The Dynamic Genetic-Hormonal Regulatory Network Controlling the Trichome Development in Leaves
Abstract
:1. Introduction
2. Genes Involved in the Initiation and Growth of Trichomes Overall in Model Species
3. Gene and Hormonal Interaction in Trichome Development
4. Regulation of the Cell Cycle and Trichome Complexity
5. Epigenetic Factors Involved in Trichome Development
miRNAs and Trichome Development
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hülskamp, M.; Misera, S.; Jürgens, G. Genetic dissection of trichome cell development in Arabidopsis. Cell 1994, 76, 555–566. [Google Scholar] [CrossRef]
- Hülskamp, M. Plant trichomes: A model for cell differentiation. Nat. Rev. Mol. Cell Biol. 2004, 5, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kurata, T.; Okada, K.; Wada, T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant. Biol. 2008, 59, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Fambrini, M.; Pugliesi, C. Usual and unusual development of the dicot leaf: Involvement of transcription factors and hormones. Plant. Cell Rep. 2013, 32, 899–922. [Google Scholar] [CrossRef] [PubMed]
- Pattanaik, S.; Patra, B.; Singh, S.K.; Yuan, L. An overview of the gene regulatory network controlling trichome development in the model plant Arabidopsis. Front. Plant. Sci. 2014, 5, 259. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Basile, A.; Fambrini, M.; Pugliesi, C. Transcription factors and hormone-mediated mechanisms regulate stomata development and responses under abiotic stresses: An overview. In Mechanism of Plant. Hormone Signaling under Stress, 1st ed.; Pandey, G., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; Volume 2, pp. 239–283. [Google Scholar]
- Doroshkov, A.V.; Kostantinov, D.K.; Afonnikov, D.A.; Gumbin, K. The evolution of gene regulatory networks controlling Arabidopsis thaliana L. trichome development. BMC Plant. Biol. 2019, 19 (Suppl. 1), 53. [Google Scholar] [CrossRef] [PubMed]
- Pesch, M.; Hülskamp, M. One, two, three… models for trichome patterning in Arabidopsis? Curr. Opin. Plant. Biol. 2009, 12, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ye, Z. Trichomes as models for studying plant cell differentiation. Cell Mol. Life Sci. 2013, 70, 1937–1948. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, L.H.; Jiao, J.; Liu, S.; Zhang, Z.; Lu, T.J. Gradient mechanical properties facilitate Arabidopsis trichome as mechanosensor. Appl. Mater. Interfaces 2016, 8, 9755–9761. [Google Scholar] [CrossRef]
- Zhou, L.H.; Liu, S.B.; Wang, P.F.; Lu, T.J.; Xu, F.; Genin, G.M.; Pickard, B.G. The Arabidopsis trichome is as active mechanosensory switch. Plant. Cell Environ. 2017, 40, 611–621. [Google Scholar] [CrossRef]
- Voirin, B.; Bayet, C.; Colson, M. Demonstration that flavone aglycones accumulate in the peltate glands of Mentha × piperita leaves. Phytochemistry 1993, 34, 85–87. [Google Scholar] [CrossRef]
- Gershenzon, J.; McConkey, M.E.; Croteau, R.B. Regulation of monoterpene accumulation in leaves of peppermint. Plant. Physiol. 2000, 122, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Gang, D.R.; Wang, J.; Dudareva, N.; Nam, K.H.; Simon, J.E.; Lewinsohn, E.; Pichersky, E. An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant. Physiol. 2001, 125, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant. Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Vendemiatti, E.; Zsögön, A.; Silva, G.F.F.E.; de Jesus, F.A.; Cutri, L.; Figueiredo, C.R.F.; Tanaka, F.A.O.; Nogueira, F.T.S.; Peres, L.E.P. Loss of type-IV glandular trichomes is a heterochronic trait in tomato and can be reverted by promoting juvenility. Plant. Sci. 2017, 259, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Maurya, S.; Chandra, M.; Yadav, R.K.; Narnoliya, L.K.; Sangwan, R.S.; Bansal, S.; Sandhu, P.; Singh, U.; Kumar, D.; Sangwan, N.S. Interspecies comparative features of trichomes in Ocimum reveal insights for biosynthesis of specialized essential oil metabolites. Protoplasma 2019. [Google Scholar] [CrossRef] [PubMed]
- Glas, J.J.; Schimmel, B.C.S.; Alba, J.M.; Escobar-Bravo, R.; Schuurink, R.C.; Kant, M.R. Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int. J. Mol. Sci. 2012, 13, 17077–17103. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Tooker, J.; Peiffer, M.; Chung, S.H.; Felton, G.W. Role of trichomes in defense against herbivores: Comparison of herbivore response to woolly and hairless trichome mutants in tomato (Solanum lycopersicum). Planta 2012, 236, 1053–1066. [Google Scholar] [CrossRef]
- Stratmann, J.W.; Bequette, C.J. Hairless but no longer clueless: Understanding glandular trichome development. J. Exp. Bot. 2016, 67, 5285–5287. [Google Scholar] [CrossRef]
- Xiao, L.; Tan, H.; Zhang, L. Artemisia annua glandular secretory trichomes: The biofactory of antimalarial agent artemisinin. Sci. Bull. 2016, 61, 26–36. [Google Scholar] [CrossRef]
- Marimoto, M. Chemical defense against insects in Heterotheca subaxillaris and three Orobanchaceae species using exudates from trichomes. Pest. Manag. Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ehleringer, J.; Björkman, O.; Mooney, H.A. Leaf pubescence: Effects on absorbance and photosynthesis in a desert shrub. Science 1976, 192, 376–377. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, D.B.; Lloyd, A.M.; Marks, M.D. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant. Sci. 2000, 5, 214–219. [Google Scholar] [CrossRef]
- Schellmann, S.; Hülskamp, M.; Uhrig, J. Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem. Soc. Trans. 2007, 35, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.-T. Molecular basis of natural variation and environmental control of trichome patterning. Front. Plant. Sci. 2014, 5, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, A.; Pan, J.; An, L.; Gan, Y.; Feng, H. The responses of trichome mutants to enhanced ultraviolet-B radiation in Arabidopsis thaliana. J. Photochem. Photobiol. B Biol. 2012, 113, 29–35. [Google Scholar] [CrossRef]
