SARS-CoV-2: Origin, Intermediate Host and Allergenicity Features and Hypotheses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Evolutionary Analysis
2.2. Key Sites Analysis
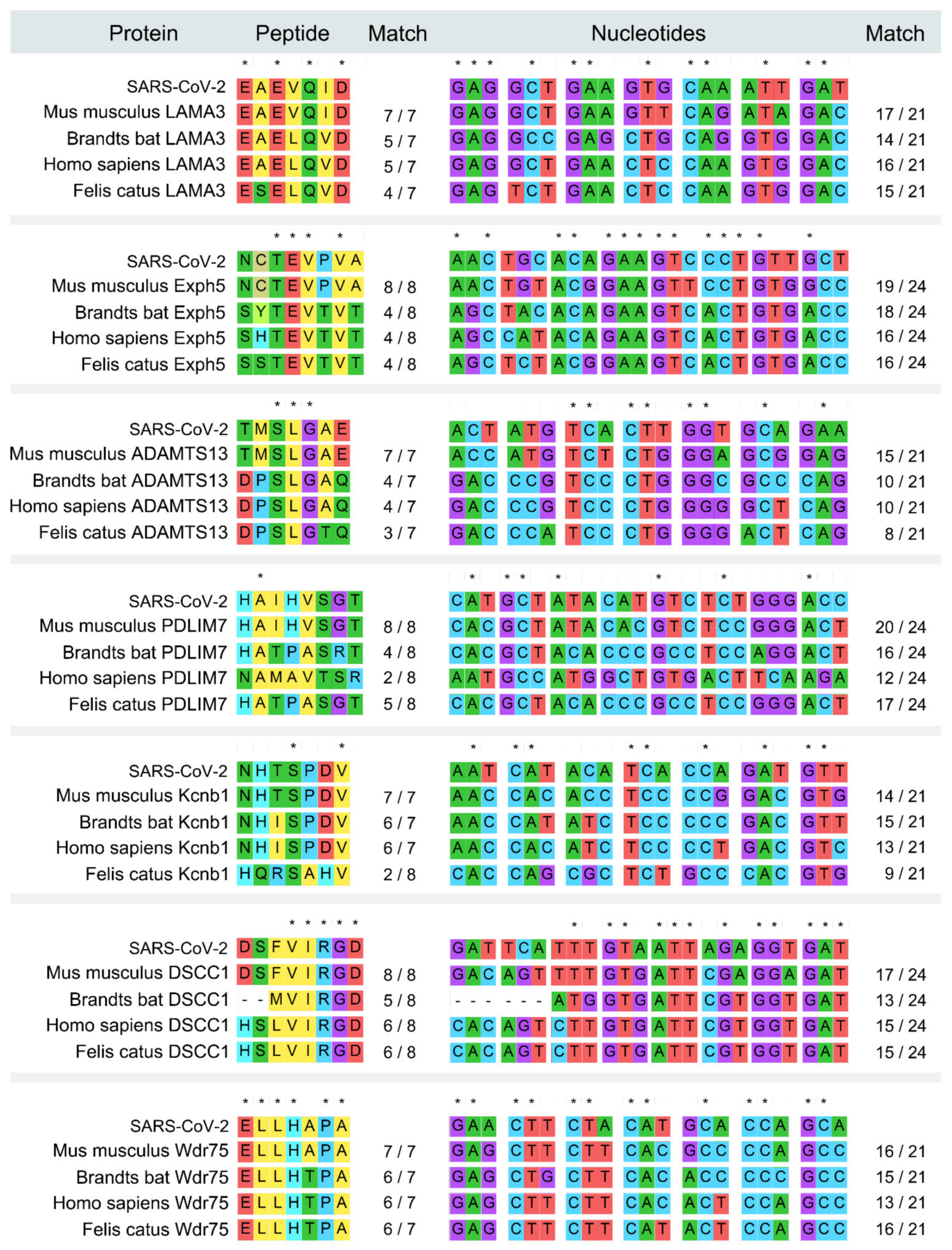
2.3. Mouse Derived Peptide Analysis
2.4. Allergenicity Assessment and Infection Population Estimating
2.5. Meteorological Parameters Analysis
3. Results
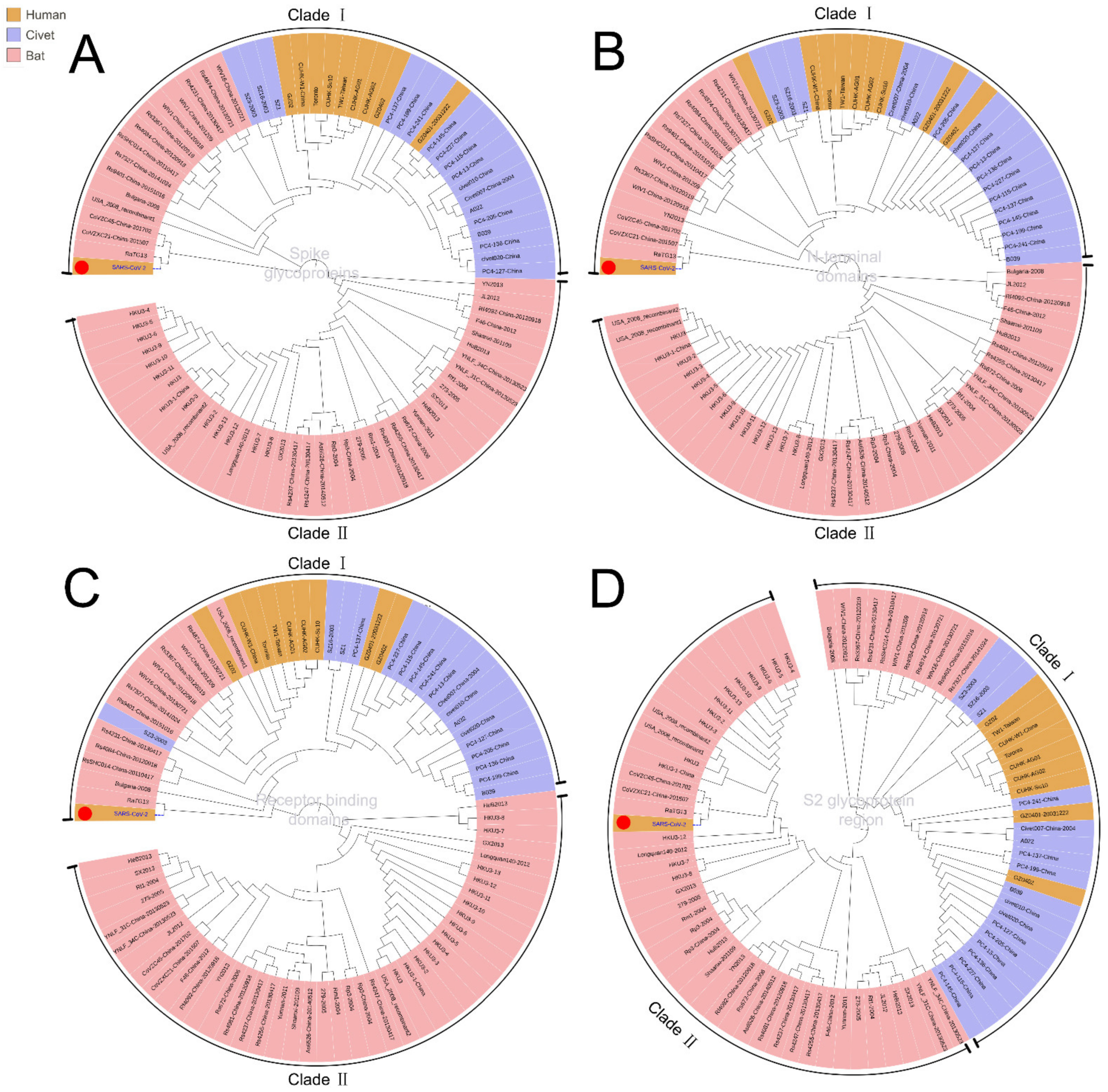
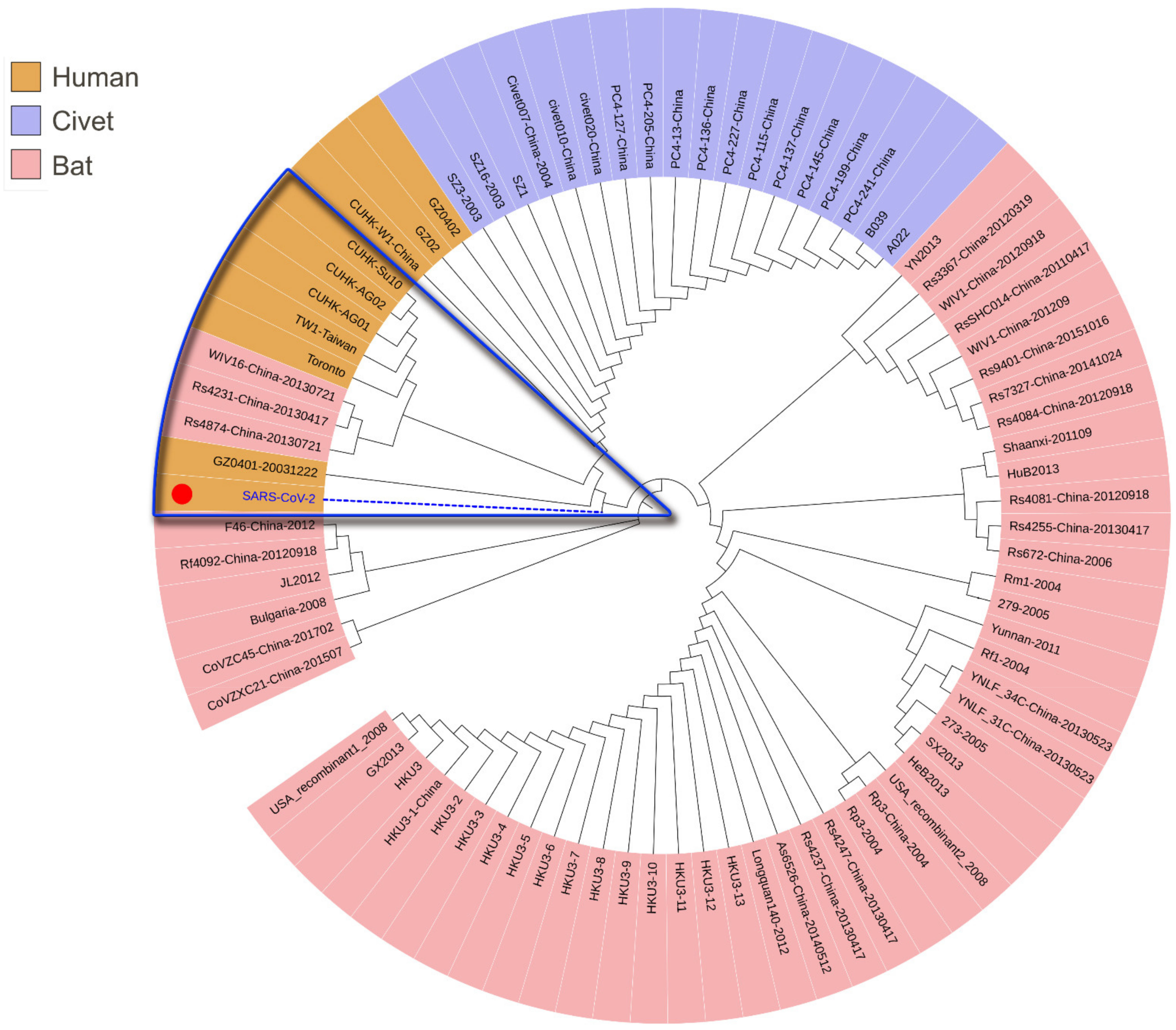
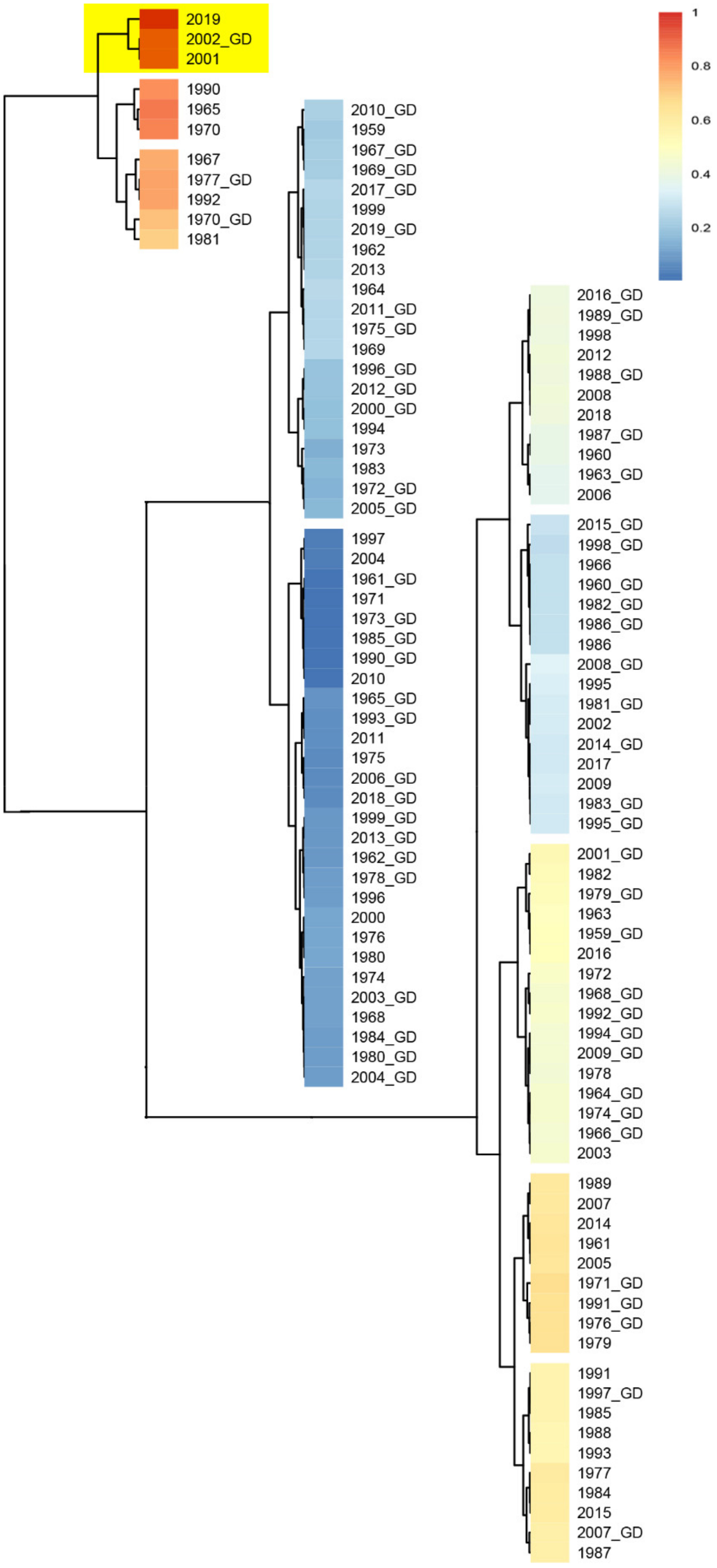
3.1. Phylogenetic Analysis of SARS-CoV-2
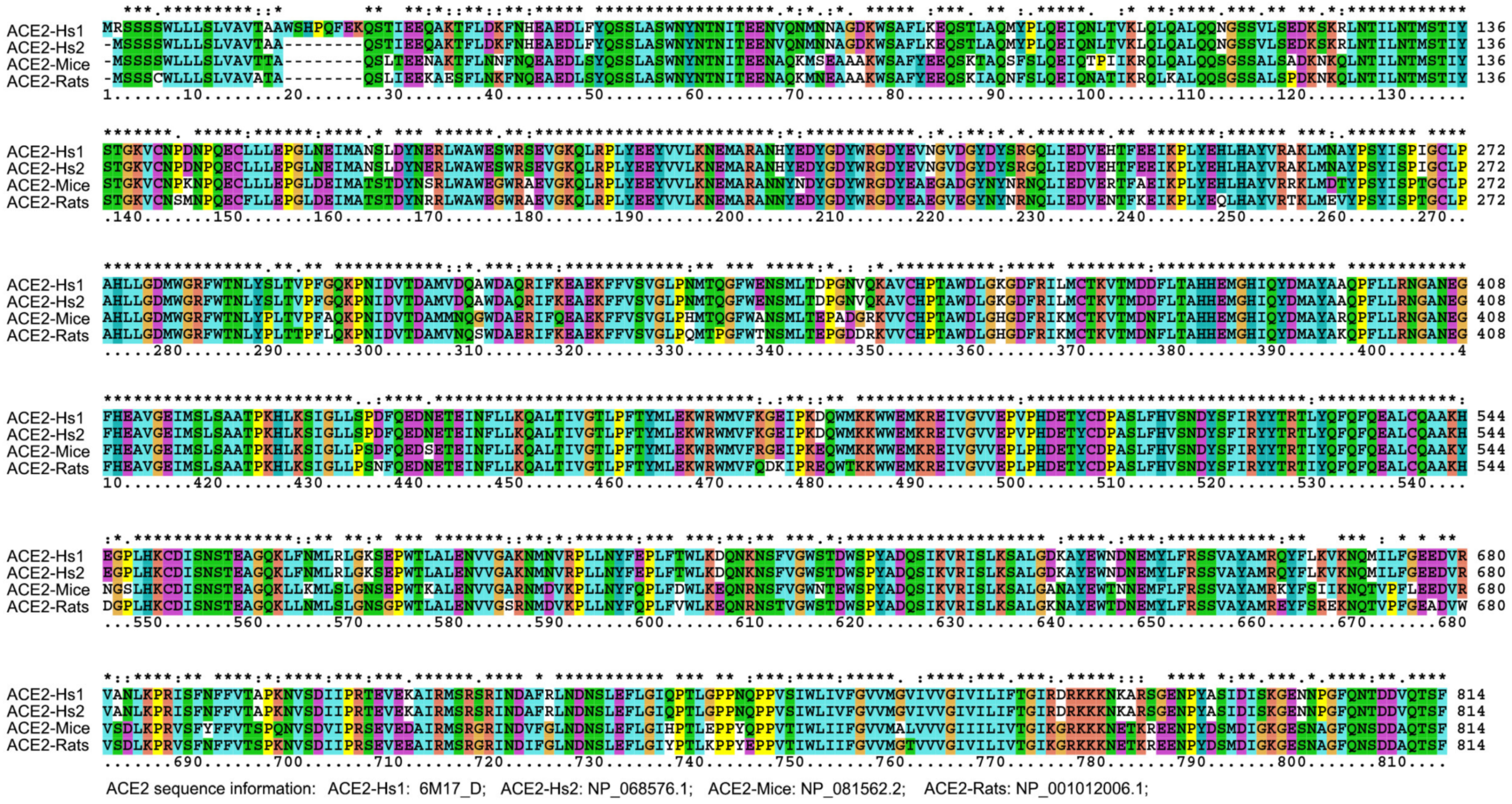
3.2. Intermediate Host Analysis
3.3. Analysis of Virus-HLA Binding Affinity for Allergenicity Assessment
3.4. Analysis of Meteorological Factors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Miłek, J.; Blicharz-Domańska, K. Coronaviruses in avian species—Review with focus on epidemiology and diagnosis in wild birds. J. Vet. Res. 2018, 62, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Li, M.; Wang, C.; Wang, P.; Fang, Z.; Tan, J.; Wu, S.; Xiao, Y.; Zhu, H. Host and infectivity prediction of Wuhan 2019 novel coronavirus using deep learning algorithm. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef]
- Lam, T.T.-Y.; Jia, N.; Zhang, Y.-W.; Shum, M.H.-H.; Jiang, J.-F.; Zhu, H.-C.; Tong, Y.-G.; Shi, Y.-X.; Ni, X.-B.; Liao, Y.-S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 9, 2013–2019. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streicker, D.G.; Lemey, P.; Velasco-Villa, A.; Rupprecht, C.E. Rates of Viral Evolution Are Linked to Host Geography in Bat Rabies. PLoS Pathog. 2012, 8, e1002720. [Google Scholar] [CrossRef] [Green Version]
