Genetic Alterations in the INK4a/ARF Locus: Effects on Melanoma Development and Progression
Abstract
:1. Introduction
2. Somatic CDKN2A Alterations in Melanoma
3. Germline CDKN2A Alterations in Familial Melanoma
4. Functional Consequences of Melanoma-Associated CDKN2A Alterations
4.1. p16INK4a Mutations
4.2. p14ARF Mutations
5. Clinical Implications of CDKN2A: Impact on Response and Resistance to Current Treatments in Melanoma
5.1. MDM2 Inhibitors
5.2. CDK Inhibitors
5.3. Epigenetic Modulators
5.4. BRAF/MEK Inhibitors
5.5. Immune Checkpoint Inhibitors
5.6. Chemotherapy
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Karimkhani, C.; Green, A.C.; Nijsten, T.; Weinstock, M.A.; Dellavalle, R.P.; Naghavi, M.; Fitzmaurice, C. The global burden of melanoma: Results from the Global Burden of Disease Study 2015. Br. J. Dermatol. 2017, 177, 134–140. [Google Scholar] [CrossRef]
- Costin, G.E.; Hearing, V.J. Human skin pigmentation: Melanocytes modulate skin color in response to stress. FASEB J. 2007, 21, 976–994. [Google Scholar] [CrossRef] [PubMed]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tyminska, A. Skin melanocytes: Biology and development. Postepy Dermatol. Alergol. 2013, 30, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Mihm, M.C., Jr. Melanoma. N. Engl. J. Med. 2006, 355, 51–65. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galuppini, F.; Dal Pozzo, C.A.; Deckert, J.; Loupakis, F.; Fassan, M.; Baffa, R. Tumor mutation burden: From comprehensive mutational screening to the clinic. Cancer Cell Int. 2019, 19, 209. [Google Scholar] [CrossRef]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.M.; Chan, M.; Harland, M.; Hayward, N.K.; Demenais, F.; Bishop, D.T.; Azizi, E.; Bergman, W.; Bianchi-Scarra, G.; Bruno, W.; et al. Features associated with germline CDKN2A mutations: A GenoMEL study of melanoma-prone families from three continents. J. Med. Genet. 2007, 44, 99–106. [Google Scholar] [CrossRef]
- de Snoo, F.A.; Hayward, N.K. Cutaneous melanoma susceptibility and progression genes. Cancer Lett. 2005, 230, 153–186. [Google Scholar] [CrossRef]
- Serrano, M.; Hannon, G.J.; Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366, 704–707. [Google Scholar] [CrossRef]
- Serrano, M.; Lee, H.; Chin, L.; Cordon-Cardo, C.; Beach, D.; DePinho, R.A. Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996, 85, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Ranade, K.; Hussussian, C.J.; Sikorski, R.S.; Varmus, H.E.; Goldstein, A.M.; Tucker, M.A.; Serrano, M.; Hannon, G.J.; Beach, D.; Dracopoli, N.C. Mutations associated with familial melanoma impair p16 INK4 function. Nature Genet. 1995, 10, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, J.; Schreiber-Agus, N.; Liegeois, N.J.; Silverman, A.; Alland, L.; Chin, L.; Potes, J.; Chen, K.; Orlow, I.; Lee, H.W.; et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 1998, 92, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xiong, Y.; Yarbrough, W.G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998, 92, 725–734. [Google Scholar] [CrossRef] [Green Version]
