Portuguese Common Bean Natural Variation Helps to Clarify the Genetic Architecture of the Legume’s Nutritional Composition and Protein Quality
Abstract
:1. Introduction
2. Results
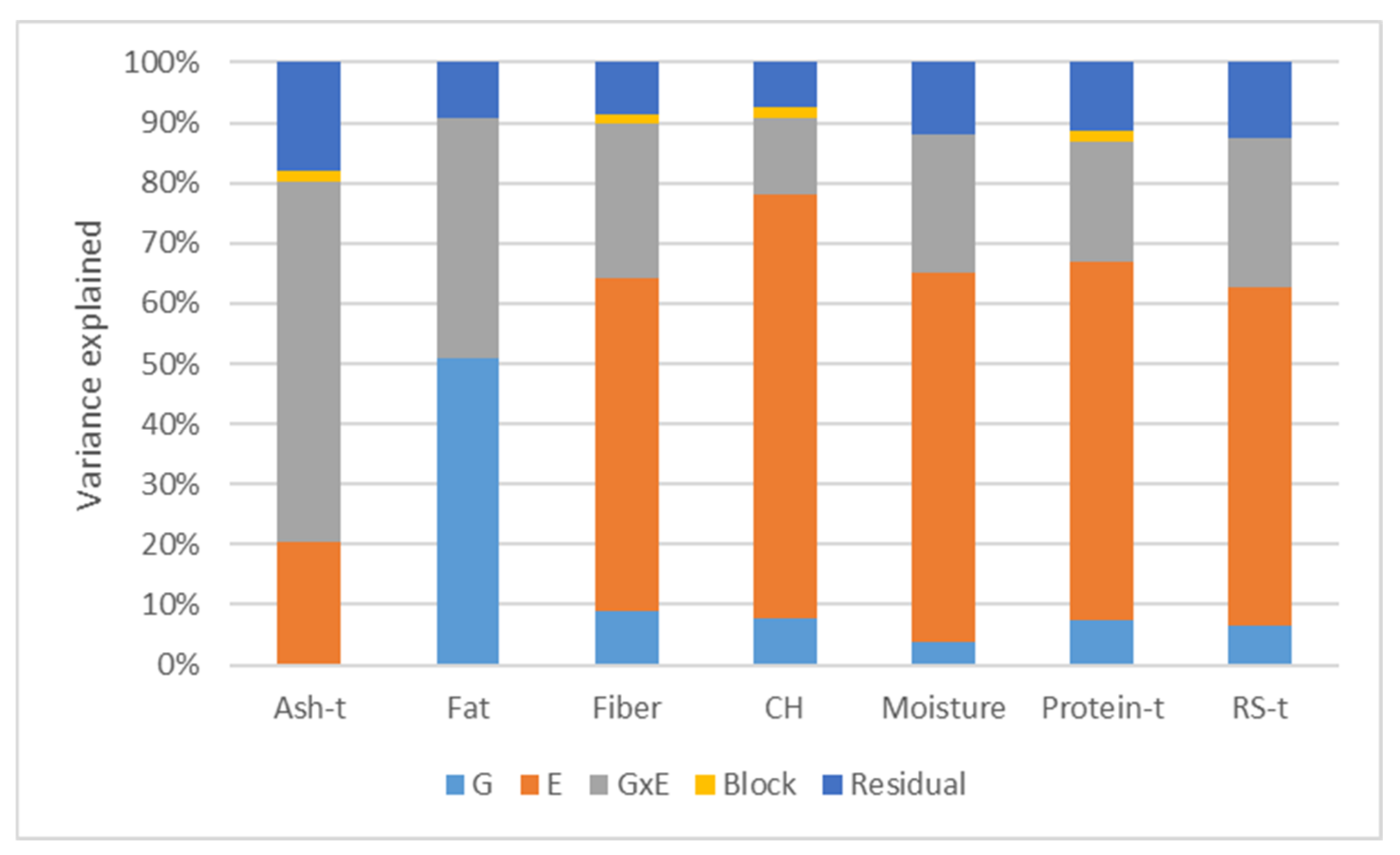
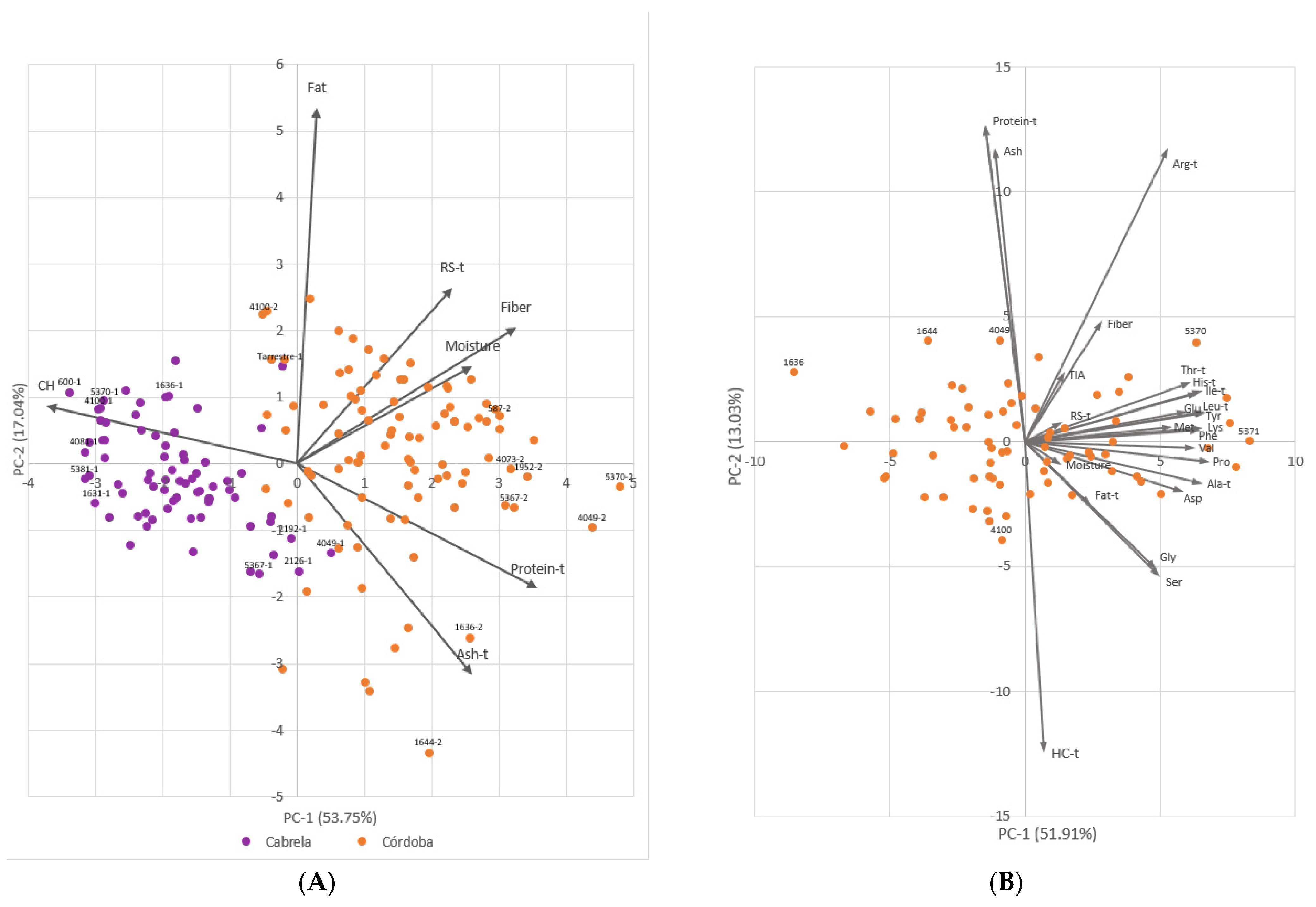
2.1. Phenotypic Trait Variation
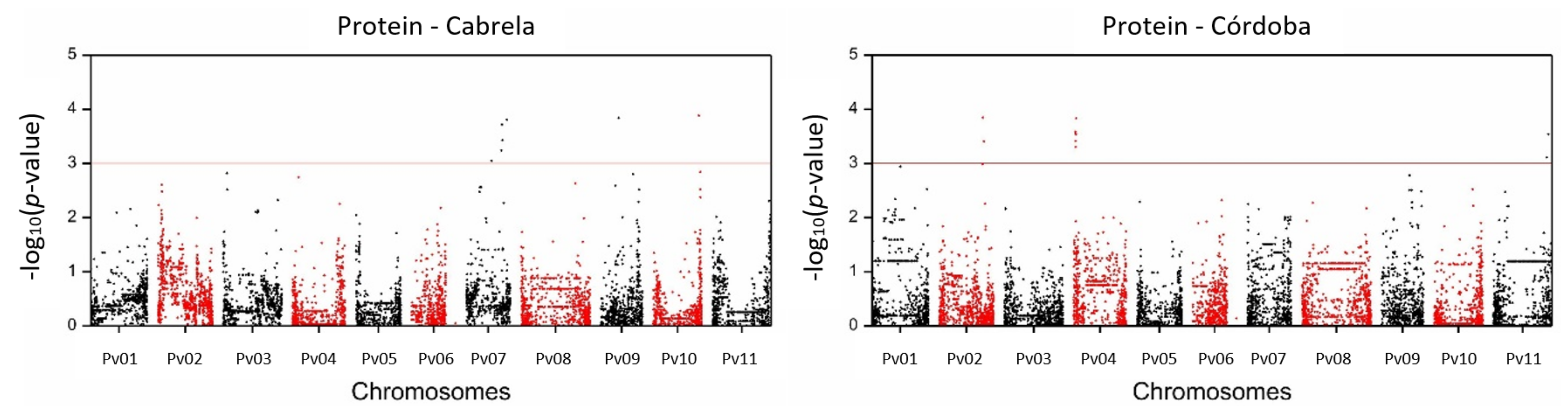
2.2. SNP-Trait Associations
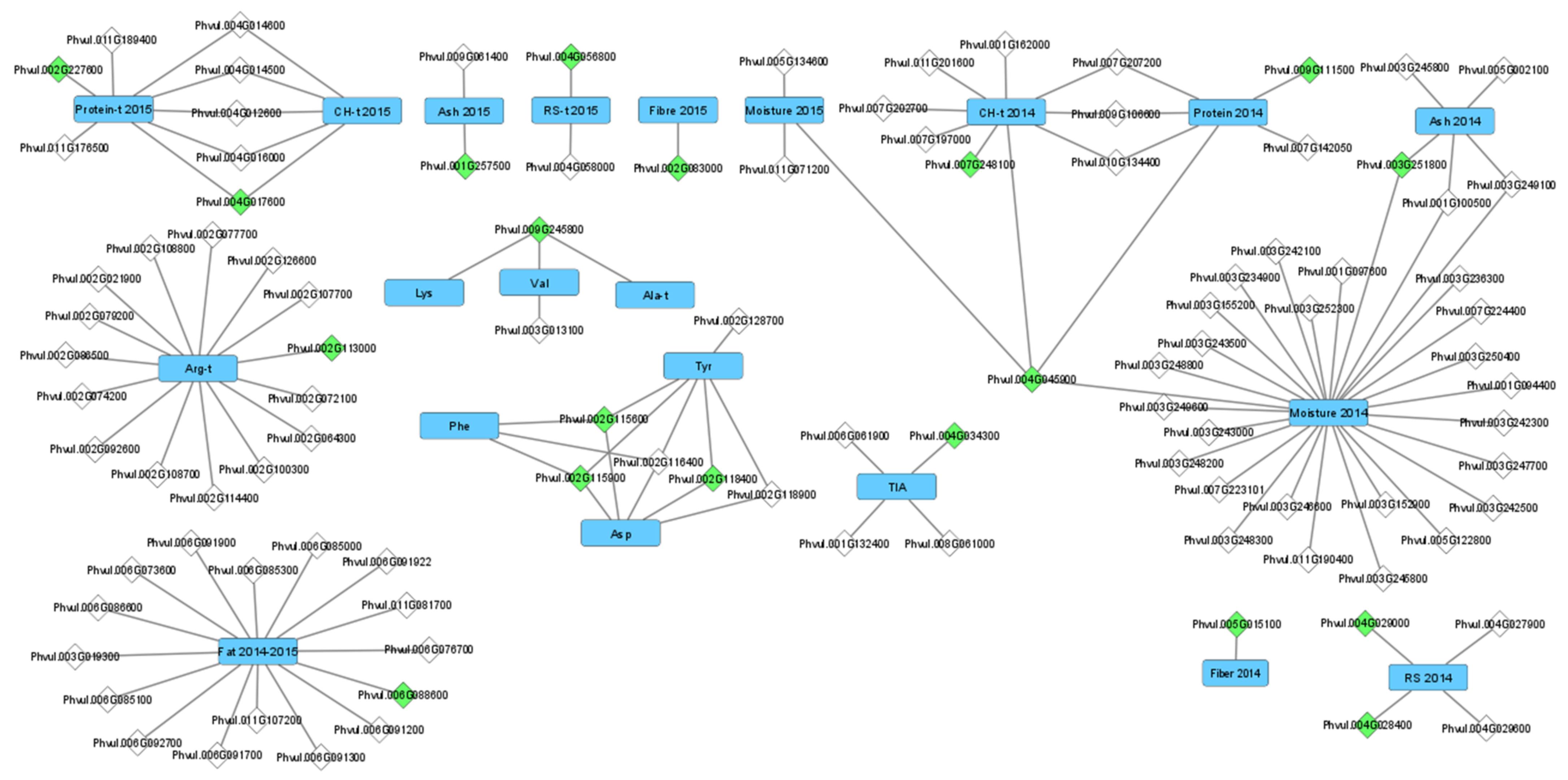
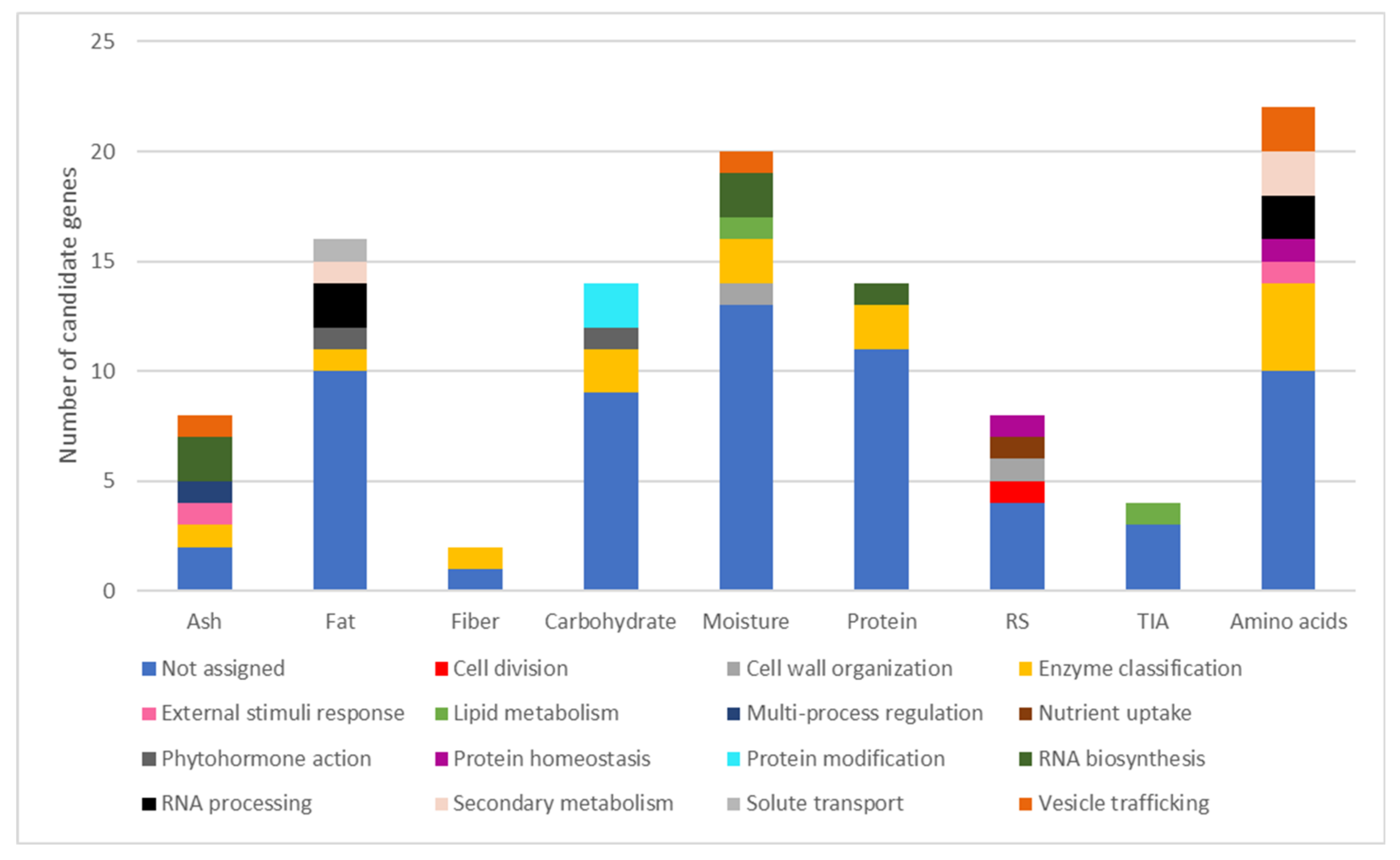
2.3. Candidate Gene Identification
3. Discussion
3.1. Genomic Regions and Candidate Genes Associated with Common Bean Nutritional Composition and Protein Quality-Related Traits
3.2. Portuguese Common Bean Germplasm Nutritional Quality Richness and the Influence of Environment
4. Materials and Methods
4.1. Plant Material and Growing Conditions
4.2. Phenotypic Data Acquisition
4.3. Phenotypic Data Analysis
4.4. Genotypic Data
4.4.1. Association-Mapping Analysis
4.4.2. Candidate Gene Identification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boye, J.; Zare, F.; Pletch, A. Pulse Proteins: Processing, Characterization, Functional Properties and Applications in Food and Feed. Food Res. Int. 2010, 43, 414–431. [Google Scholar] [CrossRef]
- Câmara, C.R.S.; Urrea, C.A.; Schlegel, V. Pinto Beans (Phaseolus vulgaris L.) as a Functional Food: Implications on Human Health. Agriculture 2013, 3, 90–111. [Google Scholar] [CrossRef]
- Rawal, V.; Navarro, D.K. The Global Economy of Pulses, 2019 ed.; FAO: Rome, Italy, 2019. [Google Scholar]
- Petry, N.; Boy, E.; Wirth, J.P.; Hurrell, R.F. Review: The Potential of the Common Bean (Phaseolus vulgaris) as a Vehicle for Iron Biofortification. Nutrients 2015, 7, 1144–1173. [Google Scholar] [CrossRef] [Green Version]
- Kalogeropoulos, N.; Chiou, A.; Ioannou, M.; Karathanos, V.T.; Hassapidou, M.; Andrikopoulos, N.K. Nutritional Evaluation and Bioactive Microconstituents (Phytosterols, Tocopherols, Polyphenols, Triterpenic Acids) in Cooked Dry Legumes Usually Consumed in the Mediterranean Countries. Food Chem. 2010, 121, 682–690. [Google Scholar] [CrossRef]
