1. Introduction
To improve the organoleptic properties of food products, the rapidly developing food industry requires increasing amounts of flavoring compounds, particularly those obtained naturally. This category includes pyrazines with varying flavor and aroma properties, with a predominant hint of nut and roast, almond, and even chocolate or grass [
1]. Pyrazines are heterocyclic organic compounds containing two nitrogen atoms in the ring, and found in plants (e.g., green peas), insects (Mediterranean fruit fly,
Ceratitis capitata), fungi, and bacteria. These substances act as odor signals, scaring predators away and effectively protecting vegetative tissues and fruits from being eaten. As a result, pyrazines are used in pesticides and insecticides. Pyrazines are also used in the production of dyes and pharmaceuticals due to their properties [
1,
2,
3,
4,
5,
6,
7]. They are formed during the thermal processing of food as a result of non-enzymatic browning (Maillard reactions), and are responsible for the characteristic smell and color. The mechanism of pyrazines formation is still unknown [
8].
The formation of pyrazines was also described during the barothermic treatment of starch raw materials used in the production of ethanol, implying that they can be extracted, for example, from volatile by-products of alcoholic fermentation after rectification [
9]. Natural, biologically produced pyrazines are classified into three types. The first class consist of pyrazines with up to four alkyl substituents, primarily methyl or ethyl, such as those produced by
Bacillus bacteria (
Figure 1).
Pyrazines with one or two branched side chains, such as 2-isopropylpyrazine or 2,6-diisopropylpyrazine, are classified in the second class, and methoxy-pyrazines, which typically have one or two alkyl substituents, such as methoxy-3-isopropylpyrazine, are classified in the third class [
10].
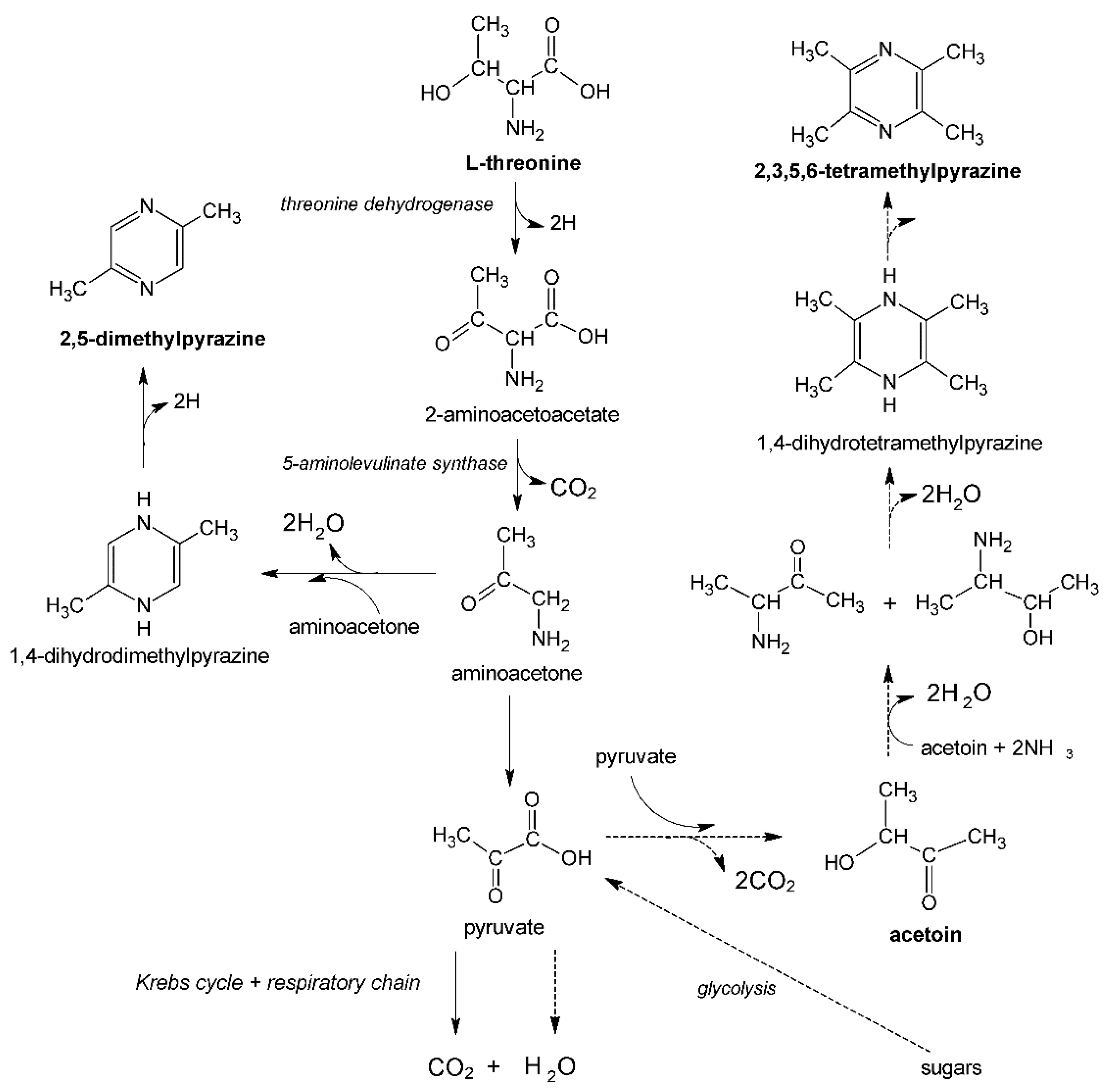
In addition to sugars as a carbon source, pyrazines precursors, such as L-threonine (2,5-dimethylpyrazine precursor) or acetoin (2,3,5,6-tetramethylpyrazine precursor), are required in pyrazines biosynthesis using
Bacillus bacteria (
Figure 2). The presence of the above-mentioned precursors in the culture medium may also stimulate the production of other first-class pyrazines (the biosynthesis mechanism of these compounds by various groups of microorganisms is still poorly understood) [
11].
