Novel Bradykinin-Potentiating Peptides and Three-Finger Toxins from Viper Venom: Combined NGS Venom Gland Transcriptomics and Quantitative Venom Proteomics of the Azemiops feae Viper
Abstract
:1. Introduction
2. Experimental Section
2.1. Specimen Collection and Tissue Preparation
2.2. cDNA Library Preparation and Sequencing
2.2.1. cDNA Synthesis and cDNA Amplification
2.2.2. Illumina Sequencing
2.3. NGS Data Analysis
2.4. Other Computational Tool
2.5. BioProject and Raw Sequence Data
2.6. Reduction, Alkylation and Digestion of the Proteins
2.7. Tryptic Peptides Desalting
2.8. Liquid Chromatography and Mass Spectrometry
2.9. Data Analysis
2.10. Characterization of Bradykinin-Potentiating Peptides by MALDI Mass Spectrometry
2.11. Peptide Synthesis
2.12. Blood Pressure Measurements
3. Results
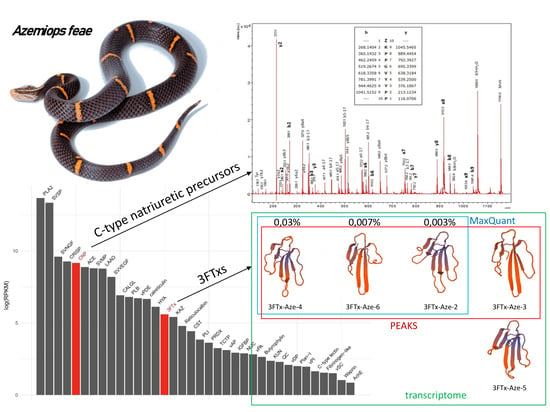
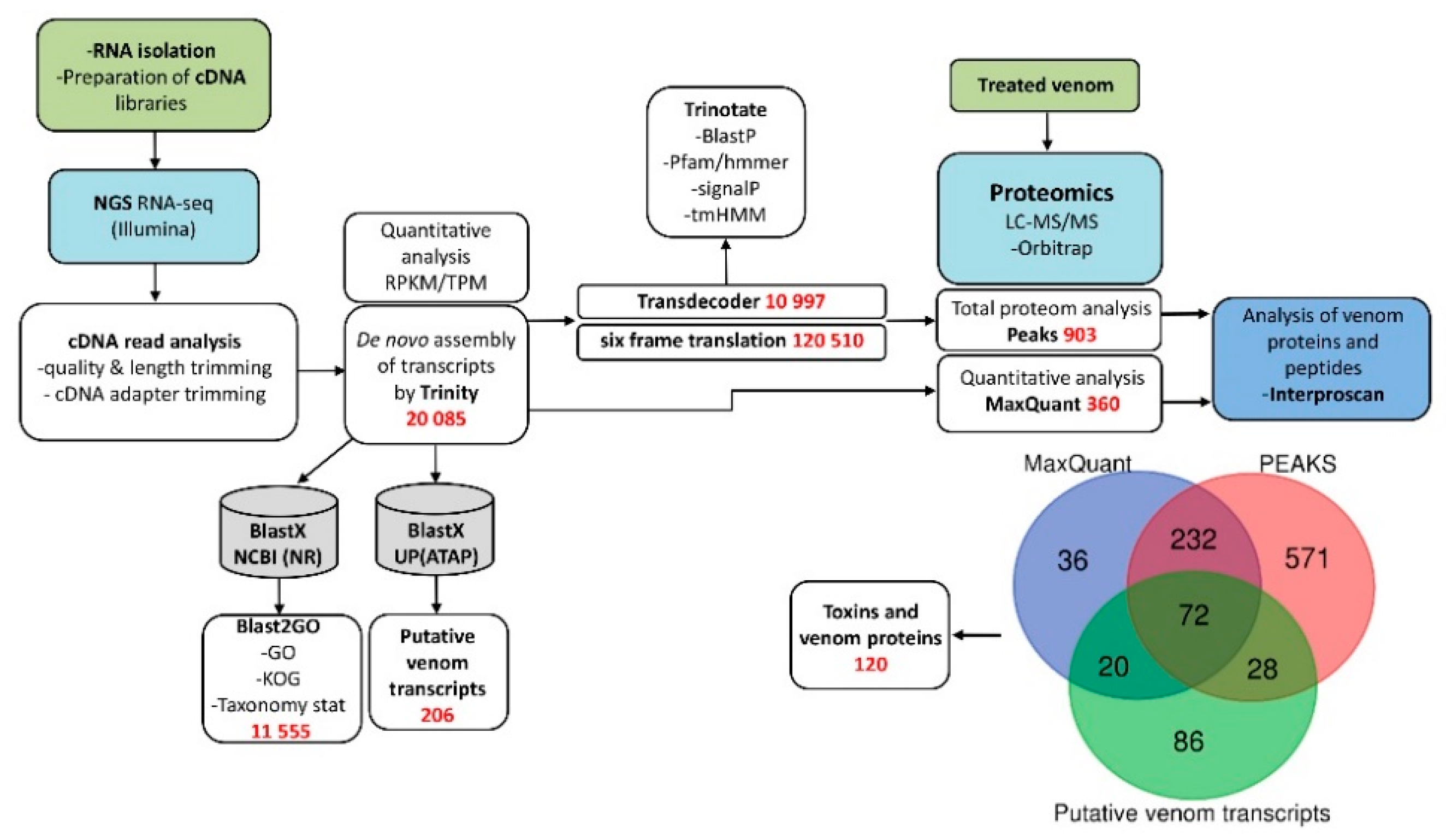
3.1. Research Outline: RNA Sequencing and Analysis of the Initial Data
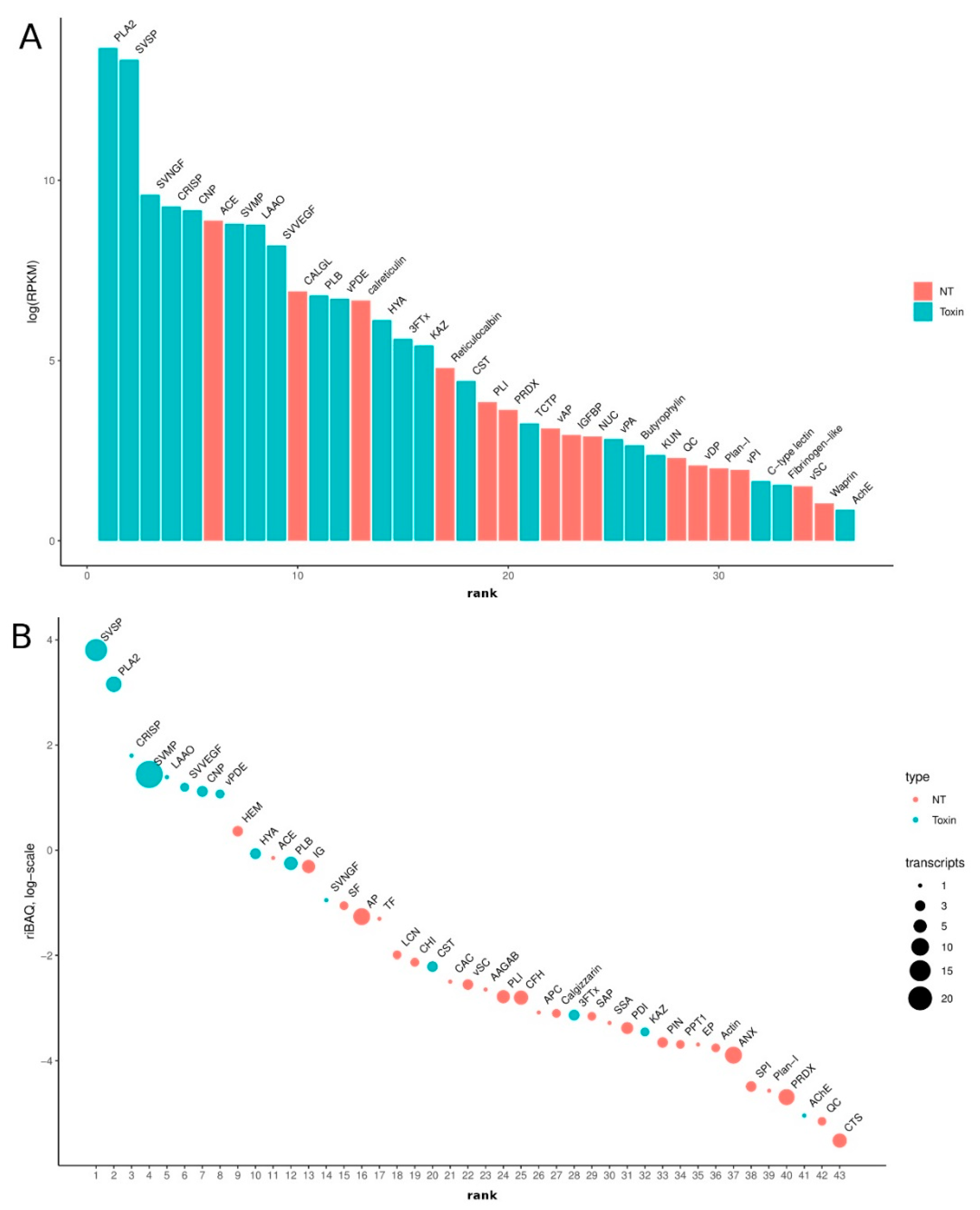
3.2. Venom-Gland Transcriptome
3.3. Venom Proteomics/Peptidomics
3.3.1. Serine Proteases
3.3.2. Phospholipases A2
3.3.3. Cysteine-Rich Seceretory Proteins
3.3.4. L-Amino Acid Oxidase
3.3.5. Snake Venom Metalloproteinase
3.3.6. Snake Venom Vascular Endothelial Growth Factor
3.3.7. Three-Finger Toxins
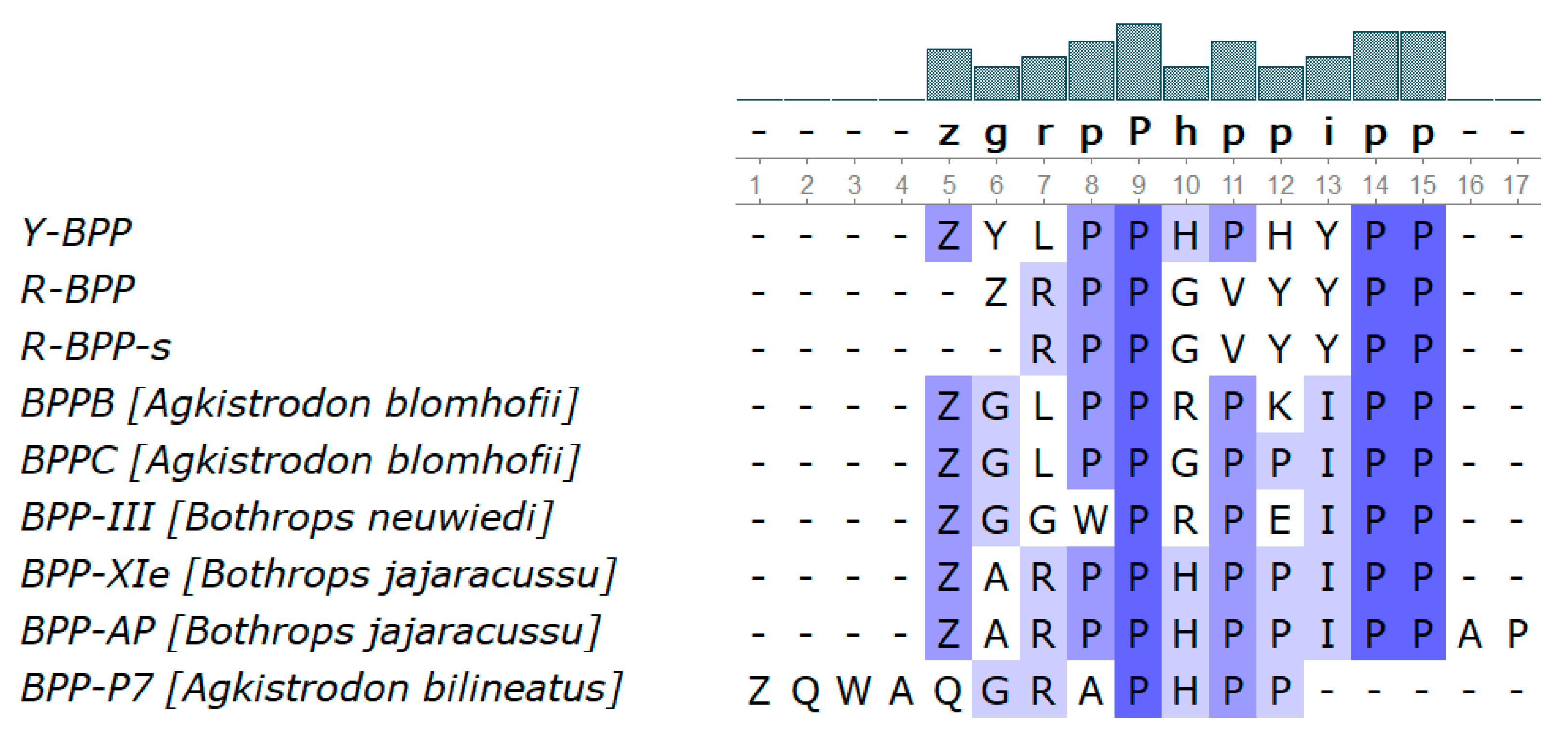
3.3.8. Bradykinin-Potentiating Peptides and Azemiopsin
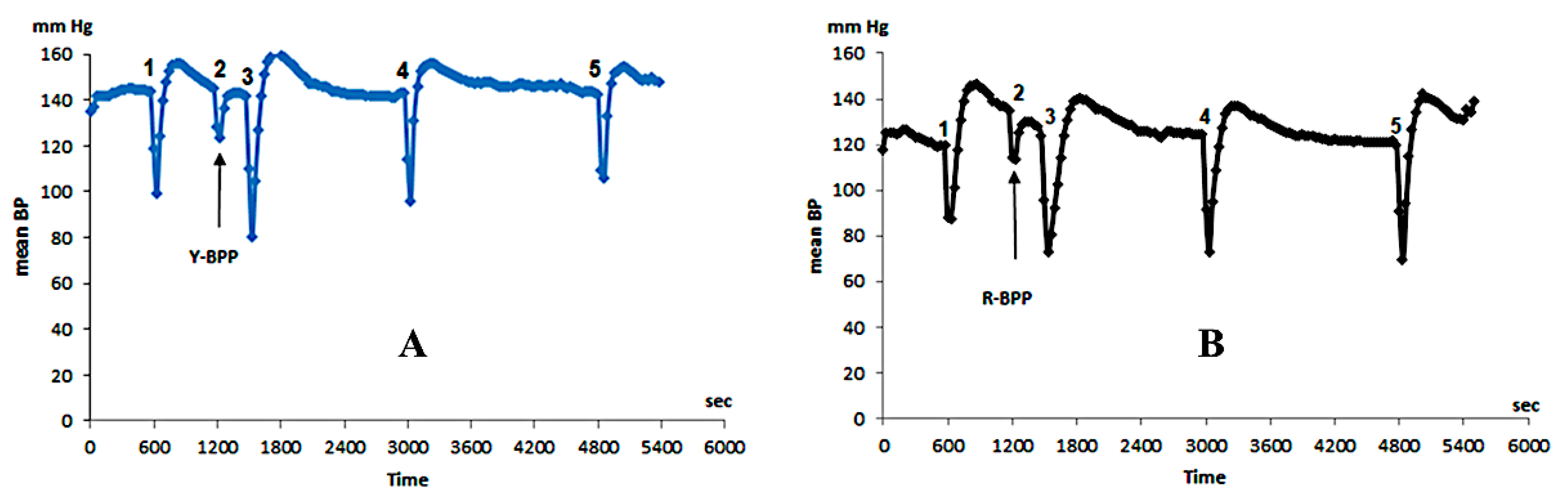
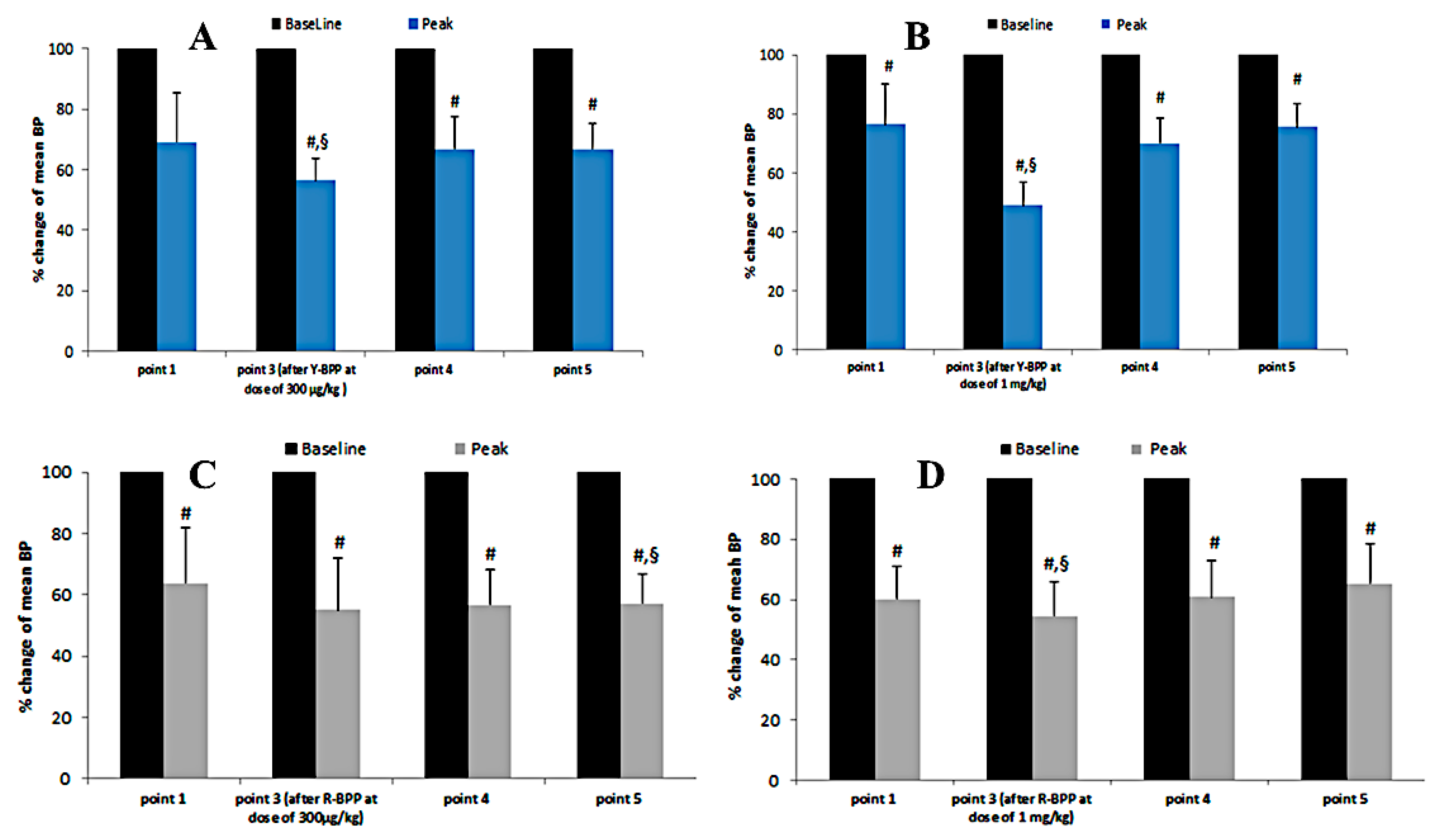
3.3.9. Biological Activity of New Bradykinin-Potentiating Peptides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef]