- Kamijo, T.; Weber, J.D.; Zambetti, G.; Zindy, F.; Roussel, M.F.; Sherr, C.J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA 1998, 95, 8292–8297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stott, F.J.; Bates, S.; James, M.C.; McConnell, B.B.; Starborg, M.; Brookes, S.; Palmero, I.; Ryan, K.; Hara, E.; Vousden, K.H. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998, 17, 5001–5014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Rizos, H.; Becker, T.M.; Holland, E.A.; Kefford, R.F.; Mann, G.J. Differential expression of p16INK4a and p16beta transcripts in B-lymphoblastoid cells from members of hereditary melanoma families without CDKN2A exon mutations. Oncogene 1997, 15, 515–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fountain, J.W.; Karayiorgou, M.; Ernstoff, M.S.; Kirkwood, J.M.; Vlock, D.R.; Titus-Ernstoff, L.; Bouchard, B.; Vijayasaradhi, S.; Houghton, A.N.; Lahti, J. Homozygous deletions within human chromosome band 9p21 in melanoma. Proc. Natl. Acad. Sci. USA 1992, 89, 10557–10561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freedberg, D.E.; Rigas, S.H.; Russak, J.; Gai, W.; Kaplow, M.; Osman, I.; Turner, F.; Randerson-Moor, J.A.; Houghton, A.; Busam, K.; et al. Frequent p16-independent inactivation of p14ARF in human melanoma. J. Natl. Cancer Inst. 2008, 100, 784–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.; McArthur, G.A. CDK4 inhibitors an emerging strategy for the treatment of melanoma. Melanoma Manag. 2015, 2, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Askarian-Amiri, M.E. Epigenetic regulation in human melanoma: Past and future. Epigenetics 2015, 10, 103–121. [Google Scholar] [CrossRef] [Green Version]
- Kostaki, M.; Manona, A.D.; Stavraka, I.; Korkolopoulou, P.; Levidou, G.; Trigka, E.A.; Christofidou, E.; Champsas, G.; Stratigos, A.J.; Katsambas, A.J.E.d. High-frequency p16 INK 4A promoter methylation is associated with histone methyltransferase SETDB 1 expression in sporadic cutaneous melanoma. Exp. Dermatol. 2014, 23, 332–338. [Google Scholar] [CrossRef]
- Jonsson, A.; Tuominen, R.; Grafström, E.; Hansson, J.; Egyhazi, S. High frequency of p16INK4A promoter methylation in NRAS-mutated cutaneous melanoma. J. Investig. Dermatol. 2010, 130, 2809–2817. [Google Scholar] [CrossRef] [Green Version]
- Straume, O.; Smeds, J.; Kumar, R.; Hemminki, K.; Akslen, L.A. Significant impact of promoter hypermethylation and the 540 C>T polymorphism of CDKN2A in cutaneous melanoma of the vertical growth phase. Am. J. Pathol. 2002, 161, 229–237. [Google Scholar] [CrossRef]
- Venza, M.; Visalli, M.; Biondo, C.; Lentini, M.; Catalano, T.; Teti, D.; Venza, I. Epigenetic regulation of p14ARF and p16INK4A expression in cutaneous and uveal melanoma. Biochim. Biophys. Acta 2015, 1849, 247–256. [Google Scholar] [CrossRef]
- Pasini, D.; Bracken, A.P.; Helin, K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle 2004, 3, 396–400. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Feng, Y.; Wang, X. Regulation of Tumor Suppressor Gene CDKN2A and Encoded p16-INK4a Protein by Covalent Modifications. Biochemistry 2018, 83, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordero, F.J.; Huang, Z.; Grenier, C.; He, X.; Hu, G.; McLendon, R.E.; Murphy, S.K.; Hashizume, R.; Becher, O.J. Histone H3.3K27M Represses p16 to Accelerate Gliomagenesis in a Murine Model of DIPG. Mol. Cancer Res. 2017, 15, 1243–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, F.; Niebel, D.; Aymans, P.; Ferring-Schmitt, S.; Dietrich, D.; Landsberg, J. H3K27me3 and EZH2 expression in melanoma: Relevance for melanoma progression and response to immune checkpoint blockade. Clin. Epigenetics 2020, 12, 24. [Google Scholar] [CrossRef] [Green Version]