The use of biological systems to produce pyrazines is currently of great interest in the food industry because consumers prefer natural ingredients in food production. Furthermore, pyrazine biosynthesis, as opposed to chemical synthesis, necessitates milder process conditions and employs substances that do not pollute the natural environment [
2,
12]. Pyrazine preparations can also be obtained by extracting these compounds, primarily through reduced pressure distillation [
13]. A high concentration of these compounds in the post-culture medium is required for efficient and, above all, cost-effective extraction of pyrazines. The use of an effective producer strain, the search for which is one of the more labor-intensive stages of development and further optimization of the biosynthesis process, is the main factor determining the high concentration of pyrazines in the solution [
1]. Finding an efficient pyrazines producer through screening tests is a prerequisite for the next steps of the biosynthetic process, which include optimizing culture conditions, metabolic modification of microorganisms, and increasing the production scale in bioreactor systems [
12,
14,
15].
So far, research has primarily focused on optimizing culture conditions (precursor concentration, temperature, aeration rate, and culture time) and metabolic modification of isolated
Bacillus strains to ensure high-yield production of 2,5-dimethylpyrazine and 2,3,5,6-tetramethylpyrazine [
11,
14,
16,
17]. There is no information in the literature about screening tests to isolate bacterial strains capable of producing alkylpyrazines, which are part of the first class of pyrazines.
The aim of this study was to evaluate Bacillus subtilis strains that synthesize alkylpyrazine from natto fermented soybeans. The study aimed to isolate an effective producer of alkylpyrazines within the B. subtilis species due to its widespread use in the food industry and GRAS status (generally recognized as safe), which predisposes this microorganism to the production of food additives. Due to the high protein content of soybeans, which can be a source of L-threonine, a precursor in alkylpyrazine biosynthesis, the use of fermented soybeans as a potential source of alkylpyrazine producing microorganisms appears justified. The scope of the study was extended to include, in addition to the most commonly reported biosynthesis of 2,5-dimethylpyrazine and 2,3,5,6-tetramethylpyrazine by B. subtilis strains, as well as the production of difficult-to-biosynthesize pyrazines such as 2-methylpyrazine, 2,3-dimethylpyrazine, 2,6-dimethylpyrazine and 2,3,5-trimethylpyrazine, which is novel in this work. The current study investigated the ability of B. subtilis bacteria isolated from natto to produce alkylpyrazines. The findings of this study can serve as a foundation for future research into the use of highly effective pyrazines producer strains. These studies should concentrate on optimizing culture conditions, medium composition, and increasing production scale through the use of bioreactor techniques. This screening study goes above and beyond the studies published by other authors. It provides a thorough examination of B. subtilis abilities to synthesize a broader range of alkylpyrazines (six different compounds) isolated from fermented foods. So far, screening studies have only revealed that isolated bacterial strains are capable of producing 2,3,5,6-tetramethylpyrazine. At the same time, the effectiveness of screening for a specific study material has only been evaluated in a limited way. The current study also aimed to evaluate the effectiveness of the screening by determining the minimum number of B. subtilis cultures isolated from fermented natto soybean that would provide a bacterial strain capable of producing the broadest profile of alkylpyrazines. These compounds can then be used in food as additives to achieve the desired sensory properties. A detailed analysis of the concentrations of six different alkylpyrazines in all isolated bacterial cultures (a first in previous studies) enabled the researchers to identify the most effective producer of the analyzed alkylpyrazines as well as the producer of a specific alkylpyrazine profile. This research strategy is an innovative approach to isolating bacterial strains with distinct metabolic features, with the potential to dedicate specific isolates to different branches of the food industry based on the expected pyrazine biosynthesis profile.
3. Results and Discussion
To test the ability of isolated B. subtilis strains to biosynthesize pyrazines, the concentration of these compounds in the media was determined after autoclaving (but before culturing the bacteria) and after the culturing was completed. The concentration of individual pyrazines resulting from autoclaving pyrazine precursor-containing media was then subtracted from the concentration of these compounds after bacteria cultivation. In this way, the ability of the isolated microorganisms to convert pyrazine precursors (L-threonine and acetoin) was determined. The analyzed samples contained up to six different compounds classified as alkylpyrazines. The following pyrazines were found in the post-culture media: 2,5-dimethylpyrazine and 2,3,5,6-tetramethylpyrazine (their precursors were added to the media), as well as 2-methylpyrazine, 2,3-dimethylpyrazine, 2,6-dimethylpyrazine, and 2,3,5-trimethylpyrazine. This demonstrated that B. subtilis isolated from fermented soybeans can produce a wide range of alkylpyrazines. However, the isolated bacterial strains showed different production efficiency of individual pyrazines: 2-methylpyrazine, 2,3-dimethylpyrazine and 2,6-dimethylpyrazine were produced in smaller amounts (µg/L), while 2,5-dimethylpyrazine, 2,3,5-trimethylpyrazine and 2,3,5,6-tetramethylpyrazine were produced in larger quantities (mg/L).