- Matson, M.J.; Yinda, C.K.; Seifert, S.N.; Bushmaker, T.; Fischer, R.J.; Van Doremalen, N.; Lloyd-Smith, J.O.; Munster, V.J. Effect of Environmental Conditions on SARS-CoV-2 Stability in Human Nasal Mucus and Sputum. Emerg. Infect. Dis. 2020, 26, 2276–2278. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Rao, Y.; Sun, Q.; Wu, X.; Jin, J.; Bi, Y.; Chen, J.; Lei, F.; Liu, Q.; Duan, Z.; et al. Identification of climate factors related to human infection with avian influenza A H7N9 and H5N1 viruses in China. Sci. Rep. 2015, 5, 18094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Tao, A. Antigenicity, Immunogenicity, Allergenicity. In Allergy Bioinformatics; Tao, A., Raz, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 8, pp. 175–186. [Google Scholar]
- Knierman, M.D.; Lannan, M.B.; Spindler, L.J.; McMillian, C.L.; Konrad, R.J.; Siegel, R.W. The human leukocyte antigen class II immunopeptidome of the SARS-CoV-2 spike glycoprotein. Cell Rep. 2020, 33, 108454. [Google Scholar] [CrossRef] [PubMed]
- Takagi, A.; Matsui, M. Identification of HLA-A* 02:01-Restricted Candidate Epitopes Derived from the Nonstructural Polyprotein 1a of SARS-CoV-2 That May Be Natural Targets of CD8 + T Cell Recognition In Vivo. J. Virol. 2021, 95. [Google Scholar] [CrossRef] [PubMed]
- Sinigaglia, F.; Guttinger, M.; Romagnoli, P.; Takacs, B. Malaria antigens and MHC restriction. Immunol. Lett. 1990, 25, 265–270. [Google Scholar] [CrossRef]
- Kazansky, D.B. MHC restriction and allogeneic immune responses. J. Immunotoxicol. 2008, 5, 369–384. [Google Scholar] [CrossRef]
- Sakuraba, A.; Haider, H.; Sato, T. Population Difference in Allele Frequency of HLA-C * 05 and Its Correlation with COVID-19 Mortality. Viruses 2020, 12, 1333. [Google Scholar] [CrossRef]
- Matzinger, P. The Danger Model: A Renewed Sense of Self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef] [Green Version]