- Ziganshin, R.H.; Kovalchuk, S.I.; Arapidi, G.P.; Starkov, V.G.; Hoang, A.N.; Thi Nguyen, T.T.; Nguyen, K.C.; Shoibonov, B.B.; Tsetlin, V.I.; Utkin, Y.N.; et al. Quantitative proteomic analysis of vietnamese krait venoms: Neurotoxins are the major components in bungarus multicinctus and phospholipases A2 in Bungarus fasciatus. Toxicon 2015, 107, 197–209. [Google Scholar] [CrossRef]
- Calvete, J.J. Snake venomics-from low-resolution toxin-pattern recognition to toxin-resolved venom proteomes with absolute quantification. Expert Rev. Proteom. 2018, 15, 555–568. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Wuster, W.; Peppin, L.; Pook, C.E.; Walker, D.E. A nesting of vipers: Phylogeny and historical biogeography of the Viperidae Squamata: Serpentes. Mol. Phylogenet. Evol. 2008, 49, 445–459. [Google Scholar] [CrossRef]
- Alencar, L.R.V.; Quental, T.B.; Grazziotin, F.G.; Alfaro, M.L.; Martins, M.; Venzon, M.; Zaher, H. Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 2016, 105, 50–62. [Google Scholar] [CrossRef]
- Orlov, N.L.; Ryabov, S.A.; Nguyen, T.T. On the taxonomy and the distribution of snakes of the genus azemiops boulenger, 1888: Description of a New Species. Russ. J. Herpetol. 2013, 20, 110–128. [Google Scholar]
- Li, J.N.; Liang, D.; Wang, Y.Y.; Guo, P.; Huang, S.; Zhang, P. A large-scale systematic framework of Chinese snakes based on a unified multilocus marker system. Mol. Phylogenet. Evol. 2020, 148, 106807. [Google Scholar] [CrossRef]
- Mebs, D.; Kuch, U.; Meier, J. Studies on venom and venom apparatus of Fea’s viper, Azemiops feae. Toxicon 1994, 32, 1275–1278. [Google Scholar] [CrossRef]