- Mecha, E.; Figueira, M.E.; Vaz Patto, M.C.; do Rosario Bronze, M. Two Sides of the Same Coin: The Impact of Grain Legumes on Human Health: Common Bean (Phaseolus vulgaris L.) as a Case Study. In Legume Seed Nutraceutical Research; Jimenez-Lopez, J.C., Clemente, A., Eds.; IntechOpen: London, UK, 2018; ISBN 978-1-78985-397-1. [Google Scholar]
- Escribano, M.R.; Santalla, M.; de Ron, A.M. Genetic Diversity in Pod and Seed Quality Traits of Common Bean Populations from Northwestern Spain. Euphytica 1997, 93, 71–81. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Araújo, S.S. Positioning Portugal into the Context of World Production and Research in Grain Legumes. Rev. Ciênc. Agrár. 2016, 39, 471–489. [Google Scholar] [CrossRef]
- Gaikwad, K.B.; Rani, S.; Kumar, M.; Gupta, V.; Babu, P.H.; Bainsla, N.K.; Yadav, R. Enhancing the Nutritional Quality of Major Food Crops Through Conventional and Genomics-Assisted Breeding. Front. Nutr. 2020, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Vaz Patto, M.C.; Amarowicz, R.; Aryee, A.N.A.; Boye, J.I.; Chung, H.-J.; Martín-Cabrejas, M.A.; Domoney, C. Achievements and Challenges in Improving the Nutritional Quality of Food Legumes. Crit. Rev. Plant Sci. 2015, 34, 105–143. [Google Scholar] [CrossRef]
- Vaz Patto, M.C. Grain Legume Protein Quality: A Hot Subject. Arbor 2016, 192, a314. [Google Scholar] [CrossRef] [Green Version]
- Boye, J.; Wijesinha-Bettoni, R.; Burlingame, B. Protein Quality Evaluation Twenty Years after the Introduction of the Protein Digestibility Corrected Amino Acid Score Method. Br. J. Nutr. 2012, 108, S183–S211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.L.; Domoney, C.; Hedley, C.L.; Casey, R.; Grusak, M.A. Can We Improve the Nutritional Quality of Legume Seeds? Plant Physiol. 2003, 131, 886–891. [Google Scholar] [CrossRef] [Green Version]
