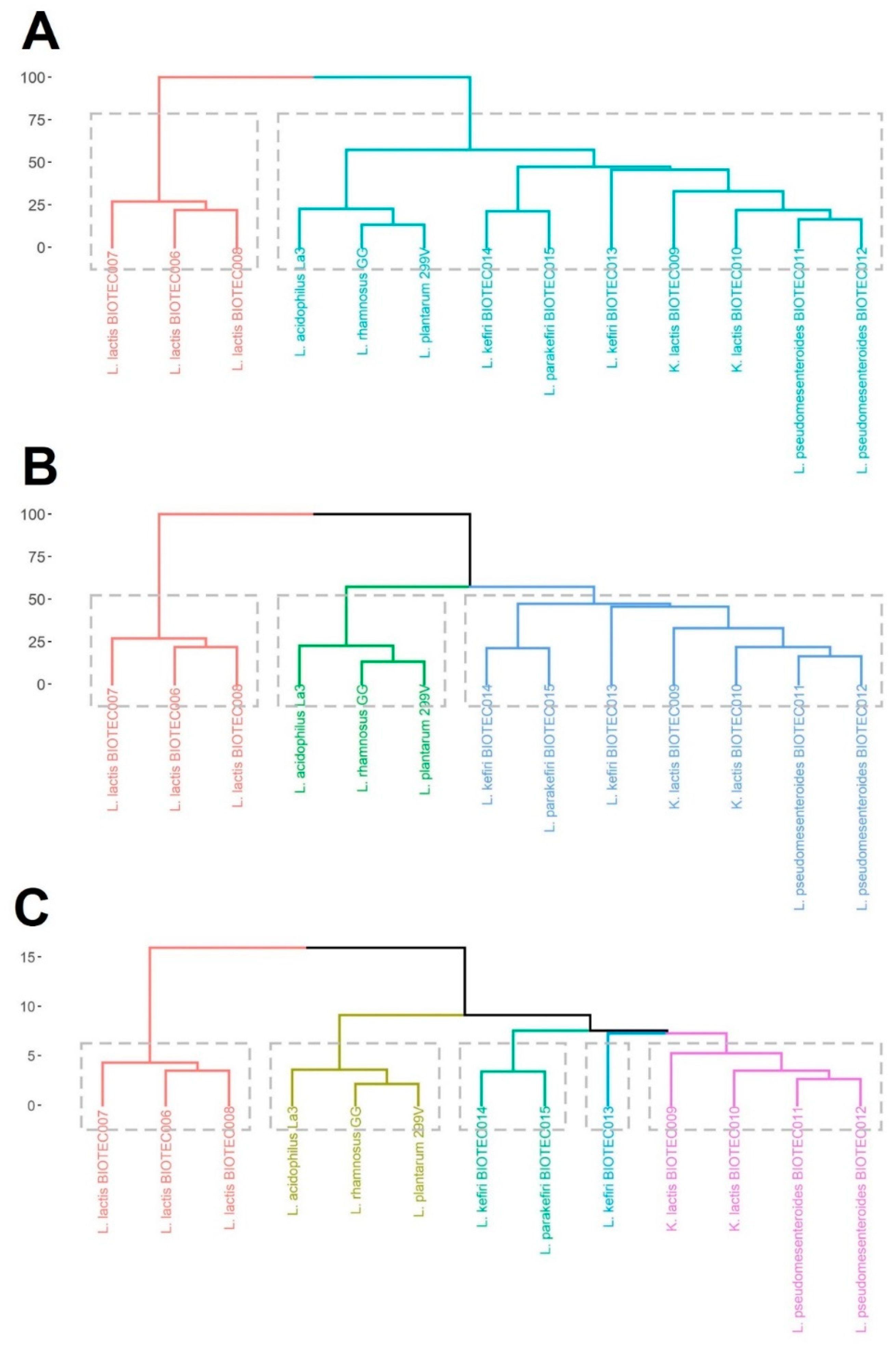
3.8. Cluster Analysis
4. Discussion
4.1. Probiotic Properties
4.2. Evaluation of Fermentability with Commercial Prebiotics
4.3. Psychobiotic Potential through GABA Production Assay
4.4. Choice of Suitable Microorganisms for Potential Functional Dairy Food Applications
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortiz, Y.; García-Amézquita, E.; Acosta, C.H.; Sepúlveda, D.R. Functional Dairy Products. In Global Food Security and Wellness; Barbosa-Cánovas, G.V., María Pastore, G., Candoğan, K., Medina Meza, I.G., Caetano da Silva Lannes, S., Buckle, K., Yada, R.Y., Rosenthal, A., Eds.; Springer: New York, NY, USA, 2017; pp. 67–103. ISBN 978-1-4939-6494-9. [Google Scholar]
- Turkmen, N.; Akal, C.; Özer, B. Probiotic Dairy-Based Beverages: A Review. J. Funct. Foods 2019, 53, 62–75. [Google Scholar] [CrossRef]
- Arslan, S. A Review: Chemical, Microbiological and Nutritional Characteristics of Kefir. CyTA—J. Food 2015, 13, 340–345. [Google Scholar] [CrossRef] [Green Version]
- Azizi, N.F.; Kumar, M.R.; Yeap, S.K.; Abdullah, J.O.; Khalid, M.; Omar, A.R.; Mohd, A.O.; Mortadza, S.A.S.; Alitheen, N.B. Kefir and Its Biological Activities. Foods 2021, 10, 1210. [Google Scholar] [CrossRef]
- Guzel-Seydim, Z.B.; Gökırmaklı, Ç.; Greene, A.K. A Comparison of Milk Kefir and Water Kefir: Physical, Chemical, Microbiological and Functional Properties. Trends Food Sci. Technol. 2021, 113, 42–53. [Google Scholar] [CrossRef]
- Chen, T.-H.; Wang, S.-Y.; Chen, K.-N.; Liu, J.-R.; Chen, M.-J. Microbiological and Chemical Properties of Kefir Manufactured by Entrapped Microorganisms Isolated from Kefir Grains. J. Dairy Sci. 2009, 92, 3002–3013. [Google Scholar] [CrossRef] [Green Version]
- Selhub, E.M.; Logan, A.C.; Bested, A.C. Fermented Foods, Microbiota, and Mental Health: Ancient Practice Meets Nutritional Psychiatry. J. Physiol. Anthropol. 2014, 33, 2. [Google Scholar] [CrossRef] [Green Version]
- Samardzic, J.; Jadzic, D.; Hencic, B.; Jancic, J.; Strac, D.S. Introductory Chapter: GABA/Glutamate Balance: A Key for Normal Brain Functioning. In GABA and Glutamate—New Developments in Neurotransmission Research; Samardzic, J., Ed.; BoD: Norderstedt, Germany, 2018; ISBN 978-953-51-3821-1. [Google Scholar]
- Del Toro-Barbosa, M.; Hurtado-Romero, A.; Garcia-Amezquita, L.E.; García-Cayuela, T. Psychobiotics: Mechanisms of Action, Evaluation Methods and Effectiveness in Applications with Food Products. Nutrients 2020, 12, 3896. [Google Scholar] [CrossRef]
- Diez-Gutiérrez, L.; San Vicente, L.; Barrón, L.J.R.; del Carmen Villarán, M.; Chávarri, M. Gamma-Aminobutyric Acid and Probiotics: Multiple Health Benefits and Their Future in the Global Functional Food and Nutraceuticals Market. J. Funct. Foods 2020, 64, 103669. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering Probiotic Microorganisms: In Vitro, in Vivo, Genetic and Omics Approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- Huebner, J.; Wehling, R.L.; Hutkins, R.W. Functional Activity of Commercial Prebiotics. Int. Dairy J. 2007, 17, 770–775. [Google Scholar] [CrossRef]