- Gonzalgo, M.L.; Bender, C.M.; You, E.H.; Glendening, J.M.; Flores, J.F.; Walker, G.J.; Hayward, N.K.; Jones, P.A.; Fountain, J.W. Low frequency of p16/CDKN2A methylation in sporadic melanoma: Comparative approaches for methylation analysis of primary tumors. Cancer Res. 1997, 57, 5336–5347. [Google Scholar] [PubMed]
- Welch, J.; Millar, D.; Goldman, A.; Heenan, P.; Stark, M.; Eldon, M.; Clark, S.; Martin, N.G.; Hayward, N.K. Lack of genetic and epigenetic changes in CDKN2A in melanocytic nevi. J. Investig. Dermatol. 2001, 117, 383–384. [Google Scholar] [CrossRef] [Green Version]
- Bishop, D.T.; Demenais, F.; Goldstein, A.M.; Bergman, W.; Bishop, J.N.; Bressac-de Paillerets, B.; Chompret, A.; Ghiorzo, P.; Gruis, N.; Hansson, J.; et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J. Natl. Cancer Inst. 2002, 94, 894–903. [Google Scholar] [CrossRef] [Green Version]
- Shain, A.H.; Yeh, I.; Kovalyshyn, I.; Sriharan, A.; Talevich, E.; Gagnon, A.; Dummer, R.; North, J.; Pincus, L.; Ruben, B.; et al. The Genetic Evolution of Melanoma from Precursor Lesions. N. Engl. J. Med. 2015, 373, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.R.; Choi, Y.D.; Kim, J.M.; Jin, S.; Shin, M.-H.; Shim, H.-J.; Lee, J.-B.; Yun, S.J. Genetic alterations in primary acral melanoma and acral melanocytic nevus in Korea: Common mutated genes show distinct cytomorphological features. J. Investig. Dermatol. 2018, 138, 933–945. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.S.; Hendricks, W.; Kiefer, J.; Schmidt, J.; Sekar, S.; Carpten, J.; Craig, D.W.; Adkins, J.; Cuyugan, L.; Manojlovic, Z.; et al. Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res. 2017, 27, 524–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussussian, C.J.; Struewing, J.P.; Goldstein, A.M.; Higgins, P.A.; Ally, D.S.; Sheahan, M.D.; Clark, W.H., Jr.; Tucker, M.A.; Dracopoli, N.C. Germline p16 mutations in familial melanoma. Nat. Genet. 1994, 8, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kamb, A.; Shattuck-Eidens, D.; Eeles, R.; Liu, Q.; Gruis, N.A.; Ding, W.; Hussey, C.; Tran, T.; Miki, Y.; Weaver-Feldhaus, J. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat. Genet. 1994, 8, 22–26. [Google Scholar] [CrossRef]
- Helgadottir, H.; Hoiom, V.; Tuominen, R.; Nielsen, K.; Jonsson, G.; Olsson, H.; Hansson, J. Germline CDKN2A Mutation Status and Survival in Familial Melanoma Cases. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, J.A.; Wachsmuth, R.C.; Harland, M.; Bataille, V.; Pinney, E.; Mac, K.P.; Baglietto, L.; Cuzick, J.; Bishop, D.T. Genotype/phenotype and penetrance studies in melanoma families with germline CDKN2A mutations. J. Investig. Dermatol. 2000, 114, 28–33. [Google Scholar] [CrossRef]
- Bahuau, M.; Vidaud, D.; Jenkins, R.B.; Bieche, I.; Kimmel, D.W.; Assouline, B.; Smith, J.S.; Alderete, B.; Cayuela, J.M.; Harpey, J.P.; et al. Germ-line deletion involving the INK4 locus in familial proneness to melanoma and nervous system tumors. Cancer Res. 1998, 58, 2298–2303. [Google Scholar]
- Randerson-Moor, J.A.; Harland, M.; Williams, S.; Cuthbert-Heavens, D.; Sheridan, E.; Aveyard, J.; Sibley, K.; Whitaker, L.; Knowles, M.; Newton Bishop, J. A germline deletion of p14ARF but not CDKN2A in a melanoma–neural system tumour syndrome family. Human Mol. Genet. 2001, 10, 55–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoude, L.G.; Wadt, K.A.; Pritchard, A.L.; Hayward, N.K. Genetics of familial melanoma: 20 years after CDKN2A. Pigment. Cell Melanoma Res. 2015, 28, 148–160. [Google Scholar] [CrossRef]