- Blair, M.W.; Astudillo, C.; Grusak, M.A.; Graham, R.; Beebe, S.E. Inheritance of Seed Iron and Zinc Concentrations in Common Bean (Phaseolus vulgaris L.). Mol. Breed. 2009, 23, 197–207. [Google Scholar] [CrossRef]
- Guzmán-Maldonado, S.H.; Martínez, O.; Acosta-Gallegos, J.A.; Guevara-Lara, F.; Paredes-López, O. Putative Quantitative Trait Loci for Physical and Chemical Components of Common Bean. Crop Sci. 2003, 43, 1029–1035. [Google Scholar] [CrossRef]
- Delfini, J.; Moda-Cirino, V.; dos Santos Neto, J.; Zeffa, D.M.; Nogueira, A.F.; Ribeiro, L.A.B.; Ruas, P.M.; Gepts, P.; Gonçalves, L.S.A. Genome-Wide Association Study for Grain Mineral Content in a Brazilian Common Bean Diversity Panel. Theor. Appl. Genet. 2021, 134, 2795–2811. [Google Scholar] [CrossRef] [PubMed]
- Gunjača, J.; Carović-Stanko, K.; Lazarević, B.; Vidak, M.; Petek, M.; Liber, Z.; Šatović, Z. Genome-Wide Association Studies of Mineral Content in Common Bean. Front. Plant Sci. 2021, 12, 305. [Google Scholar] [CrossRef]
- Casañas, F.; Pérez-Vega, E.; Almirall, A.; Plans, M.; Sabaté, J.; Ferreira, J.J. Mapping of QTL Associated with Seed Chemical Content in a RIL Population of Common Bean (Phaseolus vulgaris L.). Euphytica 2013, 192, 279–288. [Google Scholar] [CrossRef]
- Katuuramu, D.N.; Hart, J.P.; Porch, T.G.; Grusak, M.A.; Glahn, R.P.; Cichy, K.A. Genome-Wide Association Analysis of Nutritional Composition-Related Traits and Iron Bioavailability in Cooked Dry Beans (Phaseolus vulgaris L.). Mol. Breed. 2018, 38, 44. [Google Scholar] [CrossRef]
- Mitchell-Olds, T. Complex-Trait Analysis in Plants. Genome Biol. 2010, 11, 113. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A Reference Genome for Common Bean and Genome-Wide Analysis of Dual Domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alqudah, A.M.; Sallam, A.; Stephen Baenziger, P.; Börner, A. GWAS: Fast-Forwarding Gene Identification and Characterization in Temperate Cereals: Lessons from Barley—A Review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef]