- Prozherina, N.; Freiwald, V.; Rousi, M.; Oksanen, E. Interactive effect of springtime frost and elevated ozone on early growth, foliar injuries and leaf structure of birch (Betula pendula). New Phytol. 2003, 159, 623–636. [Google Scholar] [CrossRef]
- Harada, E.; Choi, Y.E. Investigation of metal exudates from tobacco glandular trichomes under heavy metal stresses using a variable pressure scanning electron microscopy system. Plant. Biotechnol. 2008, 25, 407–411. [Google Scholar] [CrossRef] [Green Version]
- Svĕtlíková, P.; Hájek, T.; Tĕšitel, J. Hydathode trichomes actively secreting water from leaves play a key role in the physiology and evolution of root-parasitic rhinanthoid Orobanchaceae. Ann. Bot. 2015, 116, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Guan, X.; Song, Q.; Chen, Z.J. Polyploidy and small RNA regulation of cotton fiber development. Trends Plant. Sci. 2014, 19, 516–528. [Google Scholar] [CrossRef]
- Telfer, A.; Bollman, K.M.; Poethig, R.S. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 1997, 124, 645–654. [Google Scholar]
- Poethig, R.S. Vegetative phase change and shoot maturation in plants. Curr. Top. Dev. Biol. 2013, 105, 125–152. [Google Scholar]
- Wang, L.; Zhou, C.M.; Mai, Y.X.; Li, L.Z.; Gao, J.; Shang, G.D.; Lian, H.; Han, L.; Zhang, T.Q.; Tang, H.B.; et al. A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. EMBO J. 2019, 38, e100063. [Google Scholar] [CrossRef]
- Walker, A.R.; Marks, M.D. Trichome initiation in Arabidopsis. In Plant Trichomes; Advances in Botanical Research; Academic Press: San Diego, CA, USA, 2000; Volume 31, pp. 219–236. [Google Scholar]
- Larkin, J.C.; Young, N.; Prigge, M.; Marks, M.D. The control of trichome spacing and number in Arabidopsis. Development 1996, 122, 997–1005. [Google Scholar]
- Greese, B.; Hülskamp, M.; Fleck, C. Quantification of variability in trichome patterns. Front. Plant. Sci. 2014, 5, 596. [Google Scholar] [CrossRef]
- McLellan, T. Correlated evolution of leaf shape and trichomes in Begonia dregei (Begoniaceae). Am. J. Bot. 2005, 92, 1616–1623. [Google Scholar] [CrossRef]
- Uematsu, K.; Kutsukake, M.; Fukatsu, T. Water-repellent plant surface structure induced by gall-forming insects for waste management. Biol. Lett. 2018, 14, 20180470. [Google Scholar] [CrossRef]
- Hülskamp, M.; Schnittger, A.; Folkers, U. Pattern formation and cell differentiation: Trichomes in Arabidopsis as a genetic model system. Int. Rev. Cytol. 1999, 186, 147–178. [Google Scholar]
- Kirik, V.; Simon, M.; Hülskamp, M.; Schiefelbein, J. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant. Mol. Biol. 2004, 55, 389–398. [Google Scholar] [CrossRef]
- Kirik, V.; Lee, M.M.; Wester, K.; Herrmann, U.; Zheng, Z.; Oppenheimer, D.; Schiefelbein, J.; Hülskamp, M. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 2005, 132, 1477–1485. [Google Scholar] [CrossRef]
- Payne, T.; Clement, J.; Arnold, D.; Lloyd, A. Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 1999, 126, 671–682. [Google Scholar]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG. Genetics 2000, 156, 1349–1362. [Google Scholar]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.M.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef]
- Marks, M.D.; Wenger, J.P.; Gilding, E.; Jilk, R.; Dixon, R.A. Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol. Plant. 2009, 2, 803–822. [Google Scholar] [CrossRef]
- Balkunde, R.; Bouyer, D.; Hülskamp, M. Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 2011, 138, 5039–5048. [Google Scholar] [CrossRef]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Walker, A.R.; Davidson, P.A.; Bolognesi-Winfield, A.J.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TEST GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant. Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef]
- Zhang, B.; Schrader, A. TRASPARENT TESTA GLABRA1-dependent regulation of flavonoid biosynthesis. Plants 2017, 6, 65. [Google Scholar] [CrossRef]
- Larkin, J.C.; Oppenheimer, D.G.; Lloyd, A.M.; Paparozzi, E.T.; Marks, M.D. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant. Cell 1994, 6, 1065–1076. [Google Scholar] [CrossRef]
- Larkin, J.C.; Walker, J.D.; Bolognesi-Winfield, A.C.; Gray, J.G.; Walker, A.R. Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 1999, 151, 1591–1604. [Google Scholar]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Ausubel, F.M.; Goodman, H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant. J. 1995, 8, 659–671. [Google Scholar] [CrossRef]
- Debeaujon, I.; Léon-Kloosterziel, K.M.; Koornneef, M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant. Physiol. 2000, 122, 403–413. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef]
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.H.; Kirik, V.; Hülskamp, M.; Nam, K.H.; Hagely, K.; Lee, M.M.; Schiefelbein, J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant. Cell 2009, 21, 1080–1094. [Google Scholar] [CrossRef]
- Balkunde, R.; Pesch, M.; Hülskamp, M. Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top. Dev. Biol. 2010, 91, 299–321. [Google Scholar]
- Rerie, W.G.; Feldmann, K.A.; Marks, M.D. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994, 8, 1388–1399. [Google Scholar] [CrossRef]
- Szymanski, D.B.; Jilk, R.A.; Pollock, S.M.; Marks, M.D. Control of GL2 expression in Arabidopsis leaves and trichomes. Development 1998, 125, 1161–1171. [Google Scholar]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant. Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Ishida, T.; Hattori, S.; Sano, R.; Inoue, K.; Shirano, Y.; Hayashi, H.; Shibata, D.; Sato, A.; Kato, T.; Tabata, S.; et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant. Cell 2007, 19, 2531–2543. [Google Scholar] [CrossRef]
- Morohashi, K.; Zhao, M.; Yang, M.; Read, B.; Lloyd, A.; Lamb, R.; Grotewold, E. Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant. Physiol. 2007, 145, 1–11. [Google Scholar] [CrossRef]
- Bouyer, D.; Geier, F.; Kragler, F.; Schnittger, A.; Pesch, M.; Wester, K.; Balkunde, R.; Timmer, J.; Fleck, C.; Hülskamp, M. Two-dimensional patterning by a trapping/depletion mechanism: The role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 2008, 6, e141. [Google Scholar] [CrossRef]