- Laud, K.; Marian, C.; Avril, M.-F.; Barrois, M.; Chompret, A.; Goldstein, A.M.; Tucker, M.A.; Clark, P.A.; Peters, G.; Chaudru, V. Comprehensive analysis of CDKN2A (p16INK4A/p14ARF) and CDKN2B genes in 53 melanoma index cases considered to be at heightened risk of melanoma. J. Med. Genet. 2006, 43, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Rizos, H.; Puig, S.; Badenas, C.; Malvehy, J.; Darmanian, A.P.; Jiménez, L.; Milà, M.; Kefford, R.F. A melanoma-associated germline mutation in exon 1 β inactivates p14ARF. Oncogene 2001, 20, 5543–5547. [Google Scholar] [CrossRef] [Green Version]
- Hewitt, C.; Lee Wu, C.; Evans, G.; Howell, A.; Elles, R.G.; Jordan, R.; Sloan, P.; Read, A.P.; Thakker, N. Germline mutation of ARF in a melanoma kindred. Hum. Mol. Genet. 2002, 11, 1273–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binni, F.; Antigoni, I.; De Simone, P.; Majore, S.; Silipo, V.; Crisi, A.; Amantea, A.; Pacchiarini, D.; Castori, M.; De Bernardo, C. Novel and recurrent p14ARF mutations in Italian familial melanoma. Clin. Genet. 2010, 77, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Mistry, S.H.; Taylor, C.; Randerson-Moor, J.A.; Harland, M.; Turner, F.; Barrett, J.H.; Whitaker, L.; Jenkins, R.B.; Knowles, M.A.; Bishop, J.A.; et al. Prevalence of 9p21 deletions in UK melanoma families. Genes Chromosomes Cancer 2005, 44, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Knappskog, S.; Geisler, J.; Arnesen, T.; Lillehaug, J.R.; Lonning, P.E. A novel type of deletion in the CDKN2A gene identified in a melanoma-prone family. Genes Chromosomes Cancer 2006, 45, 1155–1163. [Google Scholar] [CrossRef]
- Lesueur, F.; de Lichy, M.; Barrois, M.; Durand, G.; Bombled, J.; Avril, M.F.; Chompret, A.; Boitier, F.; Lenoir, G.M.; Bressac-de Paillerets, B.; et al. The contribution of large genomic deletions at the CDKN2A locus to the burden of familial melanoma. Br. J. Cancer 2008, 99, 364–370. [Google Scholar] [CrossRef] [Green Version]
- Petronzelli, F.; Sollima, D.; Coppola, G.; Martini-Neri, M.E.; Neri, G.; Genuardi, M. CDKN2A germline splicing mutation affecting both P16INK4 and P14ARF RNA processing in a melanoma/neurofibroma kindred. Genes Chromosome Cancer 2001, 31, 398–401. [Google Scholar] [CrossRef]
- Helgadottir, H.; Hoiom, V.; Jonsson, G.; Tuominen, R.; Ingvar, C.; Borg, A.; Olsson, H.; Hansson, J. High risk of tobacco-related cancers in CDKN2A mutation-positive melanoma families. J. Med. Genet. 2014, 51, 545–552. [Google Scholar] [CrossRef] [Green Version]
- Potjer, T.P.; Helgadottir, H.; Leenheer, M.; van der Stoep, N.; Gruis, N.A.; Hoiom, V.; Olsson, H.; van Doorn, R.; Vasen, H.F.A.; van Asperen, C.J.; et al. CM-Score: A validated scoring system to predict CDKN2A germline mutations in melanoma families from Northern Europe. J. Med. Genet. 2018, 55, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Potrony, M.; Puig-Butille, J.A.; Aguilera, P.; Badenas, C.; Carrera, C.; Malvehy, J.; Puig, S. Increased prevalence of lung, breast, and pancreatic cancers in addition to melanoma risk in families bearing the cyclin-dependent kinase inhibitor 2A mutation: Implications for genetic counseling. J. Am. Acad. Dermatol. 2014, 71, 888–895. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A.M.; Chan, M.; Harland, M.; Gillanders, E.M.; Hayward, N.K.; Avril, M.F.; Azizi, E.; Bianchi-Scarra, G.; Bishop, D.T.; Bressac-de Paillerets, B.; et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006, 66, 9818–9828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasen, H.F.; Gruis, N.A.; Frants, R.R.; van Der Velden, P.A.; Hille, E.T.; Bergman, W. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int. J. Cancer 2000, 87, 809–811. [Google Scholar] [CrossRef]