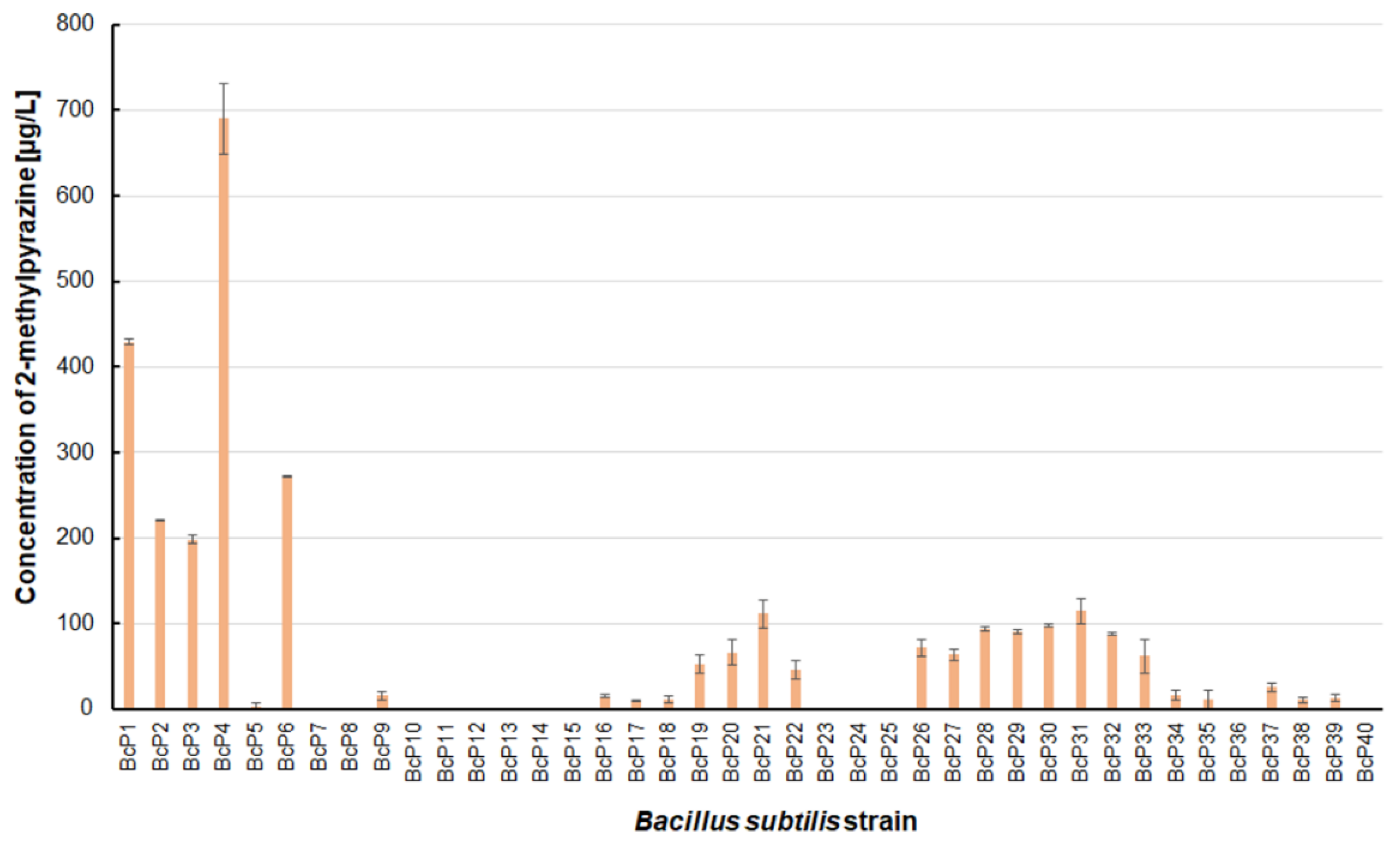
Only five isolated
B. subtilis strains could biosynthesize 2-methylpyrazine at concentrations greater than 200 µg/L. The BcP4 strain showed the highest ability to synthesize this pyrazine, giving 690.0 ± 40.5 µg/L (
Figure 3). The BcP1 strain was the second most effective producer of 2-methylpyrazine, producing it at a concentration of more than 400 µg/L. Only two of the remaining 35
B. subtilis strains (BcP21, BcP31) produced 2-methylpyrazine at concentrations greater than 100 µg/L (
Figure 3).