- Jahrling, P.B.; Hensley, L.; Martinez, M.J.; LeDuc, J.W.; Rubins, K.H.; Relman, D.; Huggins, J.W. From The Cover: Exploring the potential of variola virus infection of cynomolgus macaques as a model for human smallpox. Proc. Natl. Acad. Sci. USA 2004, 101, 15196–15200. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Liu, X.; Huang, Y.; Zou, Z.; Chen, H.; Lai, H.; Zhang, L.; Wu, Q.; Zhang, J.; Wang, S.; et al. Reduction of the Number of Major Representative Allergens: From Clinical Testing to 3-Dimensional Structures. Mediat. Inflamm. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Asero, R.; Monsalve, R.; Barber, D. Profilin sensitization detected in the office by skin prick test: A study of prevalence and clinical relevance of profilin as a plant food allergen. Clin. Exp. Allergy 2008, 38, 1033–1037. [Google Scholar] [CrossRef]
- Asero, R.; Tripodi, S.; Dondi, A.; Businco, A.D.R.; Sfika, I.; Bianchi, A.; Candelotti, P.; Caffarelli, C.; Dascola, C.P.; Ricci, G.; et al. Prevalence and Clinical Relevance of IgE Sensitization to Profilin in Childhood: A Multicenter Study. Int. Arch. Allergy Immunol. 2015, 168, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, M.; Nielsen, M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 2015, 32, 511–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, M.; Lundegaard, C.; Worning, P.; Lauemøller, S.L.; Lamberth, K.; Buus, S.; Brunak, S.; Lund, O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003, 12, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef]
- González-Galarza, F.F.; Takeshita, L.Y.; Santos, E.J.; Kempson, F.; Maia, M.H.T.; Da Silva, A.L.S.; Silva, A.L.T.; Ghattaoraya, G.; Alfirevic, A.; Jones, A.; et al. Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2014, 43, D784–D788. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, Y.; Wang, Y.; Zhou, W.; Yan, Y.; Zhu, C.; Zhang, X.; Sun, H.; Ji, W. Association of meteorological factors with childhood viral acute respiratory infections in subtropical China: An analysis over 11 years. Arch. Virol. 2013, 159, 631–639. [Google Scholar] [CrossRef]