- Borg, A.; Sandberg, T.; Nilsson, K.; Johannsson, O.; Klinker, M.; Masback, A.; Westerdahl, J.; Olsson, H.; Ingvar, C. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J. Natl. Cancer Inst. 2000, 92, 1260–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Velden, P.A.; Sandkuijl, L.A.; Bergman, W.; Pavel, S.; van Mourik, L.; Frants, R.R.; Gruis, N.A. Melanocortin-1 receptor variant R151C modifies melanoma risk in Dutch families with melanoma. Am. J. Hum. Genet. 2001, 69, 774–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demenais, F.; Mohamdi, H.; Chaudru, V.; Goldstein, A.M.; Newton Bishop, J.A.; Bishop, D.T.; Kanetsky, P.A.; Hayward, N.K.; Gillanders, E.; Elder, D.E.; et al. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: A GenoMEL study. J. Natl. Cancer Inst. 2010, 102, 1568–1583. [Google Scholar] [CrossRef]
- Yang, X.R.; Pfeiffer, R.M.; Wheeler, W.; Yeager, M.; Chanock, S.; Tucker, M.A.; Goldstein, A.M. Identification of modifier genes for cutaneous malignant melanoma in melanoma-prone families with and without CDKN2A mutations. Int. J. Cancer 2009, 125, 2912–2917. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.S.; Pfeiffer, R.M.; Wheeler, W.; Maeder, D.; Burdette, L.; Yeager, M.; Chanock, S.; Tucker, M.A.; Goldstein, A.M.; Yang, X.R. Genetic variants in DNA repair genes and the risk of cutaneous malignant melanoma in melanoma-prone families with/without CDKN2A mutations. Int. J. Cancer 2012, 130, 2062–2066. [Google Scholar] [CrossRef]
- Byeon, I.J.; Li, J.; Ericson, K.; Selby, T.L.; Tevelev, A.; Kim, H.J.; O’Maille, P.; Tsai, M.D. Tumor suppressor p16INK4A: Determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol. Cell 1998, 1, 421–431. [Google Scholar] [CrossRef]
- Harland, M.; Meloni, R.; Gruis, N.; Pinney, E.; Brookes, S.; Spurr, N.K.; Frischauf, A.M.; Bataille, V.; Peters, G.; Cuzick, J.; et al. Germline mutations of the CDKN2 gene in UK melanoma families. Hum. Mol. Genet. 1997, 6, 2061–2067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, T.M.; Rizos, H.; Kefford, R.F.; Mann, G.J. Functional impairment of melanoma-associated p16INK4a mutants in melanoma cells despite retention of cyclin-dependent kinase 4 binding. Clin. Cancer Res. 2001, 7, 3282–3288. [Google Scholar]
- Yarbrough, W.G.; Buckmire, R.A.; Bessho, M.; Liu, E.T. Biologic and biochemical analyses of p16INK4a mutations from primary tumors. J. Natl. Cancer Inst. 1999, 91, 1569–1574. [Google Scholar] [CrossRef] [Green Version]
- Walker, G.J.; Gabrielli, B.G.; Castellano, M.; Hayward, N.K. Functional reassessment of P16 variants using a transfection-based assay. Int. J. Cancer 1999, 82, 305–312. [Google Scholar] [CrossRef]
- Li, C.; Liu, T.; Liu, B.; Hernandez, R.; Facelli, J.C.; Grossman, D. A novel CDKN 2A variant (p16L117P) in a patient with familial and multiple primary melanomas. Pigment. Cell Melanoma Res. 2019, 32, 734–738. [Google Scholar] [PubMed]
- Jenkins, N.C.; Liu, T.; Cassidy, P.; Leachman, S.A.; Boucher, K.M.; Goodson, A.G.; Samadashwily, G.; Grossman, D. The p16(INK4A) tumor suppressor regulates cellular oxidative stress. Oncogene 2011, 30, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, N.C.; Jung, J.; Liu, T.; Wilde, M.; Holmen, S.L.; Grossman, D. Familial melanoma-associated mutations in p16 uncouple its tumor-suppressor functions. J. Investig. Dermatol. 2013, 133, 1043–1051. [Google Scholar] [CrossRef] [Green Version]