- Santalla, M.; Rodiño, A.; De Ron, A. Allozyme Evidence Supporting Southwestern Europe as a Secondary Center of Genetic Diversity for the Common Bean. Theor. Appl. Genet. 2002, 104, 934–944. [Google Scholar] [CrossRef]
- Leitão, S.T.; Dinis, M.; Veloso, M.M.; Šatović, Z.; Vaz Patto, M.C. Establishing the Bases for Introducing the Unexplored Portuguese Common Bean Germplasm into the Breeding World. Front. Plant Sci. 2017, 8, 1296. [Google Scholar] [CrossRef] [Green Version]
- Mecha, E.; Natalello, S.; Carbas, B.; da Silva, A.B.; Leitão, S.T.; Brites, C.; Veloso, M.M.; Rubiales, D.; Costa, J.; de Fatima Cabral, M.; et al. Disclosing the Nutritional Quality Diversity of Portuguese Common Beans—The Missing Link for Their Effective Use in Protein Quality Breeding Programs. Agronomy 2021, 11, 221. [Google Scholar] [CrossRef]
- Leitão, S.T.; Malosetti, M.; Song, Q.; van Eeuwijk, F.; Rubiales, D.; Vaz Patto, M.C. Natural Variation in Portuguese Common Bean Germplasm Reveals New Sources of Resistance Against Fusarium oxysporum f. sp. Phaseoli and Resistance-Associated Candidate Genes. Phytopathology 2020, 110, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Leitão, S.T.; Bicho, M.C.; Pereira, P.; Paulo, M.J.; Malosetti, M.; de Sousa Araújo, S.; van Eeuwijk, F.; Vaz Patto, M.C. Common Bean SNP Alleles and Candidate Genes Affecting Photosynthesis under Contrasting Water Regimes. Hortic. Res. 2021, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Considine, M.J.; Siddique, K.H.M.; Foyer, C.H. Nature’s Pulse Power: Legumes, Food Security and Climate Change. J. Exp. Bot. 2017, 68, 1815–1818. [Google Scholar] [CrossRef]