- Lin, Q.; Aoyama, T. Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: Update on the roles of the classic regulator. J. Integr. Plant. Biol. 2012, 54, 729–737. [Google Scholar]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant. Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Li, Q.; Yin, M.; Li, Y.; Fan, C.; Yang, Q.; Wu, J.; Zhang, C.; Wang, H.; Zhou, Y. Expression of Brassica napus TTG2, a regulator of trichome development, increases plant sensitivity to salt stress by suppressing the expression of auxin biosynthesis genes. J. Exp. Bot. 2015, 66, 5821–5836. [Google Scholar] [CrossRef]
- Kirik, V.; Simon, M.; Wester, K.; Schiefelbein, J.; Hülskamp, M. The ENHANCER OF TRY and CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 2001, 268, 506–513. [Google Scholar] [CrossRef]
- Schellmann, S.; Schnittger, A.; Kirik, V.; Wada, T.; Okada, K.; Beermann, A.; Thumfahrt, J.; Jürgens, G.; Hülskamp, M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002, 21, 5036–5046. [Google Scholar] [CrossRef]
- Esch, J.J.; Chen, M.A.; Hillestad, M.; Marks, M.D. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant. J. 2004, 40, 860–869. [Google Scholar] [CrossRef]
- Gan, L.; Xia, K.; Chen, J.G.; Wang, S. Functional characterization of TRICHOMELESS2, a new single repeat R3MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biology 2011, 11, 176–187. [Google Scholar] [CrossRef]
- Wang, S.; Kwak, S.H.; Zeng, Q.; Ellis, B.E.; Chen, X.Y.; Schiefelbein, J.; Chen, J.G. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 2007, 134, 3873–3882. [Google Scholar] [CrossRef]
- Wang, S.; Chen, J.G. Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant. Sci. 2014, 5, 133. [Google Scholar] [CrossRef] [Green Version]
- Digiuni, S.; Schellmann, S.; Geier, F.; Greese, B.; Pesch, M.; Wester, K.; Dartan, B.; Mach, V.; Srinivas, B.P.; Timmer, J.; et al. A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 2008, 4, 217. [Google Scholar] [CrossRef]
- Morohashi, K.; Grotewold, E. A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 2009, 5, e1000396. [Google Scholar] [CrossRef]
- Pesch, M.; Dartan, B.; Birkenbihl, R.; Somssich, I.E.; Hülskamp, M. Arabidopsis TTG2 regulates TRY expression through enhancement of activator complex-triggered activation. Plant. Cell 2014, 26, 4067–4083. [Google Scholar] [CrossRef]
- Pesch, M.; Schultheiß, I.; Klopffleisch, K.; Uhrig, J.F.; Koegl, M.; Clemen, C.S.; Simon, R.; Weidtkamp-Peters, S.; Hülskamp, M. TRANSPARENT TESTA GLABRA1 and GLABRA1 compete for binding to GLABRA3 in Arabidopsis. Plant. Physiol. 2015, 168, 584–597. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Li, Y.; Ai, Q.; Yu, D. MYB82 functions in regulation of trichome development in Arabidopsis. J. Exp. Bot. 2014, 65, 3215–3223. [Google Scholar] [CrossRef]
- Friede, A.; Zhang, B.; Herbert, S.; Pesch, M.; Schrader, A.; Hülskamp, M. The second intron is essential for the transcriptional control of the Arabidopsis thaliana GLABRA3 gene in leaves. Front. Plant. Sci. 2017, 8, 1382. [Google Scholar] [CrossRef]
- Lee, M.M.; Schiefelbein, J. Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 2001, 128, 1539–1546. [Google Scholar]
- Wang, S.; Wang, J.W.; Yu, N.; Li, C.H.; Luo, B.; Gou, J.Y.; Wang, L.J.; Chen, X.Y. Control of plant trichome development by a cotton fiber MYB gene. Plant. Cell 2004, 16, 2323–2334. [Google Scholar] [CrossRef]
- Suo, B.; Seifert, S.; Kirik, V. Arabidopsis GLASSY HAIR genes promote trichome papillae development. J. Exp. Bot. 2013, 64, 4981–4991. [Google Scholar] [CrossRef]
- Jakoby, M.J.; Falkenhan, D.; Mader, M.T.; Brininstool, G.; Wischnitzki, E.; Platz, N.; Hudson, A.; Hülskamp, M.; Larkin, J.; Schnittger, A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant. Physiol. 2008, 148, 1583–1602. [Google Scholar] [CrossRef]
- Bischoff, V.; Nita, S.; Neumetzler, L.; Schindelasch, D.; Urbain, A.; Eshed, R.; Persson, S.; Delmer, D.; Scheible, W.R. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant. Physiol. 2010, 153, 590–602. [Google Scholar] [CrossRef]
- Potikha, T.; Delmer, D.P. A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant. J. 1995, 7, 453–460. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant. Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef]
- Tian, N.; Liu, F.; Wang, P.; Yan, X.; Gao, H.; Zeng, X.; Wu, G. Overexpression of BraLTP2, a lipid transfer protein of Brassica napus, results in increased trichome density and altered concentration of secondary metabolites. Int. J. Mol. Sci. 2018, 19, 1733. [Google Scholar] [CrossRef]
- Harada, E.; Kim, J.A.; Meyer, A.J.; Hell, R.; Clemens, S.; Choi, Y.E. Expression profiling of tobacco leaf trichomes identifies genes for biotic and abiotic stresses. Plant. Cell Physiol. 2010, 51, 1627–1637. [Google Scholar] [CrossRef]
- Choi, Y.E.; Lim, S.; Kim, H.J.; Han, J.Y.; Lee, M.H.; Yang, Y.; Kim, J.A.; Kim, Y.S. Tobacco NtLTP1, a glandular-specific lipid transfer protein, is required for lipid secretion from glandular trichomes. Plant. J. 2012, 70, 480–491. [Google Scholar] [CrossRef]
- Symonds, V.V.; Hatlestad, G.; Lloyd, A.M. Natural allelic variation defines a role for ATMYC1: Trichome cell fate determination. PLoS Genet. 2011, 7, e1002069. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Zhu, D.; Cui, S.; Li, X.; Cao, Y.; Ma, L. A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J. Biol. Chem. 2012, 287, 14109–14121. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant. J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Tominaga, R.; Iwata, M.; Sano, R.; Inoue, K.; Okada, K.; Wada, T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 2008, 135, 1335–1345. [Google Scholar] [CrossRef]
- Pesch, M.; Schultheiß, I.; Digiuni, S.; Uhrig, J.F.; Hülskamp, M. Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 2013, 140, 3456–3467. [Google Scholar] [CrossRef]