- Plessas, S.; Nouska, C.; Mantzourani, I.; Kourkoutas, Y.; Alexopoulos, A.; Bezirtzoglou, E. Microbiological Exploration of Different Types of Kefir Grains. Fermentation 2016, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.P.; Hsiao, P.J.; Hong, W.S.; Dai, T.Y.; Chen, M.J. Lactobacillus Kefiranofaciens M1 Isolated from Milk Kefir Grains Ameliorates Experimental Colitis in Vitro and in Vivo. J. Dairy Sci. 2012, 95, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Diosma, G.; Romanin, D.E.; Rey-Burusco, M.F.; Londero, A.; Garrote, G.L. Yeasts from Kefir Grains: Isolation, Identification, and Probiotic Characterization. World J. Microbiol. Biotechnol. 2014, 30, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Zanirati, D.F.; Abatemarco, M.; de Cicco Sandes, S.H.; Nicoli, J.R.; Nunes, Á.C.; Neumann, E. Selection of Lactic Acid Bacteria from Brazilian Kefir Grains for Potential Use as Starter or Probiotic Cultures. Anaerobe 2015, 32, 70–76. [Google Scholar] [CrossRef] [PubMed]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; de Cadiñanos, L.P.G.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion Abilities of Dairy Lactobacillus Plantarum Strains Showing an Aggregation Phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST Static in Vitro Simulation of Gastrointestinal Food Digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Tsukatani, T.; Higuchi, T.; Matsumoto, K. Enzyme-Based Microtiter Plate Assay for γ-Aminobutyric Acid: Application to the Screening of γ-Aminobutyric Acid-Producing Lactic Acid Bacteria. Anal. Chim. Acta 2005, 540, 293–297. [Google Scholar] [CrossRef]
- Valenzuela, J.A.; Flórez, A.B.; Vázquez, L.; Vasek, O.M.; Mayo, B. Production of γ-Aminobutyric Acid (GABA) by Lactic Acid Bacteria Strains Isolated from Traditional, Starter-Free Dairy Products Made of Raw Milk. Benef. Microbes 2019, 10, 579–587. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Colton, T.; Everitt, B.; Piegorsch, W.; Ruggeri, F. Wiley StatsRef: Statistics Reference Online, 1st ed.; Balakrishnan, N., Colton, T., Everitt, B., Piegorsch, W., Ruggeri, F., Teugels, J.L., Eds.; Wiley: New York, NY, USA, 2014; ISBN 978-1-118-44511-2. [Google Scholar]
- Gradilla-Hernández, M.S.; de Anda, J.; Garcia-Gonzalez, A.; Meza-Rodríguez, D.; Yebra Montes, C.; Perfecto-Avalos, Y. Multivariate Water Quality Analysis of Lake Cajititlán, Mexico. Environ. Monit. Assess. 2020, 192, 5. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Everitt, B.; Landau, S.; Leese, M.; Stahl, D. Cluster Analysis, 5th ed.; Everitt, B., Ed.; Wiley series in probability and statistics; Wiley: Chichester, West Sussex, UK, 2011; ISBN 978-0-470-74991-3. [Google Scholar]
- Kim, E.; Cho, E.-J.; Yang, S.-M.; Kim, M.-J.; Kim, H.-Y. Novel Approaches for the Identification of Microbial Communities in Kimchi: MALDI-TOF MS Analysis and High-Throughput Sequencing. Food Microbiol. 2021, 94, 103641. [Google Scholar] [CrossRef] [PubMed]