- Magrini, M.-B.; Anton, M.; Cholez, C.; Corre-Hellou, G.; Duc, G.; Jeuffroy, M.-H.; Meynard, J.-M.; Pelzer, E.; Voisin, A.-S.; Walrand, S. Why Are Grain-Legumes Rarely Present in Cropping Systems despite Their Environmental and Nutritional Benefits? Analyzing Lock-in in the French Agrifood System. Ecol. Econ. 2016, 126, 152–162. [Google Scholar] [CrossRef]
- Mazau, D.; Esquerré-Tugayé, M.T. Hydroxyproline-Rich Glycoprotein Accumulation in the Cell Walls of Plants Infected by Various Pathogens. Physiol. Mol. Plant Pathol. 1986, 29, 147–157. [Google Scholar] [CrossRef]
- Showalter, A.M.; Bell, J.N.; Cramer, C.L.; Bailey, J.A.; Varner, J.E.; Lamb, C.J. Accumulation of Hydroxyproline-Rich Glycoprotein MRNAs in Response to Fungal Elicitor and Infection. Proc. Natl. Acad. Sci. USA 1985, 82, 6551–6555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kumar Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional Identification of an Arabidopsis Pectin Biosynthetic Homogalacturonan Galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voragen, A.G.J.; Coenen, G.-J.; Verhoef, R.P.; Schols, H.A. Pectin, a Versatile Polysaccharide Present in Plant Cell Walls. Struct. Chem. 2009, 20, 263. [Google Scholar] [CrossRef] [Green Version]
- Bernardo, R. Breeding for Quantitative Traits in Plants, 3rd ed.; Stemma Press: Woodbury, MN, USA, 2020; ISBN 978-0-9720724-3-4. [Google Scholar]
- Wirdnam, C.; Motoyama, A.; Arn-Bouldoires, E.; van Eeden, S.; Iglesias, A.; Meins, F. Altered Expression of an Ankyrin-Repeat Protein Results in Leaf Abnormalities, Necrotic Lesions, and the Elaboration of a Systemic Signal. Plant Mol. Biol. 2004, 56, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Murgia, I.; Tarantino, D.; Soave, C.; Morandini, P. Arabidopsis CYP82C4 Expression Is Dependent on Fe Availability and Circadian Rhythm, and Correlates with Genes Involved in the Early Fe Deficiency Response. J. Plant Physiol. 2011, 168, 894–902. [Google Scholar] [CrossRef]