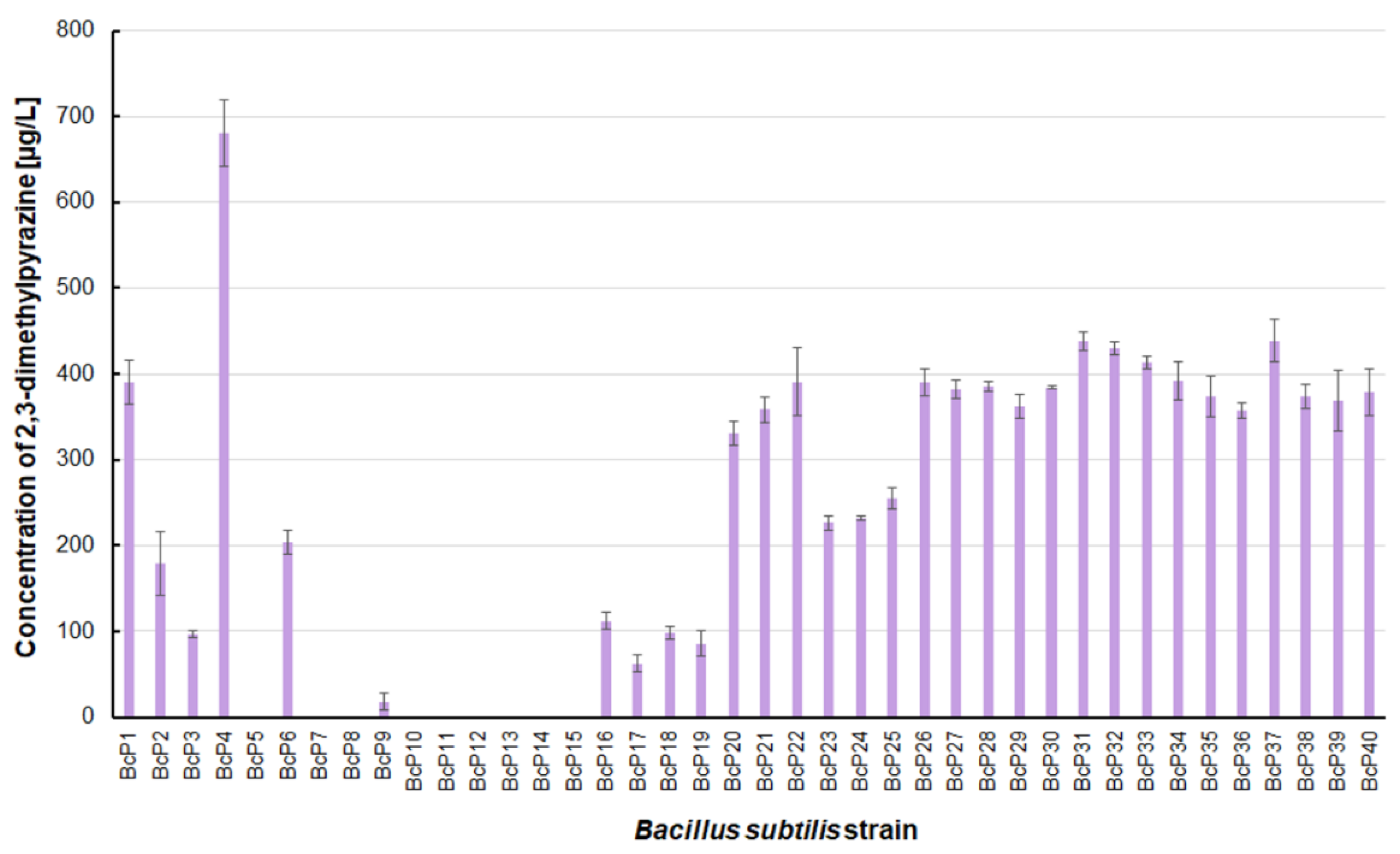
The
B. subtilis BcP4 strain also produced the most 2,3-dimethylpyrazine, with a concentration of 680.4 ± 38.6 µg/L (
Figure 4). Twenty isolates were found to be capable of producing 2,3-dimethylpyrazine at concentrations greater than 300 µg/L. Strains BcP7-8 and BcP10-15, like 2-methylpyrazine, were unable to biosynthesize 2,3-dimethylpyrazine (
Figure 4).