- Gantzias, C.; Lappa, I.K.; Aerts, M.; Georgalaki, M.; Manolopoulou, E.; Papadimitriou, K.; De Brandt, E.; Tsakalidou, E.; Vandamme, P. MALDI-TOF MS Profiling of Non-Starter Lactic Acid Bacteria from Artisanal Cheeses of the Greek Island of Naxos. Int. J. Food Microbiol. 2020, 323, 108586. [Google Scholar] [CrossRef] [PubMed]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Dlamini, Z.C.; Langa, R.L.S.; Aiyegoro, O.A.; Okoh, A.I. Safety Evaluation and Colonisation Abilities of Four Lactic Acid Bacteria as Future Probiotics. Probiotics Antimicrob. Proteins 2019, 11, 397–402. [Google Scholar] [CrossRef]
- Gad, G.F.M.; Abdel-Hamid, A.M.; Farag, Z.S.H. Antibiotic Resistance in Lactic Acid Bacteria Isolated from Some Pharmaceutical and Dairy Products. Braz. J. Microbiol. 2014, 45, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huddleston, J.R. Horizontal Gene Transfer in the Human Gastrointestinal Tract: Potential Spread of Antibiotic Resistance Genes. Infect. Drug Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerner, A.; Matthias, T.; Aminov, R. Potential Effects of Horizontal Gene Exchange in the Human Gut. Front. Immunol. 2017, 8, 1630. [Google Scholar] [CrossRef] [Green Version]
- Obioha, P.I.; Ouoba, L.I.I.; Anyogu, A.; Awamaria, B.; Atchia, S.; Ojimelukwe, P.C.; Sutherland, J.P.; Ghoddusi, H.B. Identification and Characterisation of the Lactic Acid Bacteria Associated with the Traditional Fermentation of Dairy Fermented Product. Braz. J. Microbiol. 2021, 52, 869–881. [Google Scholar] [CrossRef]
- Hu, C.; Ren, L.; Zhou, Y.; Ye, B. Characterization of Antimicrobial Activity of Three Lactobacillus Plantarum Strains Isolated from Chinese Traditional Dairy Food. Food Sci. Nutr. 2019, 7, 1997–2005. [Google Scholar] [CrossRef] [Green Version]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus Plantarum Strains as a Bio-Control Strategy against Food-Borne Pathogenic Microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef] [Green Version]
- Talib, N.; Mohamad, N.E.; Yeap, S.K.; Hussin, Y.; Aziz, M.N.M.; Masarudin, M.J.; Sharifuddin, S.A.; Hui, Y.W.; Ho, C.L.; Alitheen, N.B. Isolation and Characterization of Lactobacillus Spp. from Kefir Samples in Malaysia. Molecules 2019, 24, 2606. [Google Scholar] [CrossRef] [Green Version]
- Escobar-Ramírez, M.C.; Jaimez-Ordaz, J.; Escorza-Iglesias, V.A.; Rodríguez-Serrano, G.M.; Contreras-López, E.; Ramírez-Godínez, J.; Castañeda-Ovando, A.; Morales-Estrada, A.I.; Felix-Reyes, N.; González-Olivares, L.G. Lactobacillus Pentosus ABHEAU-05: An in Vitro Digestion Resistant Lactic Acid Bacterium Isolated from a Traditional Fermented Mexican Beverage. Rev. Argent. Microbiol. 2020, 52, 305–314. [Google Scholar] [CrossRef]
- Hurtado-Romero, A.; Del Toro-Barbosa, M.; Garcia-Amezquita, L.E.; García-Cayuela, T. Innovative Technologies for the Production of Food Ingredients with Prebiotic Potential: Modifications, Applications, and Validation Methods. Trends Food Sci. Technol. 2020, 104, 117–131. [Google Scholar] [CrossRef]