- Ma, H.; Cao, X.; Shi, S.; Li, S.; Gao, J.; Ma, Y.; Zhao, Q.; Chen, Q. Genome-Wide Survey and Expression Analysis of the Amino Acid Transporter Superfamily in Potato (Solanum tuberosum L.). Plant Physiol. Biochem. 2016, 107, 164–177. [Google Scholar] [CrossRef]
- Sehgal, A.; Sita, K.; Siddique, K.H.M.; Kumar, R.; Bhogireddy, S.; Varshney, R.K.; HanumanthaRao, B.; Nair, R.M.; Prasad, P.V.V.; Nayyar, H. Drought or/and Heat-Stress Effects on Seed Filling in Food Crops: Impacts on Functional Biochemistry, Seed Yields, and Nutritional Quality. Front. Plant Sci. 2018, 9, 1705. [Google Scholar] [CrossRef] [Green Version]
- Deryng, D.; Conway, D.; Ramankutty, N.; Price, J.; Warren, R. Global Crop Yield Response to Extreme Heat Stress under Multiple Climate Change Futures. Environ. Res. Lett. 2014, 9, 034011. [Google Scholar] [CrossRef] [Green Version]
- Sita, K.; Sehgal, A.; HanumanthaRao, B.; Nair, R.M.; Vara Prasad, P.V.; Kumar, S.; Gaur, P.M.; Farooq, M.; Siddique, K.H.M.; Varshney, R.K.; et al. Food Legumes and Rising Temperatures: Effects, Adaptive Functional Mechanisms Specific to Reproductive Growth Stage and Strategies to Improve Heat Tolerance. Front. Plant Sci. 2017, 8, 1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Soda, M.; Malosetti, M.; Zwaan, B.J.; Koornneef, M.; Aarts, M.G.M. Genotype × Environment Interaction QTL Mapping in Plants: Lessons from Arabidopsis. Trends Plant Sci. 2014, 19, 390–398. [Google Scholar] [CrossRef]
- Nosworthy, M.G.; House, J.D. Factors Influencing the Quality of Dietary Proteins: Implications for Pulses. Cereal Chem. 2017, 94, 49–57. [Google Scholar] [CrossRef]
- Montoya, C.A.; Lallès, J.-P.; Beebe, S.; Leterme, P. Phaseolin Diversity as a Possible Strategy to Improve the Nutritional Value of Common Beans (Phaseolus vulgaris). Food Res. Int. 2010, 43, 443–449. [Google Scholar] [CrossRef]