- Wei, S.; Xiang, L.; Gruber, M.G.; Feyissa, B.A.; Amyot, L.; Hannoufa, A. COP9 signalosome subunit 5A affects phenilpropanoid metabolism, trichome formation and transcription of key genes of a regulatory tri-protein complex in Arabidopsis. BMC Plant. Biol. 2018, 18, 134. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D.S.; Wuest, S.E.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwaśniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant. Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D.S.; Stewart, D.; Zheng, B.; Couplan, G.; Wellmer, F. Floral homeotic proteins modulate the genetic program for leaf development to suppress trichome formation in flowers. Development 2018, 145, dev157784. [Google Scholar] [CrossRef]
- Yan, L.; Cheng, X.; Jia, R.; Qin, Q.; Guan, L.; Du, H.; Hou, S. New phenotypic characteristics of three tmm alleles in Arabidopsis thaliana. Plant. Cell Rep. 2014, 33, 719–731. [Google Scholar] [CrossRef]
- Koornneef, M. The complex syndrome of ttg mutants. Arabidopsis Info. Serv. 1981, 18, 45–51. [Google Scholar]
- Folkers, U.; Berger, J.; Hülskamp, M. Cell morphogenesis of trichomes in Arabidopsis: Differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 1997, 124, 3779–3786. [Google Scholar]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant. Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant. J. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant. Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Wada, T.; Tachibana, T.; Shimura, Y.; Okada, K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 1997, 277, 1113–1116. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant. J. 2008, 55, 954–967. [Google Scholar] [CrossRef]
- Voges, D.; Zwickl, P.; Baumeister, W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999, 68, 1015–1068. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Bohn, S.; Beck, F.; Sakata, E.; Walzthoeni, T.; Beck, M.; Aebersold, R.; Förster, F.; Baumeister, W.; Nickell, S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. USA 2010, 107, 20992–20997. [Google Scholar] [CrossRef]
- Patra, B.; Pattanaik, S.; Yuan, L. Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant. J. 2013, 74, 435–447. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef]
- Patra, B.; Pattanaik, S.; Yuan, L. Proteolytic degradation of the flavonoid regulators, TRANSPARENT TESTA8 and TRANSPARENT TESTA GLABRA1, in Arabidopsisis mediated by the ubiquitin/26Sproteasome system. Plant. Signal. Behav. 2014, 8, e25901. [Google Scholar] [CrossRef]
- Maes, L.; Goossens, A. Hormone-mediated promotion of trichome initiation in plants is conserved but utilizes species- and trichome-specific regulatory mechanisms. Plant. Signal. Behav. 2010, 5, 205–207. [Google Scholar] [CrossRef]
- An, L.; Zhou, Z.; Yan, A.; Gan, Y. Progress on trichome development regulated by phytohormone signaling. Plant. Signal. Behav. 2011, 6, 1959–1962. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y. Trichome formation: Gibberellins on the move. Plant. Physiol. 2016, 170, 1174–1175. [Google Scholar] [CrossRef]
- Greenboim-Wainberg, Y.; Maymon, I.; Borochov, R.; Alvarez, J.; Olszewski, N.; Ori, N.; Eshed, Y.; Weiss, D. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant. Cell 2005, 17, 92–102. [Google Scholar] [CrossRef]
- Chien, J.C.; Sussex, I.M. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant. Physiol. 1996, 111, 1321–1328. [Google Scholar] [CrossRef]
- Perazza, D.; Vachon, G.; Herzog, M. Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant. Physiol. 1998, 117, 375–383. [Google Scholar] [CrossRef]
- An, L.; Zhou, Z.; Su, S.; Yan, A.; Gan, Y. GLABROUS INFLORESCENCE STEMS (GIS) is required for trichome branching through gibberellic acid signaling in Arabidopsis. Plant. Cell Physiol. 2012, 53, 457–469. [Google Scholar] [CrossRef]
- Sun, L.L.; Zhou, Z.J.; An, Y.; Zhao, Y.Q.; Meng, X.F.; Steele-King, C.; Gan, Y.B. GLABROUS INFLORESCENCE STEMS regulates trichome branching by genetically interacting with SIM in Arabidopsis. J. Zhejiang Univ. Sci. B. 2013, 14, 563–569. [Google Scholar] [CrossRef]
- Gan, Y.; Liu, C.; Yu, H.; Broun, P. Integration of cytokinin and gibberellins signaling by Arabidopsis transcription factor GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 2007, 134, 2073–2081. [Google Scholar] [CrossRef]
- Gan, Y.; Kumimoto, R.; Liu, C.; Ratcliffe, O.; Yu, H.; Broun, P. GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant. Cell 2006, 18, 1383–1395. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, A.; Zhou, Z.; Zhao, Y.; Bao, S.; Yu, H.; Gan, Y. GLABROUS INFLORESCENCE STEM3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 2015, 206, 220–230. [Google Scholar] [CrossRef]
- Zhou, Z.; An, L.; Sun, L.; Zhu, S.; Xi, W.; Broun, P.; Yu, H.; Gan, Y. Zinc Finger Protein5 is required for the control of trichome initiation by acting upstream of Zinc Finger Protein8 in Arabidopsis. Plant. Physiol. 2011, 157, 673–682. [Google Scholar] [CrossRef]
- Zhou, L.L.; Shi, M.Z.; Xie, D.Y. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta 2012, 236, 825–837. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, L.; Zhao, Y.; An, L.; Yan, A.; Meng, X.; Gan, Y. Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 2013, 198, 699–708. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Olszewski, N.E. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal-transduction. Plant. Cell 1993, 5, 887–896. [Google Scholar]
- Jacobsen, S.E.; Binkowski, K.A.; Olszewski, N.E. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 1996, 93, 9292–9296. [Google Scholar] [CrossRef]
- Tague, B.W.; Goodman, H.M. Characterization of a family of Arabidopsis zinc finger protein cDNAs. Plant. Mol. Biol. 1995, 28, 267–279. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Khan, A.R.; Liu, B.; Wu, M.; Song, G.; Ni, H.; Ying, H.; Yu, H.; Gan, Y. NbGIS regulates glaundular trichome initiation through GA signaling in tobacco. Plant. Mol. Biol. 2018, 98, 153–167. [Google Scholar] [CrossRef]