- Morin, C.W.; Stoner-Duncan, B.; Winker, K.; Scotch, M.; Hess, J.J.; Meschke, J.S.; Ebi, K.L.; Rabinowitz, P.M. Avian influenza virus ecology and evolution through a climatic lens. Environ. Int. 2018, 119, 241–249. [Google Scholar] [CrossRef]
- Mirsaeidi, M.; Motahari, H.; Khamesi, M.T.; Sharifi, A.; Campos, M.; Schraufnagel, D.E. Climate Change and Respiratory Infections. Ann. Am. Thorac. Soc. 2016, 13, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.A.; Yañez-Arenas, C.; Roberts, B.J.; Chen, C.; Plowright, R.K.; Webb, R.J.; Skerratt, L. Climatic suitability influences species specific abundance patterns of Australian flying foxes and risk of Hendra virus spillover. One Health 2016, 2, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Tamerius, J.D.; Shaman, J.; Alonso, W.; Bloom-Feshbach, K.; Uejio, C.K.; Comrie, A.; Viboud, C. Environmental Predictors of Seasonal Influenza Epidemics across Temperate and Tropical Climates. PLoS Pathog. 2013, 9, e1003194. [Google Scholar] [CrossRef]
- Sloan, C.; Moore, M.L.; Hartert, T. Impact of Pollution, Climate, and Sociodemographic Factors on Spatiotemporal Dynamics of Seasonal Respiratory Viruses. Clin. Transl. Sci. 2011, 4, 48–54. [Google Scholar] [CrossRef]
- Tarek, F.; Hassou, N.; Benchekroun, M.N.; Boughribil, S.; Hafid, J.; Bessi, H.; Ennaji, M.M. Impact of rotavirus and hepatitis A virus by worldwide climatic changes during the period between 2000 and 2013. Bioinformation 2019, 15, 194–200. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-C.; Gu, S.H.; Kim, W.-K.; Klein, T.A.; Lee, S.-H.; No, J.S.; Chong, S.-T.; Song, J.