- Mooi, W.; Peeper, D. Oncogene-induced cell senescence—halting on the road to cancer. N. Engl. J. Med. 2006, 355, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Haferkamp, S.; Becker, T.M.; Scurr, L.L.; Kefford, R.F.; Rizos, H. p16INK4a-induced senescence is disabled by melanoma-associated mutations. Aging Cell 2008, 7, 733–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damsky, W.; Micevic, G.; Meeth, K.; Muthusamy, V.; Curley, D.P.; Santhanakrishnan, M.; Erdelyi, I.; Platt, J.T.; Huang, L.; Theodosakis, N. mTORC1 activation blocks BrafV600E-induced growth arrest but is insufficient for melanoma formation. Cancer Cell 2015, 27, 41–56. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Jorapur, A.; Shain, A.H.; Lang, U.E.; Torres, R.; Zhang, Y.; McNeal, A.S.; Botton, T.; Lin, J.; Donne, M.; et al. Bi-allelic Loss of CDKN2A Initiates Melanoma Invasion via BRN2 Activation. Cancer Cell 2018, 34, 56–68.e59. [Google Scholar] [CrossRef]
- Garcia-Casado, Z.; Nagore, E.; Fernandez-Serra, A.; Botella-Estrada, R.; Lopez-Guerrero, J.A. A germline mutation of p14/ARF in a melanoma kindred. Melanoma Res. 2009, 19, 335–337. [Google Scholar] [CrossRef]
- Puszynski, K.; Gandolfi, A.; d’Onofrio, A. The pharmacodynamics of the p53-Mdm2 targeting drug Nutlin: The role of gene-switching noise. PLoS Comput. Biol. 2014, 10, e1003991. [Google Scholar] [CrossRef] [Green Version]
- Vilgelm, A.E.; Pawlikowski, J.S.; Liu, Y.; Hawkins, O.E.; Davis, T.A.; Smith, J.; Weller, K.P.; Horton, L.W.; McClain, C.M.; Ayers, G.D.; et al. Mdm2 and aurora kinase a inhibitors synergize to block melanoma growth by driving apoptosis and immune clearance of tumor cells. Cancer Res. 2015, 75, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Shattuck-Brandt, R.L.; Chen, S.-C.; Murray, E.; Johnson, C.A.; Crandall, H.; O’Neal, J.F.; Al-Rohil, R.N.; Nebhan, C.A.; Bharti, V.; Dahlman, K.B. Metastatic Melanoma Patient-derived Xenografts Respond to MDM2 Inhibition as a Single Agent or in Combination with BRAF/MEK Inhibition. Clin. Cancer Res. 2020, 15, 3803–3818. [Google Scholar] [CrossRef] [Green Version]
- Burdette-Radoux, S.; Tozer, R.G.; Lohmann, R.C.; Quirt, I.; Ernst, D.S.; Walsh, W.; Wainman, N.; Colevas, A.D.; Eisenhauer, E.A. Phase II trial of flavopiridol, a cyclin dependent kinase inhibitor, in untreated metastatic malignant melanoma. Investig. New Drugs 2004, 22, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Loo, A.; Chopra, R.; Caponigro, G.; Huang, A.; Vora, S.; Parasuraman, S.; Howard, S.; Keen, N.; Sellers, W.; et al. Abstract PR02: LEE011: An orally bioavailable, selective small molecule inhibitor of CDK4/6– Reactivating Rb in cancer. Mol. Cancer Ther. 2013, 12, PR02. [Google Scholar]
- Yadav, V.; Burke, T.F.; Huber, L.; Van Horn, R.D.; Zhang, Y.; Buchanan, S.G.; Chan, E.M.; Starling, J.J.; Beckmann, R.P.; Peng, S.B. The CDK4/6 inhibitor LY2835219 overcomes vemurafenib resistance resulting from MAPK reactivation and cyclin D1 upregulation. Mol. Cancer Ther. 2014, 13, 2253–2263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilgelm, A.E.; Saleh, N.; Shattuck-Brandt, R.; Riemenschneider, K.; Slesur, L.; Chen, S.C.; Johnson, C.A.; Yang, J.; Blevins, A.; Yan, C.; et al. MDM2 antagonists overcome intrinsic resistance to CDK4/6 inhibition by inducing p21. Sci. Transl. Med. 2019, 11, eaav7171. [Google Scholar] [CrossRef]