- Chatterjee, E.; GA Manuel, S. Effect of Fruit Pectin on Growth of Lactic Acid Bacteria. J. Probiotics Health 2016, 4, 1–4. [Google Scholar] [CrossRef]
- Figueroa-GonzáLez, I.; RodríGuez-Serrano, G.; GóMez-Ruiz, L.; GarcíA-Garibay, M.; Cruz-Guerrero, A. Prebiotic Effect of Commercial Saccharides on Probiotic Bacteria Isolated from Commercial Products. Food Sci. Technol. 2019, 39, 747–753. [Google Scholar] [CrossRef] [Green Version]
- De Souza Oliveira, R.P.; Rodrigues Florence, A.C.; Perego, P.; De Oliveira, M.N.; Converti, A. Use of Lactulose as Prebiotic and Its Influence on the Growth, Acidification Profile and Viable Counts of Different Probiotics in Fermented Skim Milk. Int. J. Food Microbiol. 2011, 145, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Ayala Monter, M.A.; Hernández Sánchez, D.; Pinto Ruiz, R.; González Muñoz, S.S.; Bárcena Gama, J.R.; Hernández Mendo, O.; Torres Salado, N. Efecto Prebiótico de Dos Fuentes de Inulina En El Crecimiento in Vitro de Lactobacillus Salivarius y Enterococcus Faecium. Rev. Mex. Cienc. Pecu. 2018, 9, 346. [Google Scholar] [CrossRef]
- Trapala, J.; Bustos-Jaimes, I.; Manzanares, P.; Bárzana, E.; Montiel, C. Purification and Characterization of an Inulinase Produced by a Kluyveromyces Marxianus Strain Isolated from Blue Agave Bagasse. Protein Expr. Purif. 2020, 176, 105718. [Google Scholar] [CrossRef]
- García Gamboa, R.; Ortiz Basurto, R.I.; Calderón Santoyo, M.; Bravo Madrigal, J.; Ruiz Álvarez, B.E.; González Avila, M. In Vitro Evaluation of Prebiotic Activity, Pathogen Inhibition and Enzymatic Metabolism of Intestinal Bacteria in the Presence of Fructans Extracted from Agave: A Comparison Based on Polymerization Degree. LWT 2018, 92, 380–387. [Google Scholar] [CrossRef]
- Redruello, B.; Saidi, Y.; Sampedro, L.; Ladero, V.; del Rio, B.; Alvarez, M.A. GABA-Producing Lactococcus Lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese. Foods 2021, 10, 633. [Google Scholar] [CrossRef]
- Bhanwar, S.; Bamnia, M.; Ghosh, M.; Ganguli, A. Use of Lactococcus Lactis to Enrich Sourdough Bread with γ-Aminobutyric Acid. Int. J. Food Sci. Nutr. 2013, 64, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Perpetuini, G.; Tittarelli, F.; Battistelli, N.; Suzzi, G.; Tofalo, R. Γ-aminobutyric Acid Production by Kluyveromyces Marxianus Strains. J. Appl. Microbiol. 2020, 129, 1609–1619. [Google Scholar] [CrossRef]
- Demirbaş, F.; İspirli, H.; Kurnaz, A.A.; Yilmaz, M.T.; Dertli, E. Antimicrobial and Functional Properties of Lactic Acid Bacteria Isolated from Sourdoughs. LWT—Food Sci. Technol. 2017, 79, 361–366. [Google Scholar] [CrossRef]
- Gómez de Cadiñanos, L.P.; García-Cayuela, T.; Martínez-Cuesta, M.C.; Peláez, C.; Requena, T. Expression of Amino Acid Converting Enzymes and Production of Volatile Compounds by Lactococcus Lactis IFPL953. Int. Dairy J. 2019, 96, 29–35. [Google Scholar] [CrossRef]