-W. Urban Rodent Surveillance, Climatic Association, and Genomic Characterization of Seoul Virus Collected at U.S. Army Garrison, Seoul, Republic of Korea, 2006–2010. Am. J. Trop. Med. Hyg. 2018, 99, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Mining coronavirus genomes for clues to the outbreak’s origins. Science 2020, 31. [Google Scholar] [CrossRef]
- König, B. Chapter 2.11—The Behaviour of the House Mouse. In The Laboratory Mouse, 2nd ed.; Hedrich, H.J., Ed.; Elsvier: Amsterdam, The Netherlands, 2012; pp. 367–381. [Google Scholar] [CrossRef]
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.; Leung, W.S.; So, R.T.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H.; et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020, 31, 107725. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.-R.; Lin, L.-F.; Duan, J.-H.; Wu, J.; Pei, F.-Q.; Lu, W.-C.; Cai, S.-W.; Zheng, H.-Y.; Yin, W.-X. Analysis on murine-like animals carrying SARS coronavirus in the hygienic units related to SARS. Chin. J. Vector Biol. Control. 2004, 15, 7–9, (In Chinese with English Abstract). [Google Scholar]
- Ng, S.K.C. Possible role of an animal vector in the SARS outbreak at Amoy Gardens. Lancet 2003, 362, 570–572. [Google Scholar] [CrossRef]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [CrossRef]
- Yuhki, N.; Beck, T.; Stephens, R.M.; Nishigaki, Y.; Newmann, K.; O’Brien, S.J. Comparative Genome Organization of Human, Murine, and Feline MHC Class II Region. Genome Res. 2003, 13, 1169–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzinger, P. The evolution of the danger theory. Expert Rev. Clin. Immunol. 2012, 8, 311–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braciale, T.J.; Morrison, L.A.; Sweetser, M.T.; Sambrook, J.; Gething, M.-J.; Braciale, V.L. Antigen Presentation Pathways to Class I and Class II MHC-Restricted T Lymphocytes. Immunol. Rev. 1987, 98, 95–114. [Google Scholar] [CrossRef]