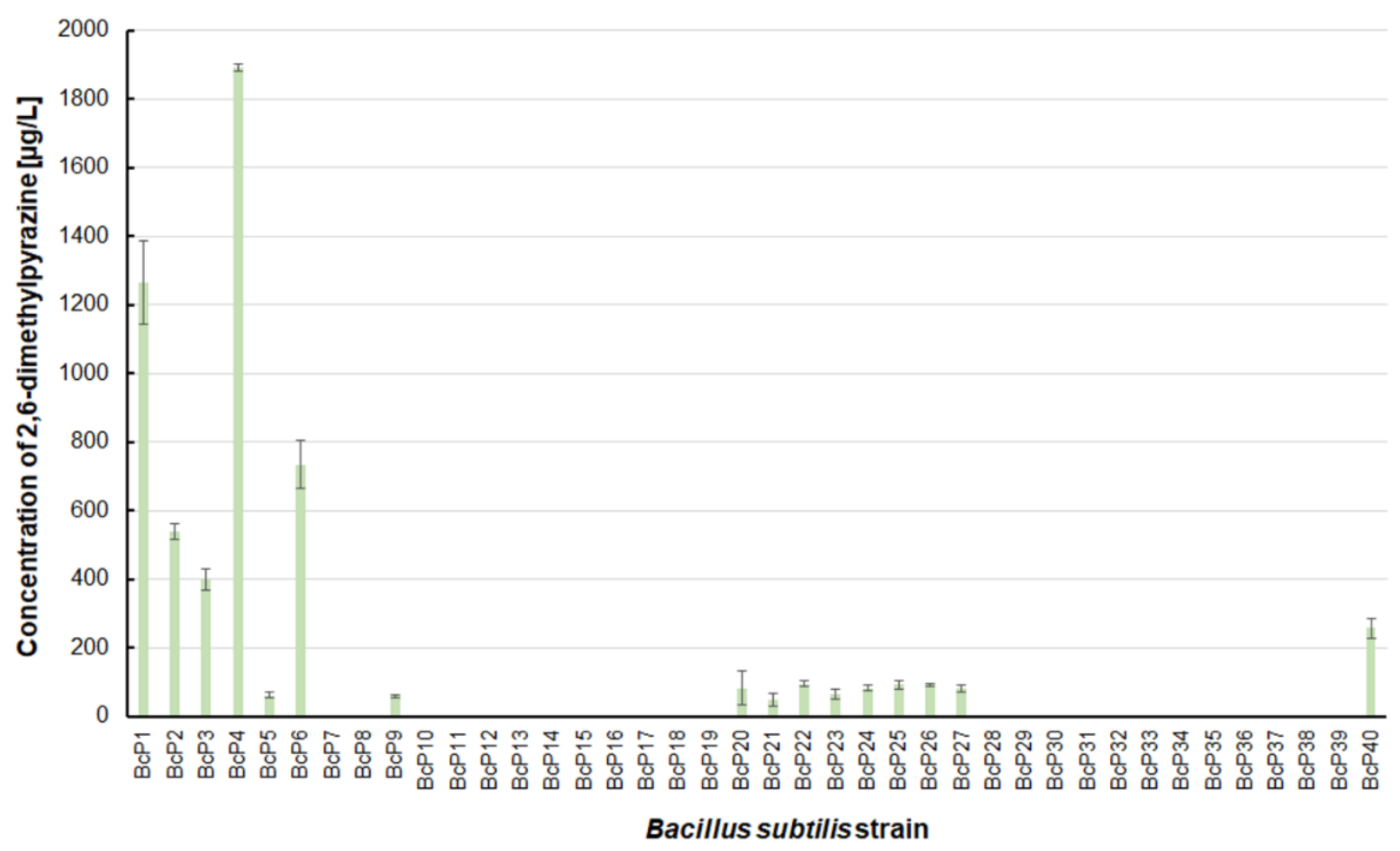
The findings for 2,6-dimethylpyrazine were comparable to those for 2-methylpyrazine (
Figure 3 and
Figure 5). The BcP4 strain was also the most effective producer of 2,6-dimethylpyrazine, but the obtained concentration was much higher, nearly 1900 µg/L. The second-best producer of this compound is also the BcP1 strain, which produced 2,6-dimethylpyrazine at 1265.2 ± 121.2 µg/L (
Figure 5). Among the isolated strains of
B. subtilis that produced pyrazines at concentrations of µg/L, the BcP4 strain was the most effective producer of 2-methylpyrazine, 2,3-dimethylpyrazine and 2,6-dimethylpyrazine (
Figure 3,
Figure 4 and
Figure 5).
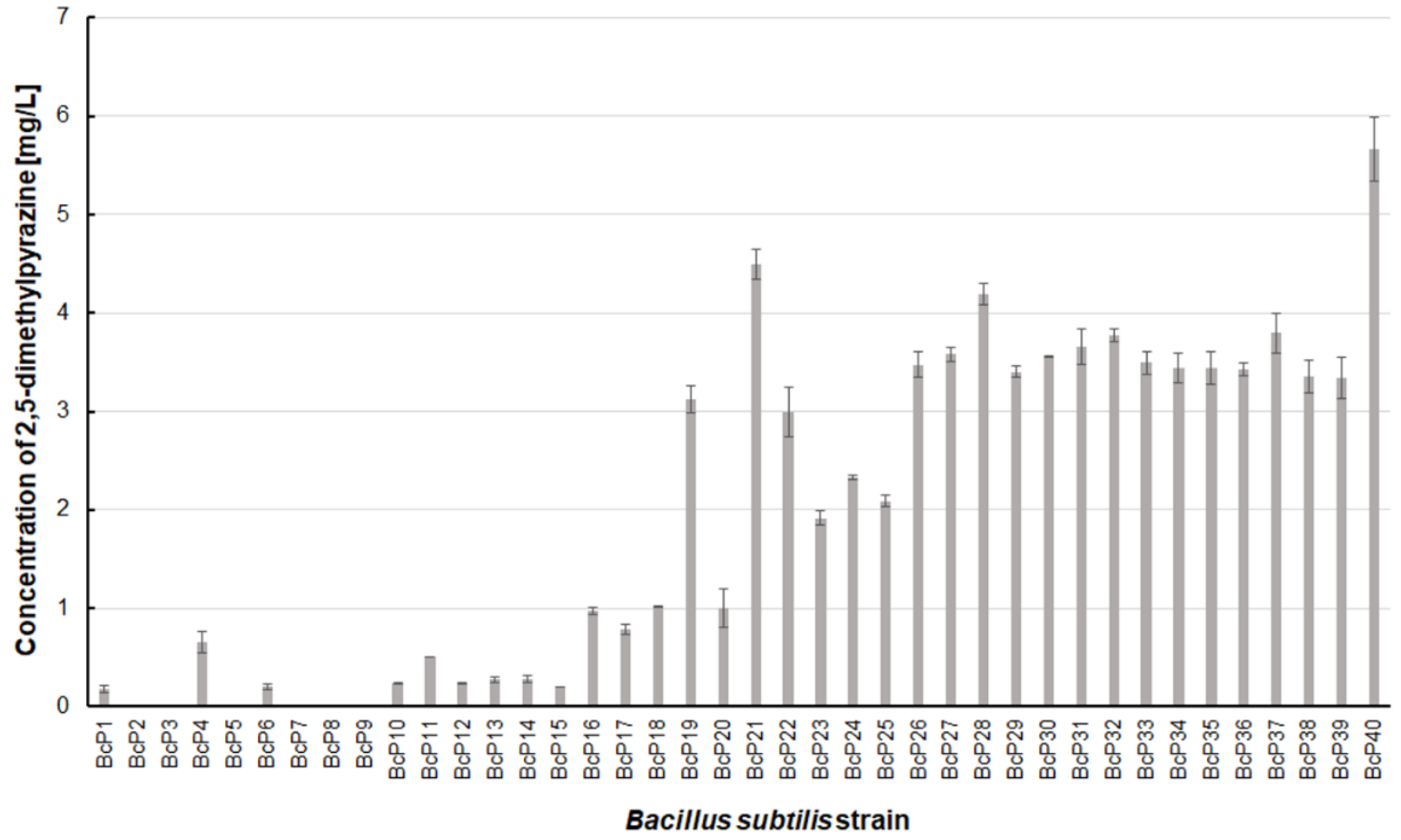
Only two
B. subtilis strains (BcP21 and BcP40) were able to produce 2,5-dimethylpyrazine at concentrations greater than 4.5 mg/L (
Figure 6). The BcP40 strain produced this compound at the highest concentration of 5.665 ± 0.326 mg/L.