- Wang, N.; Hatcher, D.W.; Tyler, R.T.; Toews, R.; Gawalko, E.J. Effect of Cooking on the Composition of Beans (Phaseolus vulgaris L.) and Chickpeas (Cicer Arietinum L.). Food Res. Int. 2010, 43, 589–594. [Google Scholar] [CrossRef]
- American Association of Cereal Chemists (AACC). Method 32-40.01 Resistant Starch Assay Procedure; AACC: St. Paul, MN, USA, 2009. [Google Scholar]
- Association of Analytical Chemists (AOAC). Method 2002.02 Resistant Starch Assay Procedure; AACC: St. Paul, MN, USA, 2002. [Google Scholar]
- VSN International. Genstat for Windows, 21st ed.; VSN International: Hemel Hempstead, UK, 2020. [Google Scholar]
- Malosetti, M.; van der Linden, C.G.; Vosman, B.; van Eeuwijk, F.A. A Mixed-Model Approach to Association Mapping Using Pedigree Information with an Illustration of Resistance to Phytophthora Infestans in Potato. Genetics 2007, 175, 879–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, M.L.; Carbas, B.; Gaspar, D.; Paulo, M.; Brites, C.; Mendes-Moreira, P.; Brites, C.M.; Malosetti, M.; van Eeuwijk, F.; Vaz Patto, M.C. Genome-Wide Association Study for Kernel Composition and Flour Pasting Behavior in Wholemeal Maize Flour. BMC Plant Biol. 2019, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Parker, C.C.; Abney, M.; Palmer, A.A. Practical Considerations Regarding the Use of Genotype and Pedigree Data to Model Relatedness in the Context of Genome-Wide Association Studies. G3 Genes Genomes Genet. 2013, 3, 1861–1867. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Yekutieli, D. The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Duggal, P.; Gillanders, E.M.; Holmes, T.N.; Bailey-Wilson, J.E. Establishing an Adjusted P-Value Threshold to Control the Family-Wide Type 1 Error in Genome Wide Association Studies. BMC Genom. 2008, 9, 516. [Google Scholar] [CrossRef] [Green Version]