- Öztürkoğlu Budak, S.; Akal, H.C. Microbial Cultures and Enzymes in Dairy Technology; Öztürkoğlu Budak, Ş., Akal, H.C., Eds.; Advances in Medical Technologies and Clinical Practice; IGI Global: Ankara, Turkey, 2018; ISBN 978-1-5225-5363-2. [Google Scholar]
| MALDI-TOF Result | Assigned Code | MALDI-TOF Score | Reliability |
|---|---|---|---|
| Lactococcus lactis | BIOTEC006 | 2.143 | Genus-level, probable species |
| Lactococcus lactis | BIOTEC007 | 2.446 | Genus-level, species level |
| Lactococcus lactis | BIOTEC008 | 2.400 | Genus-level, species level |
| Kluyveromyces lactis | BIOTEC009 | 2.102 | Genus-level, probable species |
| Kluyveromyces lactis | BIOTEC010 | 1.937 | Probable genus |
| Leuconostoc pseudomesenteroides | BIOTEC011 | 1.821 | Probable genus |
| Leuconostoc pseudomesenteroides | BIOTEC012 | 1.885 | Probable genus |
| Lentilactobacilluskefiri | BIOTEC013 | 2.006 | Genus-level, probable species |
| Lentilactobacillus kefiri | BIOTEC014 | 2.226 | Genus-level, probable species |
| Lentilactobacillus parakefiri | BIOTEC015 | 2.151 | Genus-level, probable species |
| Lactococcus lactis | BIOTEC016 | 1.861 | Probable genus |
| Penicillium commune | BIOTEC017 | 2.366 | Genus-level, species level |
| Microorganisms b | Auto-Aggregation (%) | |||
|---|---|---|---|---|
| 2 h | 6 h | 20 h | 24 h | |
| L. acidophilus La3 (−) | 4.36 ± 0.29 cd | 10.29 ± 0.10 de | 56.54 ± 0.12 c | 72.21 ± 0.53 c |
| L. rhamnosus GG (−) | 3.26 ± 0.29 f | 8.17 ± 0.19 ef | 35.17 ± 1.76 e | 53.05 ± 0.62 ef |
| L. plantarum 299V (−) | 3.54 ± 0.35 def | 8.00 ± 0.21 f | 35.86 ± 3.51 e | 46.98 ± 3.12 fg |
| L. lactis BIOTEC006 (−) | 2.97 ± 0.03 f | 12.48 ± 0.44 c | 30.85 ± 1.06 ef | 38.87 ± 2.52 gh |
| L. lactis BIOTEC007 (−) | 10.64 ± 0.15 a | 31.78 ± 1.08 b | 77.00 ± 1.94 b | 84.31 ± 1.28 b |
| L. lactis BIOTEC008 (−) | 10.09 ± 0.00 a | 32.24 ± 0.95 b | 85.93 ± 0.85 a | 94.33 ± 0.52 a |
| K. lactis BIOTEC009 (+) | 4.14 ± 0.30 de | 9.50 ± 0.54 def | 28.46 ± 0.41 f | 46.01 ± 0.38 h |
| K. lactis BIOTEC010 (+) | 5.12 ± 0.19 bc | 10.80 ± 0.38 cd | 28.82 ± 0.53 f | 39.00 ± 3.77 gh |
| L. pseudomesenteroides BIOTEC011 (+) | 3.43 ± 0.29 ef | 7.48 ± 0.46 f | 26.00 ± 0.20 f | 33.99 ± 1.42 h |
| L. pseudomesenteroides BIOTEC012 (+) | 3.50 ± 0.12 ef | 9.27 ± 0.13 def | 26.96 ± 0.28 f | 33.92 ± 0.29 h |
| L. kefiri BIOTEC013 (+) | 5.33 ± 0.17 b | 9.05 ± 0.43 def | 27.67 ± 0.29 f | 32.60 ± 3.12 h |
| L. kefiri BIOTEC014 (+) | 10.53 ± 0.07 a | 34.73 ± 0.42 a | 57.81 ± 0.04 c | 59.87 ± 0.03 de |
| L. parakefiri BIOTEC015 (−) | 2.80 ± 0.01 f | 10.44 ± 0.68 cd | 50.14 ± 2.73 d | 66.10 ± 0.65 cd |
| Microorganisms | Co-Aggregation with Pathogens (%) | ||
|---|---|---|---|
| E. coli ATCC-25922 | S. typhi BIOTEC019 | S. aureus ATCC-BAA-42 | |
| L. acidophilus La3 | 68.90 ± 1.15 c | 63.82 ± 0.07 e | 73.84 ± 1.35 efg |
| L. rhamnosus GG | 58.05 ± 0.38 de | 63.48 ± 1.13 ef | 70.83 ± 0.02 fg |
| L. plantarum 299V | 62.26 ± 0.86 cde | 53.76 ± 0.22 g | 76.55 ± 0.72 def |
| L. lactis BIOTEC006 | 83.99 ± 1.77 b | 86.41 ± 1.09 a | 84.35 ± 0.15 cd |
| L. lactis BIOTEC007 | 85.57 ± 0.81 b | 77.74 ± 0.62 bc | 86.01 ± 0.67 de |