However, as many as 20 isolated bacterial strains (BcP19, BcP21-22, BcP24-40) were able to produce 2,5-dimethylpyrazine at concentrations greater than 2 mg/L (
Figure 6). The strains (BcP1, BcP4) with the highest production capacity of 2-methylpyrazine, 2,3-dimethylpyrazine, and 2,6-dimethylpyrazine produced 2,5-dimethylpyrazine only at a concentration of 0.178 ± 0.037 mg/L and 0.653 ± 0.109 mg/L, respectively. Only five of the analyzed strains (BcP3, BcP5, BcP7-9) did not produce any 2,5-dimethylpyrazine (
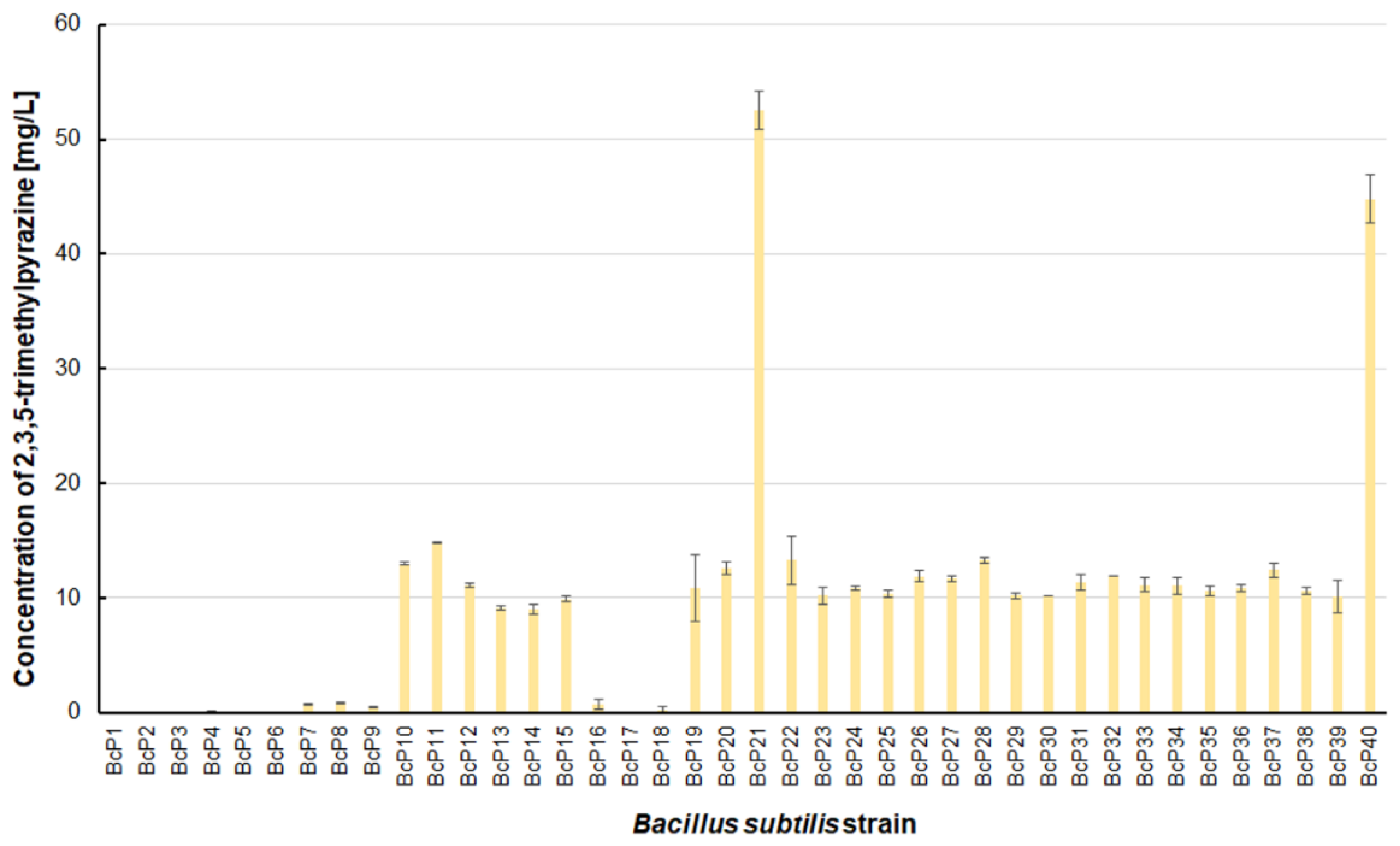
Figure 6). The BcP21 and BcP40 strains were also the best 2,3,5-trimethylpyrazine producing strains. They produced 2,3,5-trimethylpyrazine at concentrations greater than 45 mg/L, nearly four times that of the other 26 strains (BcP10-15, BcP19-20, BcP22-39), which produced ca. 10 mg of this compound per liter (