- Parham, P. Pictures of MHC restriction. Nature 1996, 384, 109–110. [Google Scholar] [CrossRef]
- Hamed, C.T.; Meiloud, G.; Veten, F.; Hadrami, M.; Ghaber, S.M.; Boussaty, E.C.; Habti, N.; Houmeida, A. HLA class I (-A, -B, -C) and class II (-DR, -DQ) polymorphism in the Mauritanian population. BMC Med. Genet. 2018, 19, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance 2020, 25, 2000180. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, L.; Hao, X.; Guo, H.; Wang, Q.; Huang, J.; He, N.; Yu, H.; Lin, X.; Pan, A.; et al. Evolving epidemiology and impact of non-pharmaceutical interventions on the outbreak of coronavirus disease 2019 in Wuhan, China. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Mo, X.; Hu, Y.; Qi, X.; Jiang, F.; Jiang, Z.; Tong, S. Epidemiology of COVID-19 Among Children in China. Pediatrics 2020, 145, e20200702. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.-Y.; Li, J.; Yang, X.-L.; Chmura, A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Aalberse, R.C. Structural biology of allergens. J. Allergy Clin. Immunol. 2000, 106, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Wentworth, D.; Gillim-Ross, L.; Espina, N.; Bernard, K.A. Mice Susceptible to SARS Coronavirus. Emerg. Infect. Dis. 2004, 10, 1293–1296. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal. Transduct. Target. Ther. 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Lai, M.M. Recombination in large RNA viruses: Coronaviruses. Semin. Virol. 1996, 7, 381–388. [Google Scholar] [CrossRef]