| L. lactis BIOTEC008 | 85.24 ± 0.74 b | 82.55 ± 1.92 ab | 83.49 ± 0.50 bcd |
| K. lactis BIOTEC009 | 83.64 ± 0.46 b | 63.57 ± 0.11 ef | 83.18 ± 0.94 abc |
| K. lactis BIOTEC010 | 62.94 ± 1.30 cde | 58.60 ± 0.15 fg | 57.63 ± 0.42 h |
| L. pseudomesenteroides BIOTEC011 | 68.58 ± 1.73 cd | 62.58 ± 0.19 ef | 71.27 ± 0.01 efg |
| L. pseudomesenteroides BIOTEC012 | 63.22 ± 0.48 e | 75.62 ± 0.33 cd | 72.45 ± 0.53 g |
| L. kefiri BIOTEC013 | 57.01 ± 0.57 e | 70.64 ± 2.41 d | 72.87 ± 0.57 fg |
| L. kefiri BIOTEC014 | 98.13 ± 0.29 a | 73.12 ± 2.92 cd | 88.38 ± 0.61 a |
| L. parakefiri BIOTEC015 | 98.76 ± 0.22 a | 84.70 ± 1.03 a | 79.79 ± 0.19 ab |
| Microorganisms | Antibiotics | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | CF | CFX | DC | CPF | GE | CLM | E | STX | PE | VA | TE | |
| L. acidophilus La3 | + | + + | + + + | + + + | + + + | + + + | − | − | + | + + + | + + + | + |
| L. rhamnosus GG | + + + | + + + | + + + | + + + | + + | + + + | + | − | + + + | + + | + + + | + + + |
| L. plantarum 299V | + + | + + + | + + + | + + + | + + | + + + | + | − | + + + | + + | + + + | + + |
| L. lactis BIOTEC006 | − | − | − | − | − | − | − | − | − | − | − | − |
| L. lactis BIOTEC007 | − | − | − | − | − | + + + | − | + + + | − | − | − | − |
| L. lactis BIOTEC008 | − | − | − | + + | + + | + + | + + + | − | − | + + + | + | − |
| K. lactis BIOTEC009 | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + |
| K. lactis BIOTEC010 | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + |
| L. pseudomesenteroides BIOTEC011 | + + | − | − | + + | − | − | + + + | − | + + | − | + + + | − |
| L. pseudomesenteroides BIOTEC012 | + + | − | − | + + | − | − | + + + | − | + + + | − | + + | − |
| L. kefiri BIOTEC013 | + | − | + | + + + | − | − | + | − | + + + | + | − | − |
| L. kefiri BIOTEC014 | − | − | − | − | + + + | − | − | − | − | + | + + | + + + |
| L. parakefiri BIOTEC015 | + + + | + + + | + + | + + + | + + | + + + | + | − | + + + | + + + | + + + | + |
| Microorganisms | Media Growth Inhibition Halos (in mm) | ||
|---|---|---|---|
| E. coli ATCC-25922 | S. typhi BIOTEC019 | S. aureus ATCC-BAA-42 | |
| L. acidophilus La3 | 18.33 ± 1.53 a | 16.67 ± 0.58 ab | 14.67 ± 0.58 b |
| L. rhamnosus GG | 18.67 ± 1.53 a | 17.67 ± 1.53 a | 14.67 ± 0.58 a |
| L. plantarum 299V | 19.00 ± 1.00 a | 18.33 ± 0.58 a | 15.33 ± 0.58 b |
| L. lactis BIOTEC006 | ND | ND | ND |
| L. lactis BIOTEC007 | ND | ND | ND |
| L. lactis BIOTEC008 | ND | ND | ND |
| K. lactis BIOTEC009 | 14.67 ± 2.08 a | 12.00 ± 1.00 a | 15.33 ± 0.58 a |
| K. lactis BIOTEC010 | 14.33 ± 1.15 a | 12.00 ± 0.00 b | 15.33 ± 0.58 a |
| L. pseudomesenteroides BIOTEC011 | 13.00± 1.00 b | 12.00 ± 0.00 b | 15.33 ± 0.58 a |
| L. pseudomesenteroides BIOTEC012 | 15.00 ± 1.00 a | 13.00 ± 0.00 b | 16.33 ± 0.58 a |
| L. kefiri BIOTEC013 | 17.00 ± 1.15 a | 14.67 ± 0.58 b | 16.00 ± 1.00 ab |
| L. kefiri BIOTEC014 | 16.33 ± 1.15 a | 14.67 ± 0.58 a | 16.33 ± 0.58 a |
| L. parakefiri BIOTEC015 | 16.00 ± 0.58 ab | 14.33 ± 0.58 b | 15.33 ± 0.58 a |