- Sosman, J.A.; Kittaneh, M.; Lolkema, M.P.J.K.; Postow, M.A.; Schwartz, G.; Franklin, C.; Matano, A.; Bhansali, S.; Parasuraman, S.; Kim, K. A phase 1b/2 study of LEE011 in combination with binimetinib (MEK162) in patients with NRAS-mutant melanoma: Early encouraging clinical activity. J. Clin. Oncol. 2014, 32, 9009. [Google Scholar] [CrossRef]
- Schuler, M.H.; Ascierto, P.A.; De Vos, F.Y.F.L.; Postow, M.A.; Van Herpen, C.M.L.; Carlino, M.S.; Sosman, J.A.; Berking, C.; Long, G.V.; Weise, A.; et al. Phase 1b/2 trial of ribociclib+binimetinib in metastatic NRAS-mutant melanoma: Safety, efficacy, and recommended phase 2 dose (RP2D). J. Clin. Oncol. 2017, 35, 9519. [Google Scholar] [CrossRef]
- Taylor, M.; Sosman, J.; Gonzalez, R.; Carlino, M.S.; Kittaneh, M.; Lolkema, M.P.; Miller, W.; Marino, A.; Zhang, V.; Bhansali, S.G.; et al. Phase Ib/Ii Study of Lee011 (Cdk4/6 Inhibitor) and Lgx818 (Braf Inhibitor) in Braf-Mutant Melanoma. Ann. Oncol. 2014, 25, iv374. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Bechter, O.; Wolter, P.; Lebbe, C.; Elez, E.; Miller, W.H.; Long, G.V.; Omlin, A.G.; Siena, S.; Calvo, E.; et al. A phase Ib/II dose-escalation study evaluating triple combination therapy with a BRAF (encorafenib), MEK (binimetinib), and CDK 4/6 (ribociclib) inhibitor in patients (Pts) with BRAF V600-mutant solid tumors and melanoma. J. Clin. Oncol. 2017, 35, 9518. [Google Scholar] [CrossRef]
- Wiedemeyer, W.R.; Dunn, I.F.; Quayle, S.N.; Zhang, J.; Chheda, M.G.; Dunn, G.P.; Zhuang, L.; Rosenbluh, J.; Chen, S.; Xiao, Y.; et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc. Natl. Acad. Sci. USA 2010, 107, 11501–11506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsumi, Y.; Iehara, T.; Miyachi, M.; Yagyu, S.; Tsubai-Shimizu, S.; Kikuchi, K.; Tamura, S.; Kuwahara, Y.; Tsuchiya, K.; Kuroda, H.; et al. Sensitivity of malignant rhabdoid tumor cell lines to PD 0332991 is inversely correlated with p16 expression. Biochem. Biophys. Res. Commun. 2011, 413, 62–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konecny, G.E.; Winterhoff, B.; Kolarova, T.; Qi, J.; Manivong, K.; Dering, J.; Yang, G.; Chalukya, M.; Wang, H.J.; Anderson, L.; et al. Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin. Cancer Res. 2011, 17, 1591–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfel, T.; Hauer, M.; Schneider, J.; Serrano, M.; Wolfel, C.; Klehmann-Hieb, E.; De Plaen, E.; Hankeln, T.; Meyer zum Buschenfelde, K.H.; Beach, D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 1995, 269, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Young, R.J.; Waldeck, K.; Martin, C.; Foo, J.H.; Cameron, D.P.; Kirby, L.; Do, H.; Mitchell, C.; Cullinane, C.; Liu, W.; et al. Loss of CDKN2A expression is a frequent event in primary invasive melanoma and correlates with sensitivity to the CDK4/6 inhibitor PD0332991 in melanoma cell lines. Pigment. Cell Melanoma Res. 2014, 27, 590–600. [Google Scholar] [CrossRef]
- Zhao, R.; Choi, B.Y.; Lee, M.H.; Bode, A.M.; Dong, Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine 2016, 8, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Long, G.V.; Eroglu, Z.; Infante, J.; Patel, S.; Daud, A.; Johnson, D.B.; Gonzalez, R.; Kefford, R.; Hamid, O.; Schuchter, L.; et al. Long-Term Outcomes in Patients With BRAF V600-Mutant Metastatic Melanoma Who Received Dabrafenib Combined With Trametinib. J. Clin. Oncol. 2018, 36, 667–673. [Google Scholar] [CrossRef]