- Tang, X.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020, 7, 1012–1023. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Li, H.; Wu, X.; Zhong, Y.; Zhang, K.; Zhang, Y.-P.; Boerwinkle, E.; Fu, Y.-X. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004, 4, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wang, R.; Qu, G.; Wang, Y.; Liu, P.; Zhu, Y.; Fei, D.; Ren, L.; Zhou, Y.; Liu, L. Autopsy and general observation of the cadaver died from the novel coronavirus SARS-CoV-2. J. Forensic Med. 2020, 36, 1–3. (In Chinese) [Google Scholar]
- Ding, Y.; Wang, H.; Shen, H.; Li, Z.; Geng, J.; Han, H.; Cai, J.; Li, X.; Kang, W.; Weng, D.; et al. The clinical pathology of severe acute respiratory syndrome (SARS): A report from China. J. Pathol. 2003, 200, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Russell, C.D.; Millar, J.E.; Baillie, J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020, 395, 473–475. [Google Scholar] [CrossRef] [Green Version]
- Fujii, T.; Iwamoto, A.; Nakamura, T. Current concepts in SARS treatment. J. Infect. Chemother. 2004, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B.; Li, Q.; Wen, L.; Zhang, R. Clinical Features of 69 Cases With Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020, 71, 769–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.; Deming, D.; Paddock, C.D.; Cheng, A.; Yount, B.; Vogel, L.; Herman, B.D.; Sheahan, T.; Heise, M.; Genrich, G.L.; et al. A Mouse-Adapted SARS-Coronavirus Causes Disease and Mortality in BALB/c Mice. PLoS Pathog. 2007, 3, e5. [Google Scholar] [CrossRef]
- Menachery, V.D.; Yount, B.L.; Debbink, K.; Agnihothram, S.; Gralinski, L.E.; Plante, J.A.; Graham, R.L.; Scobey, T.; Ge, X.-Y.; Donaldson, E.F.; et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015, 21, 1508–1513. [Google Scholar] [CrossRef] [PubMed]


| HLA Class. | Alleles | No. of High Affinity Peptides | % of High Affinity Peptides | No. of Ultrahigh Affinity Peptides | % of Individuals That Have the Specific HLA Allele in the Population of | |