| Microorganisms | LOG (CFU/mL) | |||
|---|---|---|---|---|
| Initial Phase | Oral Phase | Gastric Phase | Intestinal Phase | |
| L. acidophilus La3 | 9.41 ± 0.01 b | 9.62 ± 0.01 a | 8.88 ± 0.02 c | 9.15 ± 0.01 d |
| L. rhamnosus GG | 9.59 ± 0.16 a | 9.54 ± 0.09 a | 8.56 ± 0.03 b | 8.77 ± 0.08 b |
| L. plantarum 299V | 9.55 ± 0.07 b | 9.79 ± 0.02 a | 8.60 ± 0.02 d | 8.85 ± 0.02 c |
| L. lactis BIOTEC006 | 8.39 ± 0.12 a | 8.38 ± 0.12 a | 4.84 ± 0.08 b | 4.75 ± 0.21 b |
| L. lactis BIOTEC007 | 8.00 ± 0.00 b | 8.30 ± 0.00 a | 6.95 ± 0.07 c | 6.86 ± 0.05 c |
| L. lactis BIOTEC008 | 9.35 ± 0.49 a | 8.65 ± 0.07 a | 5.45 ± 0.21 b | 5.22 ± 0.21 b |
| K. lactis BIOTEC009 | 8.74 ± 0.06 a | 8.94 ± 0.14 a | 6.75 ± 0.21 b | 6.60 ± 0.00 b |
| K. lactis BIOTEC010 | 8.96 ± 0.05 a | 9.09 ± 0.02 a | 6.99 ± 0.12 b | 6.75 ± 0.21 b |
| L. pseudomesenteroides BIOTEC011 | 9.24 ± 0.09 a | 9.05 ± 0.08 a | 6.78 ± 0.17 b | 6.95 ± 0.04 b |
| L. pseudomesenteroides BIOTEC012 | 8.80 ± 0.02 a | 9.02 ± 0.03 a | 6.90 ± 0.00 b | 6.99 ± 0.12 b |
| L. kefiri BIOTEC013 | 9.09 ± 0.12 a | 9.15 ± 0.21 a | 6.58 ± 0.00 b | 6.56 ± 0.17 b |
| L. kefiri BIOTEC014 | 8.09 ± 0.04 a | 7.84 ± 0.00 b | 6.46 ± 0.06 c | 6.50 ± 0.00 c |
| L. parakefiri BIOTEC015 | 8.09 ± 0.05 a | 7.54 ± 0.01 a | 5.86 ± 0.07 b | 5.60 ± 0.01 c |
| Microorganisms | Glucose | Lactulose | Agave Inulin | Citric Pectin | |
|---|---|---|---|---|---|
| L. acidophilus La3 | ODmax | 1.78 ± 0.01 Bb | 1.87 ± 0.00 Aa | 1.14 ± 0.00 Db | 1.27± 0.04 Cb |
| μmax | 0.21 ± 0.00 Bcd | 0.19 ± 0.00 Ca | 0.22 ± 0.00 Aa | 0.09 ± 0.00 De | |
| lag | 1.68 ± 0.10 Cij | 2.04 ± 0.02 Bg | 1.01 ± 0.03 Df | 16.76 ± 0.07 Ac | |
| L. rhamnosus GG | ODmax | 1.87 ± 0.02 Aa | 1.87 ± 0.01 Aa | 1.10 ± 0.01 Cc | 1.38 ± 0.02 Ba |
| μmax | 0.21 ± 0.00 Bbcd | 0.18 ± 0.00 Cb | 0.25 ± 0.00 Aa | 0.17 ± 0.00 Dc | |
| lag | 3.01 ± 0.09 Ah | 3.22 ± 0.01 Af | 1.25 ± 0.04 Cef | 2.46 ± 0.39 Bf | |
| L. plantarum 299v | ODmax | 1.81 ± 0.00 Ab | 1.84 ± 0.01 Aa | 1.25 ± 0.01 Ba | 1.26 ± 0.03 Bb |
| μmax | 0.19 ± 0.00 Ae | 0.19 ± 0.00 Aab | 0.23 ± 0.05 Aa | 0.18 ± 0.00 Ab | |
| lag | 1.26 ± 0.08 Aj | 2.03 ± 0.03 Ag | 1.55 ± 0.01 Ae | 1.80 ± 0.63 Af | |
| L. lactis BIOTEC006 | ODmax | 0.67 ± 0.03 Bf | 0.89 ± 0.01 Agh | 0.45 ± 0.00 Ch | 0.11 ± 0.02 Dh |
| μmax | 0.08 ± 0.03 Bh | 0.07 ± 0.01 Cf | 0.10 ± 0.00 Abc | 0.00 ± 0.02 Dg | |
| lag | 11.87 ± 0.03 Bb | 6.64 ± 0.01 Cc | 3.87 ± 0.00 Cc | 38.90 ± 0.02 Aa | |
| L. lactis BIOTEC007 | ODmax | 0.35 ± 0.01 Cg | 0.76 ± 0.01 Ai | 0.39 ± 0.01 Bi | 0.15 ± 0.00 Dh |
| μmax | 0.01 ± 0.00 Ci | 0.03 ± 0.00 Ah | 0.02 ± 0.00 Bd | 0.00 ± 0.00 Dg | |
| lag | 13.10 ± 0.11 Da | 11.02 ± 0.07 Ab | 10.74 ± 0.02 Bb | 32.04 ± 0.71 Cb | |
| L. lactis BIOTEC008 | ODmax | 0.74 ± 0.00 Be | 0.83 ± 0.00 Ahi | 0.46 ± 0.00 Ch | 0.10 ± 0.01 Dh |
| μmax | 0.11 ± 0.00 Ag | 0.06 ± 0.00 Cf | 0.09 ± 0.00 Bc | 0.00 ± 0.00 Dg | |
| lag | 7.03 ± 0.05 Bd | 5.76 ± 0.02 Cd | 3.20 ± 0.01 Dd | 39.57 ± 0.78 Aa | |
| K. lactis BIOTEC009 | ODmax | 1.01 ± 0.01 Ad | 0.96 ± 0.02 Bfg | 0.55 ± 0.01 Dg | 0.88 ± 0.02 Cde |