- Debono, J.; Bos, M.H.A.; Coimbra, F.; Ge, L.; Frank, N.; Kwok, H.F.; Fry, B.G. Basal but divergent: Clinical implications of differential coagulotoxicity in a clade of Asian vipers. Toxicol. In Vitro Int. J. Publ. Assoc. BIBRA 2019, 58, 195–206. [Google Scholar] [CrossRef]
- Fry, B.G.; Wuster, W.; Ryan Ramjan, S.F.; Jackson, T.; Martelli, P.; Kini, R.M. Analysis of colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun. Mass Spectrom. RCM 2003, 17, 2047–2062. [Google Scholar] [CrossRef]
- Tsai, I.H.; Wang, Y.M.; Huang, K.F. Structures of azemiops feae venom phospholipases and cys-rich-secretory protein and implications for taxonomy and toxinology. Toxicon 2016, 114, 31–39. [Google Scholar] [CrossRef]
- Brust, A.; Sunagar, K.; Undheim, E.A.; Vetter, I.; Yang, D.C.; Casewell, N.R.; Jackson, T.N.; Koludarov, I.; Alewood, P.F.; Hodgson, W.C.; et al. Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol. Cell. Proteom. MCP 2013, 12, 651–663. [Google Scholar] [CrossRef] [Green Version]
- Sunagar, K.; Jackson, T.N.; Undheim, E.A.; Ali, S.A.; Antunes, A.; Fry, B.G. Three-fingered RAVERs: Rapid accumulation of variations in exposed residues of snake venom toxins. Toxins 2013, 5, 2172–2208. [Google Scholar] [CrossRef] [Green Version]
- Utkin, Y.N.; Weise, C.; Kasheverov, I.E.; Andreeva, T.V.; Kryukova, E.V.; Zhmak, M.N.; Starkov, V.G.; Hoang, N.A.; Bertrand, D.; Ramerstorfer, J.; et al. Azemiopsin from azemiops feae viper venom, a novel polypeptide ligand of nicotinic acetylcholine receptor. J. Biol. Chem. 2012, 287, 27079–27086. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.Y.; Machleder, E.M.; Chenchik, A.; Li, R.; Siebert, P.D. Reverse transcriptase template switching: A SMART approach for full-length cDNA library construction. Bio. Tech. 2001, 30, 892–897. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Method. 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. RCM 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteom. Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Method. 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Shin, J.B.; Krey, J.F.; Hassan, A.; Metlagel, Z.; Tauscher, A.N.; Pagana, J.M.; Sherman, N.E.; Jeffery, E.D.; Spinelli, K.J.; Zhao, H.; et al. Molecular architecture of the chick vestibular hair bundle. Nat. Neurosci. 2013, 16, 365–374. [Google Scholar] [CrossRef] [Green Version]
- Suckau, D.; Resemann, A.; Schürenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A novel MALDI-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef]
- Shelukhina, I.V.; Zhmak, M.N.; Lobanov, A.V.; Ivanov, I.A.; Garifulina, A.I.; Kravchenko, I.N.; Rasskazova, E.A.; Salmova, M.A.; Tukhovskaya, E.A.; Rykov, V.A.; et al. Azemiopsin, a selective peptide antagonist of muscle nicotinic acetylcholine receptor: Preclinical evaluation as a local muscle relaxant. Toxins 2018, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Fry, B.G.; Vidal, N.; Norman, J.A.; Vonk, F.J.; Scheib, H.; Ramjan, S.F.; Kuruppu, S.; Fung, K.; Hedges, S.B.; Richardson, M.K.; et al. Early evolution of the venom system in lizards and snakes. Nature 2006, 439, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Sousa, L.F.; Nicolau, C.A.; Peixoto, P.S.; Bernardoni, J.L.; Oliveira, S.S.; Portes-Junior, J.A.; Mourao, R.H.; Lima-dos-Santos, I.; Sano-Martins, I.S.; Chalkidis, H.M.; et al. Comparison of phylogeny, venom composition and neutralization by antivenom in diverse species of bothrops complex. PLoS Negl. Trop. Dis. 2013, 7, e2442. [Google Scholar] [CrossRef] [Green Version]
- Nirthanan, S.; Gopalakrishnakone, P.; Gwee, M.C.; Khoo, H.E.; Kini, R.M. Non-conventional toxins from elapid venoms. Toxicon 2003, 41, 397–407. [Google Scholar] [CrossRef]
- Utkin, Y.; Sunagar, K.; Jackson, T.N.W.; Reeks, T.; Fry, B.G. Three-finger toxins (3FTxs). In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 215–227. [Google Scholar]
- Makarova, Y.V.; Kryukova, E.V.; Shelukhina, I.V.; Lebedev, D.S.; Andreeva, T.V.; Ryazantsev, D.Y.; Balandin, S.V.; Ovchinnikova, T.V.; Tsetlin, V.I.; Utkin, Y.N.; et al. The first recombinant viper three-finger toxins: Inhibition of muscle and neuronal nicotinic acetylcholine receptors. Doklad. Biochem. Biophys. 2018, 479, 127–130. [Google Scholar] [CrossRef]
- Weinstein, S.A.; Schmidt, J.J.; Bernheimer, A.W.; Smith, L.A. Characterization and amino acid sequences of two lethal peptides isolated from venom of Wagler’s pit viper, Trimeresurus wagleri. Toxicon 1991, 29, 227–236. [Google Scholar] [CrossRef]
- Zainal Abidin, S.A.; Rajadurai, P.; Chowdhury, M.E.; Ahmad Rusmili, M.R.; Othman, I.; Naidu, R. Proteomic characterization and comparison of malaysian tropidolaemus wagleri and cryptelytrops purpureomaculatus cenom using shotgun-proteomics. Toxins 2016, 8, 