- Kim, S.Y.; Hyoung, S.; So, W.M.; Shin, J.S. The novel transcription factor TRP interacts with ZFP5, a trichome initiation-related transcription factor, and negatively regulates trichome initiation through gibberellic acid signaling. Plant. Mol. Biol. 2018, 96, 315–326. [Google Scholar] [CrossRef]
- Castillejo, C.; Pelaz, S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Osnato, M.; Castillejo, C.; Matías-Hernández, L.; Pelaz, S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 2012, 3, 808. [Google Scholar] [CrossRef] [PubMed]
- Matías-Hernández, L.; Aguilar Jaramillo, A.E.; Osnato, M.; Weinstain, R.; Shani, E.; Suárez-López, P.; Pelaz, S. TEMPRANILLO reveals the mesophyll as crucial for epidermal trichome formation. Plant. Physiol. 2016, 170, 1624–1639. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Yu, F.; Du, C.; Li, X.; Zhao, X.; Liu, X. RPN1a, a subunit of the 26S proteasome, controls trichome development in Arabidopsis. Plant. Physiol. Biochem. 2015, 88, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Maes, L.; Inzé, D.; Goossens, A. Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant. Physiol. 2008, 148, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Traw, B.M.; Bergelson, J. Interactive effects of jasmonic acid, salicylic acid, and gibberellins on induction of trichomes in Arabidopsis. Plant. Physiol. 2003, 133, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant. Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant. Biol. 2001, 4, 301–308. [Google Scholar] [CrossRef]
- Yan, T.; Chen, M.; Shen, Q.; Li, L.; Fu, X.; Pan, Q.; Tang, Y.; Shi, P.; Lv, Z.; Jiang, W.; et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2017, 213, 1145–1155. [Google Scholar] [CrossRef]
- Noorden, R.V. Demand for malaria drug soars. Nature 2010, 466, 672–673. [Google Scholar] [CrossRef]
- Tan, H.; Xiao, L.; Gao, S.; Li, Q.; Chen, J.; Xiao, Y.; Ji, Q.; Chen, R.; Chen, W.; Zhang, L. TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant. 2015, 8, 1396–1411. [Google Scholar] [CrossRef]
- War, A.R.; Hussain, B.; Sharma, H.C. Induced resistance in groundnut by jasmonic acid and salicylic acid through alteration of trichome density and oviposition by Helicoverpa armigera (Lepidoptera: Noctuidae). AoB Plants 2013, 5, plt053. [Google Scholar] [CrossRef]
- Bowling, S.A.; Clarke, J.D.; Liu, Y.; Klessig, D.F.; Dong, X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant. Cell 1997, 9, 1573–1584. [Google Scholar]
- Brininstool, G.; Kasili, R.; Simmons, L.A.; Kirik, V.; Hülskamp, M.; Larkin, J.C. Constitutive Expressor of Pathogenesis-Related Genes5 affects cell wall biogenesis and trichome development. BMC Plant. Biol. 2008, 8, 58. [Google Scholar] [CrossRef]
- Völz, R.; Kim, S.K.; Mi, J.; Mariappan, K.G.; Guo, X.; Bigeard, J.; Alejandro, S.; Pflieger, D.; Rayapuram, N.; Al-Babili, S.; et al. The Trihelix transcription factor GT2-like 1 (GTL1) promotes salicylic acid metabolism, and regulates bacterial-triggered immunity. PLoS Genet. 2018, 14, e1007708. [Google Scholar] [CrossRef]
- Pandey, S.; Goel, R.; Bhardwaj, A.; Asif, M.H.; Sawant, S.V.; Misra, P. Transcriptome analysis provides insight into prickle development and its link to defense and secondary metabolism in Solanum viarum Dunal. Sci. Rep. 2018, 8, 17092. [Google Scholar] [CrossRef]
- Plett, J.M.; Mathur, J.; Regan, S. Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 3923–3933. [Google Scholar] [CrossRef]
- Laxmi, A.; Paul, L.K.; Peters, J.L.; Khurana, J.P. Arabidopsis constitutive photomorphogenic mutant, bls1, displays altered brassinosteroid response and sugar sensitivity. Plant. Mol. Biol. 2004, 56, 185–201. [Google Scholar] [CrossRef]
- Campos, M.L.; de Almeida, M.; Rossi, M.L.; Martinelli, A.P.; Litholdo, C.G.; Figueira, A.; Rampelotti-Ferreira, F.T.; Vendramim, J.D.; Benedito, V.A.; Peres, L.E. Brassinosteroids interact negatively with jasmonates in the formation of anti-herbivory traits in tomato. J. Exp. Bot. 2009, 60, 4347–4361. [Google Scholar] [CrossRef] [Green Version]
- Koka, C.V.; Cerny, R.E.; Gardner, R.G.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Yoschida, S.; Clouse, S.D. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant. Physiol. 2000, 122, 85–98. [Google Scholar] [CrossRef]
- Li, L.; Li, C.; Howe, G.A. Genetic analysis of wound signaling in tomato. Evidence for a dual role of jasmonic acid in defense and female fertility. Plant. Physiol. 2001, 127, 1414–1417. [Google Scholar] [CrossRef]
- D’Amato, F. Nuclear Cytology in Relation to Development; Cambridge University Press: Cambridge, UK, 1977. [Google Scholar]
- Vermeer, J.; Peterson, R.L. Glandular trichomes on the inflorescence of Chrysanthemum morifolium cv. Dramatic (Compositae). 2 Ultrastructure and histochemistry. Can. J. Bot. 1979, 57, 714–729. [Google Scholar] [CrossRef]
- Bramsiepe, J.; Wester, K.; Weinl, C.; Roodbarkelari, F.; Kasili, R.; Larkin, J.C.; Hülskamp, M.; Schnittger, A. Endoreplication controls cell fate maintenance. PLoS Genet. 2010, 6, e1000996. [Google Scholar] [CrossRef]
- Desvoyes, B.; Ramirez-Parra, E.; Xie, Q.; Chua, N.H.; Gutierrez, C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant. Physiol. 2006, 140, 67–80. [Google Scholar] [CrossRef]
- Lammens, T.; Boudolf, V.; Kheibarshekan, L.; Zalmas, L.P.; Gaamouche, T.; Maes, S.; Vanstraelen, M.; Kondorosi, E.; La Thangue, N.B.; Govaerts, W.; et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA 2008, 105, 14721–14726. [Google Scholar] [CrossRef]
- Larson-Rabin, Z.; Li, Z.; Masson, P.H.; Day, C.D. FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant. Physiol. 2009, 149, 874–884. [Google Scholar] [CrossRef]
- Esch, J.J.; Chen, M.; Sanders, M.; Hillestad, M.; Ndkium, S.; Idelkope, B.; Neizer, J.; Marks, M.D. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 2003, 130, 5885–5894. [Google Scholar] [CrossRef]
- Dewitte, W.; Murray, J.A. The plant cell cycle. Annu. Rev. Plant. Biol. 2003, 54, 235–264. [Google Scholar] [CrossRef]
- Vandepoele, K.; Raes, J.; De Veylder, L.; Rouze, P.; Rombauts, S.; Inzé, D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant. Cell 2002, 14, 903–916. [Google Scholar] [CrossRef]