| μmax | 0.08 ± 0.00 Ch | 0.12 ± 0.00 Bde | 0.11 ± 0.01 Bbv | 0.19 ± 0.00 Aa | |
| lag | 9.57 ± 0.30 Bc | 1.37 ± 0.15 Ci | 1.16 ± 0.08 Cf | 12.59 ± 0.16 Ade | |
| K. lactis BIOTEC010 | ODmax | 1.28 ± 0.01 Ac | 1.07 ± 0.00 Bd | 0.69 ± 0.00 Df | 0.87 ± 0.01 Cde |
| μmax | 0.14 ± 0.01 Bf | 0.12 ± 0.00 Cd | 0.10 ± 0.00 Dbc | 0.17 ± 0.00 Abc | |
| lag | 5.02 ± 0.25 Bf | 1.59 ± 0.08 Chi | 0.00 ± 0.00 Dg | 12.23 ± 0.10 Ae | |
| L. pseudomesenteroides BIOTEC011 | ODmax | 1.86 ± 0.00 Aa | 0.99 ± 0.01 Bef | 0.53 ± 0.00 Dg | 0.82 ± 0.02 Ce |
| μmax | 0.22 ± 0.00 Abcd | 0.11 ± 0.00 De | 0.13 ± 0.00 Cb | 0.16 ± 0.00 Bd | |
| lag | 1.99 ± 0.02 Bi | 1.81 ± 0.05 Cgh | 1.29 ± 0.01 Def | 12.78 ± 0.10 Ade | |
| L. pseudomesenteroides BIOTEC012 | ODmax | 1.81 ± 0.00 Ab | 1.03 ± 0.08 Bde | 0.54 ± 0.01 Cg | 0.94 ± 0.02 Bc |
| μmax | 0.22 ± 0.00 Abc | 0.12 ± 0.02 Cd | 0.11 ± 0.00 Cbc | 0.19 ± 0.01 Ba | |
| lag | 5.60 ± 0.05 Be | 1.55 ± 0.29 Chi | 1.30 ± 0.01 Cef | 12.08 ± 0.21 Ae | |
| L. kefiri BIOTEC013 | ODmax | 1.87 ± 0.02 Aa | 1.70 ± 0.01 Bb | 1.12 ± 0.01 Cbc | 0.90 ± 0.02 Dcd |
| μmax | 0.21 ± 0.01 Ade | 0.13 ± 0.00 Cd | 0.08 ± 0.00 Dc | 0.16 ± 0.00 Bd | |
| lag | 5.50 ± 0.32 De | 17.27 ± 0.15 Aa | 14.81 ± 0.40 Ba | 12.17 ± 0.12 Ce | |
| L. kefiri BIOTEC014 | ODmax | 1.88 ± 0.01 Aa | 1.23 ± 0.02 Bc | 0.79 ± 0.00 Ce | 0.23 ± 0.00 Dg |
| μmax | 0.24 ± 0.00 Aa | 0.14 ± 0.00 Bc | 0.08 ± 0.01 Cc | 0.02 ± 0.00 Df | |
| lag | 4.96 ± 0.00 Af | 4.29 ± 0.06 Be | 1.54 ± 0.00 Ce | 4.13 ± 0.31 Bf | |
| L. parakefiri BIOTEC015 | ODmax | 1.88 ± 0.04 Aa | 1.64 ± 0.02 Bb | 0.95 ± 0.04 Cd | 0.35 ± 0.01 Df |
| μmax | 0.22 ± 0.00 Ab | 0.04 ± 0.00 Bg | 0.03 ± 0.00 Cd | 0.01 ± 0.00 Dg | |
| lag | 4.42 ± 0.11 Bg | 0.00 ± 0.00 Cj | 0.00 ± 0.00 Cg | 14.84 ± 1.23 Acd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hurtado-Romero, A.; Del Toro-Barbosa, M.; Gradilla-Hernández, M.S.; Garcia-Amezquita, L.E.; García-Cayuela, T. Probiotic Properties, Prebiotic Fermentability, and GABA-Producing Capacity of Microorganisms Isolated from Mexican Milk Kefir Grains: A Clustering Evaluation for Functional Dairy Food Applications. Foods 2021, 10, 2275. https://doi.org/10.3390/foods10102275
Hurtado-Romero A, Del Toro-Barbosa M, Gradilla-Hernández MS, Garcia-Amezquita LE, García-Cayuela T. Probiotic Properties, Prebiotic Fermentability, and GABA-Producing Capacity of Microorganisms Isolated from Mexican Milk Kefir Grains: A Clustering Evaluation for Functional Dairy Food Applications. Foods. 2021; 10(10):2275. https://doi.org/10.3390/foods10102275
Chicago/Turabian StyleHurtado-Romero, Alejandra, Mariano Del Toro-Barbosa, Misael Sebastián Gradilla-Hernández, Luis Eduardo Garcia-Amezquita, and Tomás García-Cayuela. 2021. "Probiotic Properties, Prebiotic Fermentability, and GABA-Producing Capacity of Microorganisms Isolated from Mexican Milk Kefir Grains: A Clustering Evaluation for Functional Dairy Food Applications" Foods 10, no. 10: 2275. https://doi.org/10.3390/foods10102275