|---|---|---|---|---|---|---|
| Chinese | American | |||||
| I | B*15:03 | 97 | 7.67 | 1 | 0.18 | 2.69 |
| I | A*02:03 | 52 | 4.11 | 1 | 8.12 | 1.07 |
| I | B*15:17 | 51 | 4.03 | 3 | 0.81 | 0.90 |
| I | A*24:03 | 35 | 2.77 | 2 | 0.43 | 0.57 |
| I | A*30:01 | 26 | 2.06 | 1 | 14.27 | / |
| II | DRB3*03:01 | 998 | 79.59 | 64 | / | 12.72 |
| II | DRB1*10:01 | 805 | 64.19 | 30 | 2.75 | 3.41 |
| II | DRB1*09:01 | 555 | 44.26 | 9 | 24.28 | 4.94 |
| II | DRB1*16:02 | 528 | 42.11 | 14 | 5.15 | 1.96 |
| II | DRB1*13:02 | 481 | 38.36 | 38 | 7.58 | 9.56 |
| II | DRB1*01:01 | 375 | 29.90 | 8 | 4.14 | 10.75 |
| II | DRB1*11:01 | 175 | 13.96 | 2 | 8.49 | / |
| ACE2 Comparison | Max Score | Query Coverage | E Value | % of Identities |
|---|---|---|---|---|
| ACE2-Hs1 vs. ACE2-Hs2 | 1673 | 99% | 0 | 99.01 |
| ACE2-Hs1 vs. ACE2-mouse | 1361 | 98% | 0 | 81.05 |
| ACE2-Hs1 vs. ACE2-rat | 1353 | 96% | 0 | 82.49 |
| ACE2-Hs2 vs. ACE2-mouse | 1369 | 98% | 0 | 81.86 |
| ACE2-Hs2 vs. ACE2-rat | 1360 | 98% | 0 | 82.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Xie, J.; Guo, Y.; Sun, W.; He, Y.; Liu, K.; Yan, J.; Tao, A.; Zhong, N. SARS-CoV-2: Origin, Intermediate Host and Allergenicity Features and Hypotheses. Healthcare 2021, 9, 1132. https://doi.org/10.3390/healthcare9091132
Huang Y, Xie J, Guo Y, Sun W, He Y, Liu K, Yan J, Tao A, Zhong N. SARS-CoV-2: Origin, Intermediate Host and Allergenicity Features and Hypotheses. Healthcare. 2021; 9(9):1132. https://doi.org/10.3390/healthcare9091132
Chicago/Turabian StyleHuang, Yuyi, Junmou Xie, Yuhe Guo, Weimin Sun, Ying He, Kequn Liu, Jie Yan, Ailin Tao, and Nanshan Zhong. 2021. "SARS-CoV-2: Origin, Intermediate Host and Allergenicity Features and Hypotheses" Healthcare 9, no. 9: 1132. https://doi.org/10.3390/healthcare9091132