- Shen, W.H. The plant E2F-Rb pathway and epigenetic control. Trends Plant. Sci. 2002, 7, 505–511. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant. Physiol. 2002, 128, 833–843. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant. Physiol 2003, 132, 2012–2022. [Google Scholar] [CrossRef]
- Breuer, C.; Morohashi, K.; Kawamura, A.; Takahashi, N.; Ishida, T.; Umeda, M.; Grotewold, E.; Sugimoto, K. Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J. 2012, 31, 4488–4501. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, N.; Kajihara, T.; Okamura, C.; Kim, Y.; Katagiri, Y.; Okushima, Y.; Matsunaga, S.; Hwang, I.; Umeda, M. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 2013, 23, 1812–1817. [Google Scholar] [CrossRef]
- Vlieghe, K.; Boudolf, V.; Beemster, G.T.S.; Maes, S.; Magyar, Z.; Atanassova, A.; De Almeida Engler, J.; De Groodt, R.; Inzé, D.; De Veylder, L. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 2005, 15, 59–63. [Google Scholar] [CrossRef]
- Schnittger, A.; Weinl, C.; Bouyer, D.; Schöbinger, U.; Hülskamp, M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant. Cell 2003, 15, 303–315. [Google Scholar] [CrossRef]
- Wang, G.; Kong, H.; Sun, Y.; Zhang, X.; Zhang, W.; Altman, N.; dePamphilis, C.W. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like protein. Plant. Physiol. 2004, 135, 1084–1099. [Google Scholar] [CrossRef]
- Hochegger, H.; Takeda, S.; Hunt, T. Cyclin-dependent kinases and cell-cycle transitions: Does one fit all? Nat. Rev. Mol. Cell Biol. 2008, 9, 910–916. [Google Scholar] [CrossRef]
- Ilgenfritz, H.; Bouyer, D.; Schnittger, A.; Malthur, J.; Kirik, V.; Schwab, B.; Chua, N.H.; Jürgens, G.; Hülskamp, M. The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant. Physiol. 2003, 131, 643–655. [Google Scholar] [CrossRef]
- Churchman, M.L.; Brown, M.L.; Kato, N.; Kirik, V.; Hülskamp, M.; Inzé, D.; De Veylder, L.; Walker, J.D.; Zheng, Z.; Oppenheimer, D.G.; et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant. Cell 2006, 18, 3145–3157. [Google Scholar] [CrossRef]
- Walker, J.D.; Oppenheimer, D.G.; Concienne, J.; Larkin, J.C. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 2000, 127, 3931–3940. [Google Scholar]
- Kasili, R.; Walker, J.D.; Simmons, L.A.; Zhou, J.; De Veylder, L.; Larkin, J.C. SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 2010, 185, 257–268. [Google Scholar] [CrossRef]
- Peeper, D.S.; Parker, L.L.; Ewen, M.E.; Toebes, M.; Hall, F.L.; Xu, M.; Zantema, A.; van der Eb, A.J.; Piwnica-Worms, H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993, 12, 1947–1955. [Google Scholar] [CrossRef]
- Schnittger, A.; Schöbinger, U.; Stierhof, Y.-D.; Hülskamp, M. Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr. Biol. 2002, 12, 415–420. [Google Scholar] [CrossRef]
- Marks, M.D.; Gilding, E.; Wenger, J.P. Genetic interaction between glabra3-shapeshifter and siamese in Arabidopsis thaliana converts trichome precursors into cells with meristematic activity. Plant. J. 2007, 52, 352–361. [Google Scholar] [CrossRef]
- Wenger, J.P.; Marks, M.D. E2F and retinoblastoma related proteins may regulate GL1 expression in developing Arabidopsis trichomes. Plant. Signal. Behav. 2008, 3, 420–422. [Google Scholar] [CrossRef]
- Huchelmann, A.; Boutry, M.; Hachez, C. Plant glandular trichomes: Natural cell factories of high biotechnological interest. Plant. Physiol. 2017, 175, 6–22. [Google Scholar] [CrossRef]
- Grebe, M. The pattering of epidermal hairs in Arabidopsis updated. Curr. Opin. Plant. Biol. 2012, 15, 31–37. [Google Scholar] [CrossRef]
- Xi, A.; Yang, X.; Deng, M.; Chen, Y.; Shao, J.; Zhao, J.; An, L. Isolation and identification of two new alleles of STICHEL in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 499, 605–610. [Google Scholar] [CrossRef]
- Kasili, R.; Huang, C.C.; Walker, J.D.; Simmons, L.A.; Zhou, J.; Faulk, C.; Hülskamp, M.; Larkin, J.C. BRANCHLESS TRICHOMES links cell shape and cell cycle control in Arabidopsis trichomes. Development 2011, 138, 2379–2388. [Google Scholar] [CrossRef]
- Perazza, D.; Herzog, M.; Hülskamp, M.; Brown, S.; Dorne, A.M.; Bonneville, J.-M. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 1999, 152, 461–476. [Google Scholar]
- El Refy, A.; Perazza, D.; Zekraoui, L.; Valay, J.G.; Bechtold, N.; Hülskamp, M.; Herzog, M.; Bonneville, J.-M. The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol. Genet. Genom. 2003, 270, 403–414. [Google Scholar] [CrossRef]
- Downes, B.P.; Stupar, R.M.; Gingerich, D.J.; Vierstra, R.D. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant. J. 2003, 35, 729–742. [Google Scholar] [CrossRef]
- Iwata, E.; Ikeda, S.; Matsunaga, S.; Kurata, M.; Yoshioka, Y.; Criqui, M.C.; Genschik, P.; Ito, M. GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant. Cell 2011, 23, 4382–4393. [Google Scholar] [CrossRef]
- Heyman, J.; Polyn, S.; Eekhout, T.; De Veylder, L. Tissue-specific control of the endocycle by the anaphase promoting complex/cyclosom inhibitor UVI4 and DEL1. Plant. Physiol. 2017, 175, 303–313. [Google Scholar] [CrossRef]
- Oppenheimer, D.G.; Pollock, M.A.; Vacik, J.; Szymanski, D.B.; Ericson, B.; Feldmann, K.; Marks, M.D. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 6261–6266. [Google Scholar] [CrossRef]
- Mathur, J.; Chua, N.H. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant. Cell 2000, 12, 465–477. [Google Scholar] [CrossRef]
- Kirik, V.; Grini, P.E.; Mathur, J.; Klinkhammer, I.; Adler, K.; Bechtold, N.; Herzog, M.; Bonneville, J.-M.; Hülskamp, M. The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the α/β-tubulin monomer balance. Plant. Cell 2002, 14, 2265–2276. [Google Scholar] [CrossRef]
- Abe, T.; Thitamadee, S.; Hashimoto, T. Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant. Cell Physiol. 2004, 45, 211–220. [Google Scholar] [CrossRef]
- Mitchison, T.; Kirschner, M.W. Dynamic instability of microtubule growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef]