Figure 7). In contrast, the BcP1–4 strains did not biosynthesize 2,3,5-trimethylpyrazine despite being good producers of 2-methylpyrazine, 2,3-dimethylpyrazine, and 2,6-dimethylpyrazine (
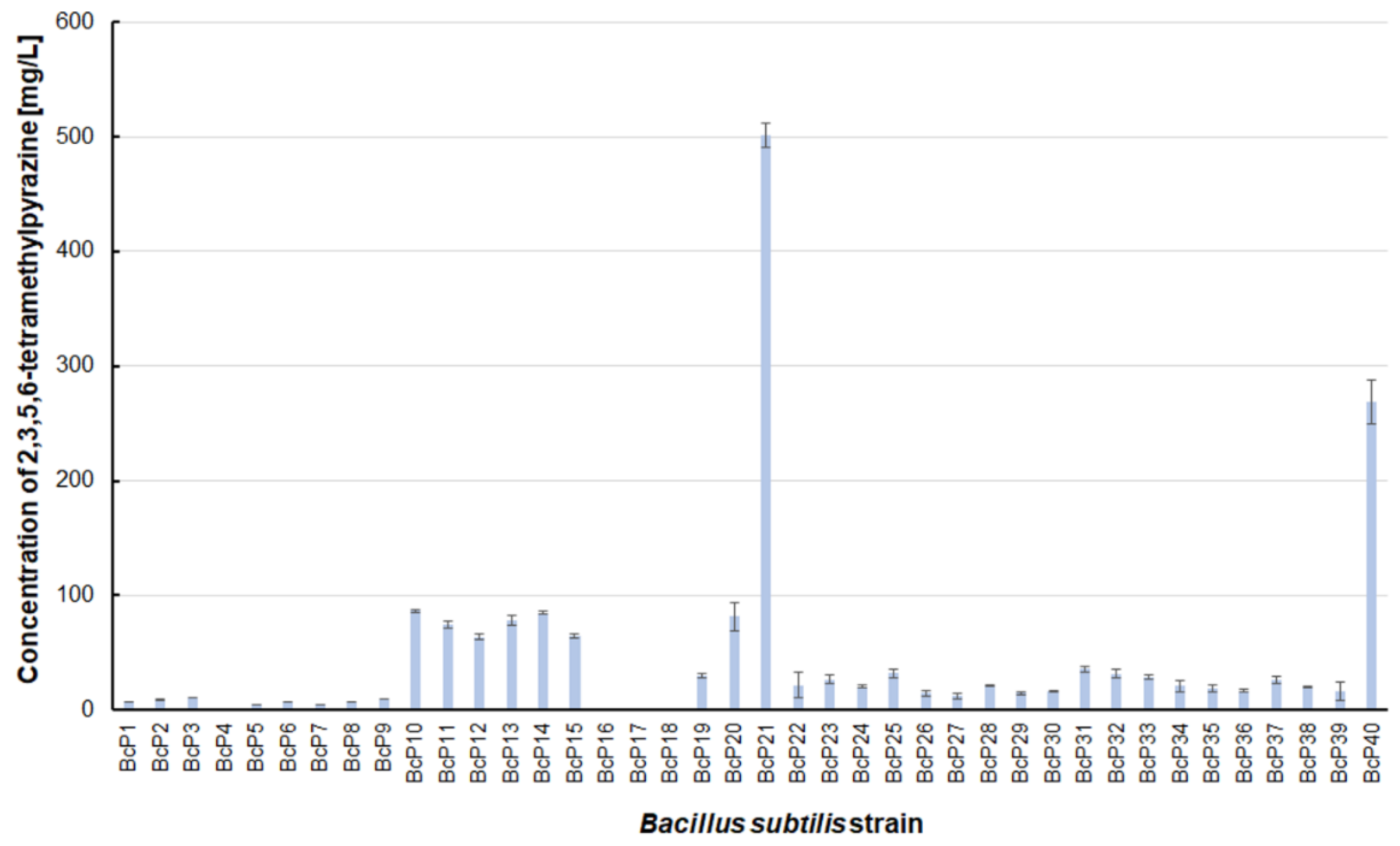
Figure 7). The highest production efficiency was observed for 2,3,5,6-tetramethylpyrazine (ca. 500 mg/L). This compound was most efficiently synthesized by the
B. subtilis BcP21 strain (
Figure 8). The second best producer of this compound was the BcP40 strain, but it provided ca. 270 mg of 2,3,5,6-tetramethylpyrazine per liter. No other bacterial strain isolated from fermented soybeans synthesized 2,3,5,6-tetramethylpyrazine in concentrations greater than 100 mg/L (
Figure 8). Only four strains did not synthesize any 2,3,5,6-tetramethylpyrazine. Among them was the BcP4 strain, which had the highest production capacity for 2-methylpyrazine, 2,3-dimethylpyrazine, and 2,6-dimethylpyrazine (
Figure 8).