299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.H.; Tan, K.Y.; Yap, M.K.; Tan, N.H. Venomics of Tropidolaemus wagleri, the sexually dimorphic temple pit viper: Unveiling a deeply conserved atypical toxin arsenal. Sci. Rep. 2017, 7, 43237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenta, J.; Stach, Z.; Stourac, P.; Kadanka, Z.; Michalek, P. Neurological symptoms following the Fea’s viper (Azemiops feae) bite. Clin. Toxicol. 2015, 53, 1150–1151. [Google Scholar] [CrossRef]
- Harris, R.J.; Zdenek, C.N.; Debono, J.; Harrich, D.; Fry, B.G. Evolutionary interpretations of nicotinic acetylcholine receptor targeting venom effects by a clade of asian Viperidae snakes. Neurotox. Res. 2020, 38, 312–318. [Google Scholar] [CrossRef]
- Fry, B.G.; Wuster, W.; Kini, R.M.; Brusic, V.; Khan, A.; Venkataraman, D.; Rooney, A.P. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evolut. 2003, 57, 110–129. [Google Scholar] [CrossRef] [Green Version]
- Thakur, R.; Chattopadhyay, P.; Mukherjee, A.K. Biochemical and pharmacological characterization of a toxic fraction and its cytotoxin-like component isolated from Russell’s viper (Daboia russelii russelii) venom. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2015, 168, 55–65. [Google Scholar] [CrossRef]
- Nirthanan, S.; Charpantier, E.; Gopalakrishnakone, P.; Gwee, M.C.; Khoo, H.E.; Cheah, L.S.; Bertrand, D.; Kini, R.M. Candoxin, a novel toxin from Bungarus candidus, is a reversible antagonist of muscle (alphabetagammadelta) but a poorly reversible antagonist of neuronal α 7 nicotinic acetylcholine receptors. J. Biol. Chem. 2002, 277, 17811–17820. [Google Scholar] [CrossRef] [Green Version]