- Mandelkow, E.; Mandelkow, E.-M. Microtubules and microtubule-associated proteins. Curr. Opin. Cell Biol. 1995, 7, 72–81. [Google Scholar] [CrossRef]
- Burk, D.H.; Liu, B.; Zhong, R.; Morrison, W.H.; Ye, Z.H. A katanin like protein regulates normal cell wall biosynthesis and cell elongation. Plant. Cell 2001, 13, 807–827. [Google Scholar] [CrossRef]
- Abe, T.; Hashimoto, T. Altered microtubule dynamics by expression of modified-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant. J. 2005, 43, 191–204. [Google Scholar] [CrossRef]
- Thitamadee, S.; Tuchihara, K.; Hashimoto, T. Microtubule basis for left-handed helical growth in Arabidopsis. Nature 2002, 417, 193–196. [Google Scholar] [CrossRef]
- Tian, J.; Han, L.; Feng, Z.; Wang, G.; Liu, W.; Ma, Y.; Yu, Y.; Kong, Z. Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. eLife 2015, 4, e09351. [Google Scholar] [CrossRef]
- Chen, L.; Peng, L.; Tian, J.; Wang, X.; Kong, Z.; Mao, T.; Yuan, M.; Li, Y. TCS1, a microtubule-binding protein, interacts with KCBP/ZWICHEL to regulate trichome cell shape in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006266. [Google Scholar] [CrossRef]
- Liang, S.; Yang, X.; Deng, M.; Zhao, J.; Shao, J.; Qi, Y.; Liu, X.; Yu, F.; An, L. A new allele of the SPIKE1 locus reveals distinct regulation of trichome and pavement cell development and plant growth. Front. Plant. Sci. 2019, 10, 16. [Google Scholar] [CrossRef]
- Kiyokawa, E.; Hashimoto, Y.; Kobayashi, S.; Sugimura, H.; Kurata, T.; Matsuda, M. Activation of Rac1 by a CrkSH3-binding protein, DOCK180. Genes Dev. 1998, 12, 3331–3336. [Google Scholar] [CrossRef]
- Bompard, G.; Caron, E. Regulation of WASP/WAVE proteins: Making a long story short. J. Cell Biol. 2004, 166, 957–962. [Google Scholar] [CrossRef]
- Perez-Rodriguez, M.; Jaffe, F.W.; Butelli, E.; Glover, B.J.; Martin, C. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Anthirrhinum majus flowers. Development 2005, 132, 359–379. [Google Scholar] [CrossRef]
- Brockington, S.F.; Alvarez-Fernandez, R.; Landis, J.B.; Alcorn, K.; Walker, R.H.; Thomas, M.M.; Hileman, L.C.; Glover, B.J. Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation. Mol. Biol. Evol. 2013, 30, 526–540. [Google Scholar] [CrossRef]
- Shi, P.; Fu, X.; Shen, Q.; Liu, M.; Pan, Q.; Tang, Y.; Jiang, W.; Lv, Z.; Yan, T.; Ma, Y.; et al. The role of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol. 2018, 217, 261–276. [Google Scholar] [CrossRef]
- Serna, L.; Martin, C. Trichomes: Different regulatory networks lead to convergent structures. Trends Plant. Sci. 2006, 11, 1360–1385. [Google Scholar] [CrossRef]
- Yang, C.; Li, H.; Zhang, J.; Luo, Z.; Gong, P.; Zhang, C.; Li, J.; Wang, T.; Zhang, Y.; Lu, Y.; et al. A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl. Acad. Sci. USA 2011, 108, 11836–11841. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; van Herwijnn, Z.O.; Dräger, D.B.; Sui, C.; Haring, M.A.; Schuurink, R.C. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant. Cell 2018, 30, 2988–3005. [Google Scholar] [CrossRef]
- Lloyd, A.M.; Walbot, V.; Davis, R.W. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 1992, 258, 1773–1775. [Google Scholar] [CrossRef]
- Abe, M.; Katsumata, H.; Komeda, Y.; Takahashi, T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 2003, 130, 635–643. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant. Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, F.; Tang, Y.; Yuan, Y.; Deng, W.; Li, Z. Auxin response gene SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant. Cell Physiol. 2015, 56, 2110–2124. [Google Scholar]
- Pikaard, C.S.; Mittelsten Scheid, O. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, 12. [Google Scholar] [CrossRef]
- Deng, X.; Qiu, Q.; Cao, X. The seekers: How epigenetic modifying enzymes find their hidden genomic targets in Arabidopsis. Curr. Opin. Plant. Biol. 2018, 45, 75–81. [Google Scholar] [CrossRef]
- Exner, V.; Taranto, P.; Schonrock, N.; Gruissem, W.; Hennig, L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 2006, 133, 4163–4172. [Google Scholar] [CrossRef] [Green Version]
- Exner, V.; Gruissem, W.; Hennig, L. Control of trichome branching by Chromatin Assembly Factor-I. BMC Plant. Biol. 2008, 8, 54–66. [Google Scholar] [CrossRef]
- Smith, S.; Stillman, B. Purification and characterization of CAF-1, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 1989, 58, 15–25. [Google Scholar] [CrossRef]
- Tyler, J.K.; Adams, C.R.; Chen, S.R.; Kobayashi, R.; Ramakaka, R.T.; Kodanaga, J.T. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 1999, 402, 555–560. [Google Scholar] [CrossRef]
- Kaya, H.; Shibahara, K.-I.; Taoka, K.-I.; Iwabuchi, M.; Stillman, B.; Araki, T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 2001, 104, 131–142. [Google Scholar] [CrossRef]
- Pavlištová, V.; Dvořáčková, M.; Jež, M.; Mozgová, I.; Mokroš, P.; Fajkus, J. Phenotypic reversion in fas mutants of Arabidopsis thaliana by reintroduction of FAS genes: Variable recovery of telomeres with major spatial rearrangements and transcriptional reprogramming of 45S rDNA genes. Plant. J. 2016, 88, 411–424. [Google Scholar] [CrossRef]
- Jarillo, J.A.; Piñeiro, M.; Cubas, P.; Martínez-Zapater, J.M. Chromatin remodeling in plant development. Int. J. Dev. Biol. 2009, 53, 1581–1596. [Google Scholar] [CrossRef] [Green Version]
- Berr, A.; Shafiq, S.; Shen, W.H. Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 2011, 1809, 567–576. [Google Scholar] [CrossRef]
- Candau, R.; Zhou, J.; Allis, C.D.; Berger, S.L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997, 16, 555–565. [Google Scholar] [CrossRef]
- Wang, T.; Jia, Q.; Wang, W.; Hussain, S.; Ahmed, S.; Adnan; Zhou, D.-X.; Ni, S.; Wang, S. GCN5 modulates trichome initiation in Arabidopsis by manipulating histone acetylation of core trichome initiation regulator genes. Plant. Cell Rep. 2019, 38, 755–765. [Google Scholar] [CrossRef]