The BcP21 strain was the best producer of pyrazines (in total) of all the strains selected from fermented soybeans, producing these compounds at a total concentration of approximately ca. 560 mg/L (
Table 1). The second most effective strain was BcP40, capable of producing pyrazines at a level of 320 mg/L. None of the other analyzed strains produced pyrazines in concentrations greater than 100 mg/L (
Table 1). The dominant compound among the produced pyrazines was 2,3,5,6-tetramethylpyrazine. When analyzing the results of screening tests, one should also consider the profile of the obtained pyrazine derivatives. Some strains (BcP1, BcP4) could produce 2-methylpyrazine, 2,3-dimethylpyrazine, and 2,6-dimethylpyrazine, whereas others (BcP21, BcP40) could produce 2,5-dimethylpyrazine, 2,3,5-trimethylpyrazine and 2,3,5,6-tetramethylpyrazine. While conducting further research on the optimization of pyrazines biosynthesis, it is necessary to select the strains capable of producing the desired profile of compounds.
Isolating effective target compound producers from the natural environment is a critical step in the development of a biosynthetic process. Screening is typically the most time-consuming stage of research, with the goal of selecting a microorganism capable of producing a diverse range of bioproducts. For years, researchers have been looking for microorganisms capable of pyrazine biosynthesis, but most have paid little attention to screening and have focused primarily on optimizing culture conditions [
11,
21]. Other authors did not analyze the ability of bacteria to synthesize a variety of alkylpyrazines (e.g., six, as in this study), instead focusing on the ability to produce 2,3,5,6-tetramethylpyrazine [
22,
23]. Only a few authors provided information about the efficacy of screening tests. Zhu et al. (2010) isolated 300 bacterial strains from starter cultures (Daqu) used in the production of Maotai-flavor liquor, a Chinese distillate produced from sorghum. One hundred of them were used in the search for a pyrazine producer. Daqu proved to be a good source of pyrazines producers, as the isolated strain of
B. subtilis was able to produce up to 1.1 g of 2,3,5,6-tetramethylpyrazine per liter of the culture medium after optimizing the medium composition and culture conditions [
22]. It should be noted, however, that in contrast to this study, only the production of 2,3,5,6-tetramethylpyrazine was examined. Besson et al. (1997) analyzed only seven strains of
B. subtilis isolated from fermented soybeans, the best of which produced pyrazines at concentrations of 118 mg/kg. However, the analysis included only 2,5-dimethylpyrazine, trimethylpyrazine, and 2,3,5,6-tetramethylpyrazine [
16]. A review of the literature revealed a scarcity of studies evaluating the ability of microorganisms isolated from their natural environment to produce a diverse range of pyrazines. Due to the large differences in the metabolism of these compounds in different strains of
B. subtilis, our study suggests that the efficiency of production of various alkylpyrazines should be investigated. Adams and De Kimpe (2007) investigated the utility of
Bacillus bacteria in the production of 2,5-dimethylpyrazine and trimethylpyrazine. However, four different strains of
B. cereus only produced these compounds at a maximum concentration of 56 µg/kg, indicating that these microorganisms were not effective producers of pyrazines [
17]. Many methods are used to optimize the culture medium or modify the cellular metabolism in order to increase the production of pyrazines by
B. subtilis. Under process conditions similar to those used in our work, a study using
B. subtilis strain producing pyrazines (2,5-dimethylpyrazine, trimethylpyrazine, and 2,3,5,6-tetramethylpyrazine) at a total concentration of 410 mg/L increased the concentration of the analyzed compounds to around ca. 1500 mg/L thanks to optimization of the substrate composition (nitrogen and carbon source) and culture conditions (pH, temperature and aeration) conditions (pH, temperature and aeration) [
11]. Mutagenesis using chemical compounds such as nitrosoguanidine is another method for increasing pyrazines production.
Bacillus mutants produced up to 4 g/L of 2,3,5,6-tertramethylpyrazine after optimizing the medium composition. [
24]. Another possibility is to alter acetoin metabolism by inhibiting the 2,3-butanediol (2,3-BD) dehydrogenase gene, which is responsible for the production of 2,3-BD and the accumulation of the acetoin precursor during the early growth phase [
14]. The isolated
B. subtilis strains were found to be useful for the production of a wide range of alkylpyrazines in this study. However, further optimization of the culture conditions and medium composition is required so that the selected pyrazines can be produced in concentrations that ensure economic profitability.