- Utkin, Y.N.; Kukhtina, V.V.; Kryukova, E.V.; Chiodini, F.; Bertrand, D.; Methfessel, C.; Tsetlin, V.I. Weak toxin from Naja kaouthia is a nontoxic antagonist of alpha 7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001, 276, 15810–15815. [Google Scholar] [CrossRef] [Green Version]
- Lyukmanova, E.N.; Shenkarev, Z.O.; Shulepko, M.A.; Paramonov, A.S.; Chugunov, A.O.; Janickova, H.; Dolejsi, E.; Dolezal, V.; Utkin, Y.N.; Tsetlin, V.I.; et al. Structural insight into specificity of interactions between nonconventional three-finger weak toxin from Naja kaouthia (WTX) and muscarinic acetylcholine receptors. J. Biol. Chem. 2015, 290, 23616–23630. [Google Scholar] [CrossRef] [Green Version]
- Mordvintsev, D.Y.; Polyak, Y.L.; Rodionov, D.I.; Jakubik, J.; Dolezal, V.; Karlsson, E.; Tsetlin, V.I.; Utkin, Y.N. Weak toxin WTX from Naja kaouthia cobra venom interacts with both nicotinic and muscarinic acetylcholine receptors. FEBS J. 2009, 276, 5065–5075. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, S.C.; Favreau, P.; Cheneval, O.; Laing, G.D.; Wilkinson, M.C.; Miller, R.L.; Stocklin, R.; Harrison, R.A. Molecular characterisation of endogenous snake venom metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2008, 365, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Yee, K.T.; Pitts, M.; Tongyoo, P.; Rojnuckarin, P.; Wilkinson, M.C. Snake venom metalloproteinases and their peptide inhibitors from myanmar Russell’s viper venom. Toxins 2016, 9, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tashima, A.K.; Zelanis, A.; Kitano, E.S.; Ianzer, D.; Melo, R.L.; Rioli, V.; Sant’anna, S.S.; Schenberg, A.C.; Camargo, A.C.; Serrano, S.M.; et al. Peptidomics of three bothrops snake venoms: Insights into the molecular diversification of proteomes and peptidomes. Mol. Cell. Proteomic. MCP 2012, 11, 1245–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babenko, V.V.; Ziganshin, R.H.; Weise, C.; Dyachenko, I.; Shaykhutdinova, E.; Murashev, A.N.; Zhmak, M.; Starkov, V.; Hoang, A.N.; Tsetlin, V.; et al. Novel Bradykinin-Potentiating Peptides and Three-Finger Toxins from Viper Venom: Combined NGS Venom Gland Transcriptomics and Quantitative Venom Proteomics of the Azemiops feae Viper. Biomedicines 2020, 8, 249. https://doi.org/10.3390/biomedicines8080249
Babenko VV, Ziganshin RH, Weise C, Dyachenko I, Shaykhutdinova E, Murashev AN, Zhmak M, Starkov V, Hoang AN, Tsetlin V, et al. Novel Bradykinin-Potentiating Peptides and Three-Finger Toxins from Viper Venom: Combined NGS Venom Gland Transcriptomics and Quantitative Venom Proteomics of the Azemiops feae Viper. Biomedicines. 2020; 8(8):249. https://doi.org/10.3390/biomedicines8080249
Chicago/Turabian StyleBabenko, Vladislav V., Rustam H. Ziganshin, Christoph Weise, Igor Dyachenko, Elvira Shaykhutdinova, Arkady N. Murashev, Maxim Zhmak, Vladislav Starkov, Anh Ngoc Hoang, Victor Tsetlin, and et al. 2020. "Novel Bradykinin-Potentiating Peptides and Three-Finger Toxins from Viper Venom: Combined NGS Venom Gland Transcriptomics and Quantitative Venom Proteomics of the Azemiops feae Viper" Biomedicines 8, no. 8: 249. https://doi.org/10.3390/biomedicines8080249