- Resende, R.T.; Resende, M.D.V.; Silva, F.F.; Azevedo, C.F.; Takahashi, E.K.; Silva-Junior, O.B.; Grattapaglia, D. Regional Heritability Mapping and Genome-Wide Association Identify Loci for Complex Growth, Wood and Disease Resistance Traits in Eucalyptus. New Phytol. 2017, 213, 1287–1300. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Schwacke, R.; Ponce-Soto, G.Y.; Krause, K.; Bolger, A.M.; Arsova, B.; Hallab, A.; Gruden, K.; Stitt, M.; Bolger, M.E.; Usadel, B. MapMan4: A Refined Protein Classification and Annotation Framework Applicable to Multi-Omics Data Analysis. Mol. Plant 2019, 12, 879–892. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
| Ash | Fat | Fiber | CH | Moisture | Protein | RS | |
|---|---|---|---|---|---|---|---|
| Cabrela | 3.16 ± 0.08 a (2.6) | 1.44 ± 0.24 a (16.4) | 5.75 ± 0.45 a (7.8) | 60.57 ± 1.53 a (2.5) | 13.55 ± 0.47 a (3.5) | 21.28 ± 1.44 a (6.8) | 30.74 ± 3.49 a (11.4) |
| Córdoba | 3.25 ± 0.13 b (4.1) | 1.49 ± 0.32 a (21.7) | 6.77 ± 0.72 b (10.7) | 56.65 ± 1.75 b (3.1) | 14.48 ± 0.51 b (3.5) | 24.14 ± 1.66 b (6.9) | 45.23 ± 11.36 b (25.1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, F.A.; Leitão, S.T.; Correia, V.; Mecha, E.; Rubiales, D.; Bronze, M.R.; Vaz Patto, M.C. Portuguese Common Bean Natural Variation Helps to Clarify the Genetic Architecture of the Legume’s Nutritional Composition and Protein Quality. Plants 2022, 11, 26. https://doi.org/10.3390/plants11010026
Mendes FA, Leitão ST, Correia V, Mecha E, Rubiales D, Bronze MR, Vaz Patto MC. Portuguese Common Bean Natural Variation Helps to Clarify the Genetic Architecture of the Legume’s Nutritional Composition and Protein Quality. Plants. 2022; 11(1):26. https://doi.org/10.3390/plants11010026
Chicago/Turabian StyleMendes, Francisco A., Susana T. Leitão, Verónica Correia, Elsa Mecha, Diego Rubiales, Maria Rosário Bronze, and Maria Carlota Vaz Patto. 2022. "Portuguese Common Bean Natural Variation Helps to Clarify the Genetic Architecture of the Legume’s Nutritional Composition and Protein Quality" Plants 11, no. 1: 26. https://doi.org/10.3390/plants11010026