- Kotak, J.; Saisana, M.; Gegas, V.; Pechlivani, N.; Kaldis, A.; Papoutsoglou, P.; Makris, A.; Burns, J.; Kendig, A.L.; Sheikh, M.; et al. The histone acetyltransferase GCN5 and the transcriptional coactivator ADA2b affect leaf development and trichome morphogenesis in Arabidopsis. Planta 2018, 248, 613–628. [Google Scholar] [CrossRef]
- Perron, M.P.; Provost, P. Protein interactions and complexes in human microRNA biogenesis and function. Front. Biosci. 2008, 13, 2537–2547. [Google Scholar] [CrossRef]
- Wightman, B.H.I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediated temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Wang, Q.; Cobb, G.P.; Anderson, T.A. Computational identification of microRNAs and their targets. Comput. Biol. Chem. 2006, 30, 395–407. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNA. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorshid, H.R.K.; Fard, S.S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Singh, N.; Srivastava, S.; Shasany, A.K.; Sharma, A. Identification of miRNAs and their targets involved in the secondary metabolic pathways of Mentha spp. Comput. Biol. Chem. 2016, 64, 154–162. [Google Scholar] [CrossRef]
- Singh, S.K.; Dhawan, S.S. Analyzing trichome and spatio-temporal expression of a cysteine protease gene Macunain in Macuna pruriens L. (DC). Protoplasma 2018, 255, 575–584. [Google Scholar] [CrossRef]
- Rodriguez, E.; Towers, G.; Mitchell, J. Biological activities of sesquiterpene lactones. Phytochemistry 1976, 15, 1573–1580. [Google Scholar] [CrossRef]
- Harada, A.; Sakata, K.; Ina, H.; Ina, K. Isolation and identification of xanthatin as an anti-attaching repellent against Blue Mussel. Agric. Biol. Chem. 1985, 49, 1887–1888. [Google Scholar] [CrossRef]
- Saxena, V.; Mondal, S. A xanthanolide from Xanthium strumarium. Phytochemistry 1994, 35, 1080–1082. [Google Scholar] [CrossRef]
- Fan, R.; Li, Y.; Li, C.; Zhang, Y. Differential microRNA analysis of glandular trichomes and young leaves in Xanthium strumarium L. reveals their putative roles in regulating terpenoid biosynthesis. PLoS ONE 2015, 10, e0139002. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant. Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef]
- Yu, N.; Cai, W.J.; Wang, S.; Shan, C.M.; Wang, L.J.; Chen, X.Y. Temporal control of trichome distribution by MicroRNA156-targeted SPL genes in Arabidopsis thaliana. Plant. Cell 2010, 22, 2322–2335. [Google Scholar] [CrossRef]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Wang, J.W.; Park, M.Y.; Wang, L.J.; Koo, Y.; Chen, X.Y.; Weigel, D.; Poethig, R.S. miRNA control of vegetative phase change in trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Shikata, M.; Koyama, T.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant. Cell Physiol. 2009, 50, 2133–2145. [Google Scholar] [CrossRef]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant. Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Wei, S.; Yu, B.; Gruber, M.Y.; Khachatourians, G.G.; Hegedus, D.D.; Hannoufa, A. Enhanced seed carotenoid levels and branching in transgenic Brassica napus expressing the Arabidopsis miR156b gene. J. Agric. Food Chem. 2010, 58, 9572–9578. [Google Scholar] [CrossRef]
- Fu, C.; Sunkar, R.; Zhou, C.; Shen, H.; Zhang, J.Y.; Matts, J.; Wolf, J.; Mann, D.G.J.; Stewart, C.N., Jr.; Tang, Y.; et al. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant. Biotechnol. J. 2012, 10, 443–452. [Google Scholar] [CrossRef]
- Aung, B.; Gruber, M.Y.; Amyot, L.; Omari, K.; Bertrand, A.; Hannoufa, A. MicroRNA156 as a promising tool for alfalfa improvement. Plant. Biotechnol. J. 2015, 13, 779–790. [Google Scholar] [CrossRef]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banrjee, A.K. MicroRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant. Physiol 2014, 164, 1011–1027. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant. Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Gao, R.; Austin, R.S.; Amyot, L.; Hannoufa, A. Comparative transcriptome investigation of global gene expression changes caused by miR156 overexpression in Medicago sativa. BMC Genomics 2016, 17, 658. [Google Scholar] [CrossRef]
- Zhang, T.Q.; Wang, J.W.; Zhou, C.M. The role of miR156 in developmental transitions in Nicotiana tabacum. Sci. China Life Sci. 2015, 58, 253–260. [Google Scholar] [CrossRef]
- Xue, X.-Y.; Zhao, B.; Chao, L.-M.; Chen, D.-Y.; Cui, W.-R.; Mao, Y.-B.; Wang, L.-J.; Chen, X.-Y. Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet. 2014, 10, e1004246. [Google Scholar] [CrossRef]
- Guan, X.; Pang, M.; Nah, G.; Shi, X.; Ye, W.; Stelly, D.M.; Chen, Z.J. miR828 and miR858 regulate homeologous MYB2 gene function in Arabidopsis trichome and cotton fibre development. Nat. Commun. 2014, 5, 3050. [Google Scholar] [CrossRef]
- Xie, F.; Wang, Q.; Sun, R.; Zhang, B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015, 66, 789–804. [Google Scholar] [CrossRef]
- McCaskill, D.; Gershenzon, J.; Croteau, R. Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha piperita L.). Planta 1992, 187, 445–454. [Google Scholar] [CrossRef]
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef]
- Sun, G. MicroRNAs and their diverse functions in plants. Plant. Mol. Biol. 2012, 80, 17–36. [Google Scholar] [CrossRef]
- Liu, H.; Liu, S.; Jiao, J.; Lu, T.J.; Xu, F. Trichomes as a natural biophysical barrier for plants and their bioinspired applications. Soft Matter. 2017, 13, 5096–5106. [Google Scholar] [CrossRef]
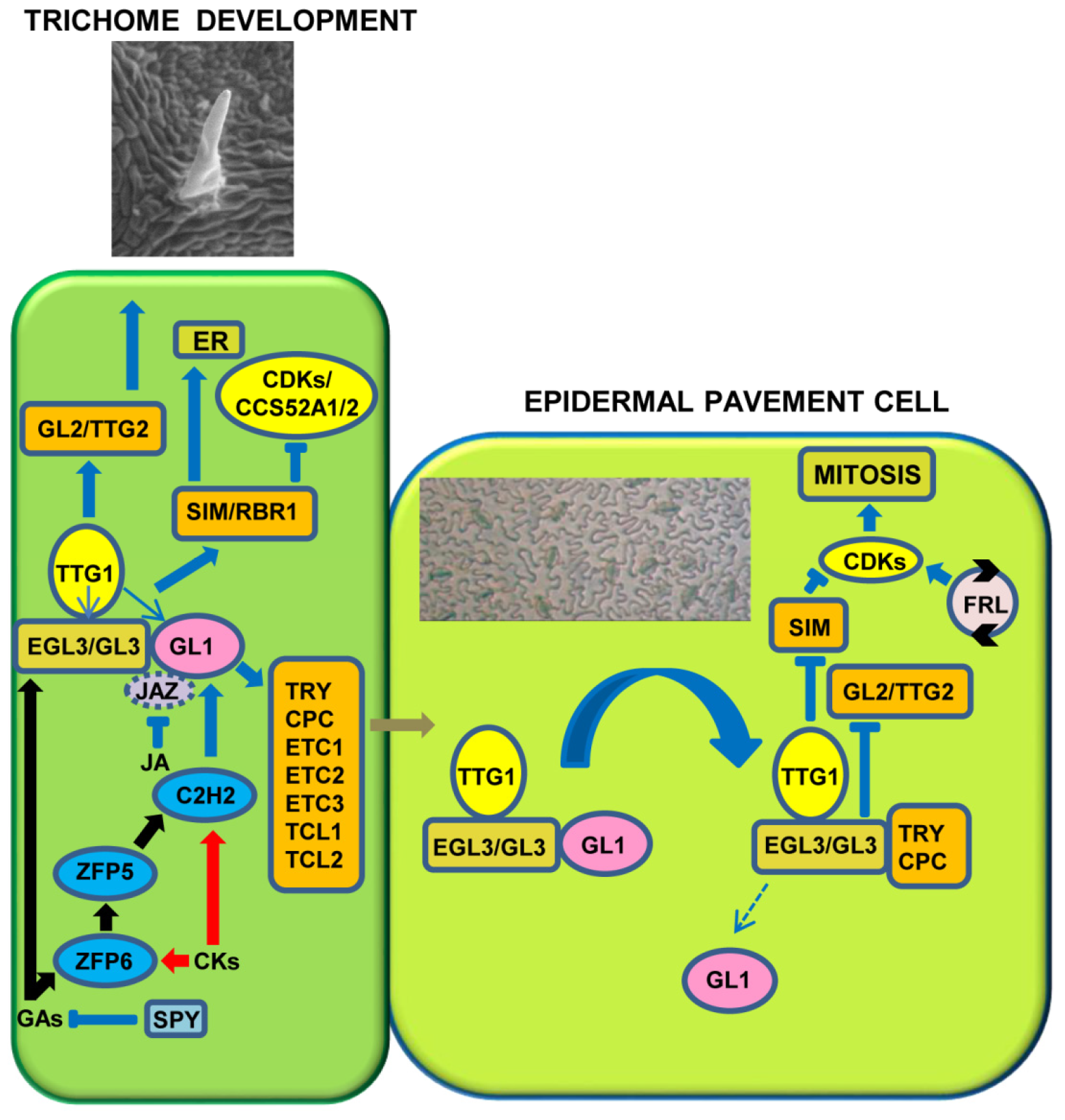
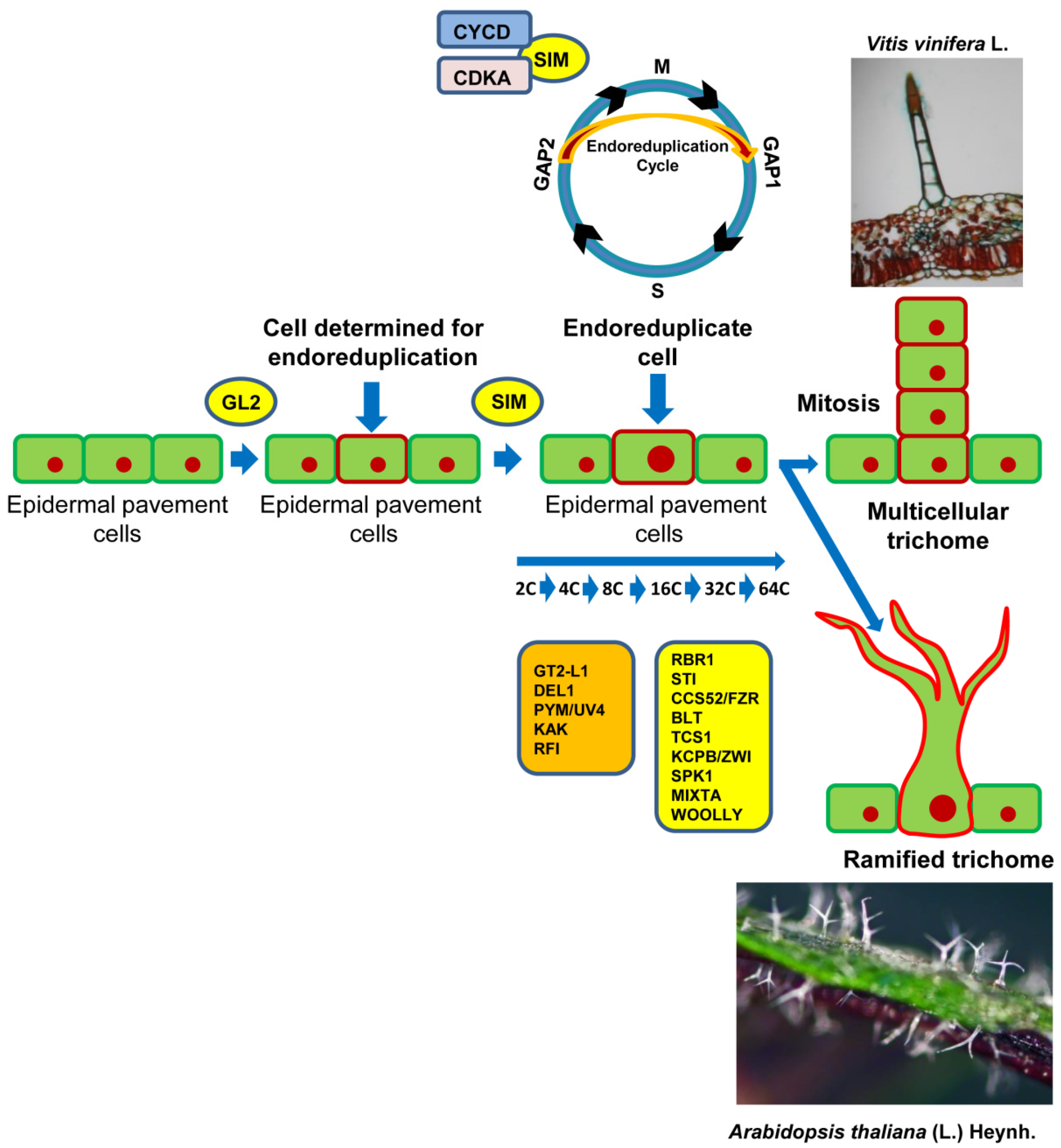
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fambrini, M.; Pugliesi, C. The Dynamic Genetic-Hormonal Regulatory Network Controlling the Trichome Development in Leaves. Plants 2019, 8, 253. https://doi.org/10.3390/plants8080253
Fambrini M, Pugliesi C. The Dynamic Genetic-Hormonal Regulatory Network Controlling the Trichome Development in Leaves. Plants. 2019; 8(8):253. https://doi.org/10.3390/plants8080253
Chicago/Turabian StyleFambrini, Marco, and Claudio Pugliesi. 2019. "The Dynamic Genetic-Hormonal Regulatory Network Controlling the Trichome Development in Leaves" Plants 8, no. 8: 253. https://doi.org/10.3390/plants8080253