- Smalley, K.S.; Lioni, M.; Dalla Palma, M.; Xiao, M.; Desai, B.; Egyhazi, S.; Hansson, J.; Wu, H.; King, A.J.; Van Belle, P. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E–mutated melanomas. Mol. Cancer Ther. 2008, 7, 2876–2883. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Hugo, W.; Kong, X.; Hong, A.; Koya, R.C.; Moriceau, G.; Chodon, T.; Guo, R.; Johnson, D.B.; Dahlman, K.B.; et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014, 4, 80–93. [Google Scholar] [CrossRef] [Green Version]
- Hugo, W.; Shi, H.; Sun, L.; Piva, M.; Song, C.; Kong, X.; Moriceau, G.; Hong, A.; Dahlman, K.B.; Johnson, D.B.; et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015, 162, 1271–1285. [Google Scholar] [CrossRef] [Green Version]
- Moriceau, G.; Hugo, W.; Hong, A.; Shi, H.; Kong, X.; Yu, C.C.; Koya, R.C.; Samatar, A.A.; Khanlou, N.; Braun, J.; et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015, 27, 240–256. [Google Scholar] [CrossRef] [Green Version]
- Nathanson, K.L.; Martin, A.M.; Wubbenhorst, B.; Greshock, J.; Letrero, R.; D’Andrea, K.; O’Day, S.; Infante, J.R.; Falchook, G.S.; Arkenau, H.T.; et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436). Clin. Cancer Res. 2013, 19, 4868–4878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conway, C.; Beswick, S.; Elliott, F.; Chang, Y.M.; Randerson-Moor, J.; Harland, M.; Affleck, P.; Marsden, J.; Sanders, D.S.; Boon, A.; et al. Deletion at chromosome arm 9p in relation to BRAF/NRAS mutations and prognostic significance for primary melanoma. Genes Chromosomes Cancer 2010, 49, 425–438. [Google Scholar] [CrossRef] [Green Version]
- Hodi, F.S.; Corless, C.L.; Giobbie-Hurder, A.; Fletcher, J.A.; Zhu, M.; Marino-Enriquez, A.; Friedlander, P.; Gonzalez, R.; Weber, J.S.; Gajewski, T.F.; et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J. Clin. Oncol. 2013, 31, 3182–3190. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.T.; Roussel, M.F.; Sherr, C.J. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2006, 103, 6688–6693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schadendorf, D.; Hodi, F.S.; Robert, C.; Weber, J.S.; Margolin, K.; Hamid, O.; Patt, D.; Chen, T.T.; Berman, D.M.; Wolchok, J.D. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J. Clin. Oncol. 2015, 33, 1889–1894. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Ribas, A.; Schachter, J.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.M.; Lotem, M.; et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): Post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019, 20, 1239–1251. [Google Scholar] [CrossRef]
- Hodi, F.S.; Chesney, J.; Pavlick, A.C.; Robert, C.; Grossmann, K.F.; McDermott, D.F.; Linette, G.P.; Meyer, N.; Giguere, J.K.; Agarwala, S.S.; et al. Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2-year overall survival outcomes in a multicentre, randomised, controlled, phase 2 trial. Lancet Oncol. 2016, 17, 1558–1568. [Google Scholar] [CrossRef] [Green Version]
- Hodi, F.S.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018, 19, 1480–1492. [Google Scholar] [CrossRef]
- Wargo, J.A.; Reddy, S.M.; Reuben, A.; Sharma, P. Monitoring immune responses in the tumor microenvironment. Curr. Opin. Immunol. 2016, 41, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, I.; Guroji, P.; DeBrot, A.H.; Manapragada, P.P.; Katiyar, S.K.; Elmets, C.A.; Yusuf, N. Loss of INK4a/Arf gene enhances ultraviolet radiation-induced cutaneous tumor development. Exp. Dermatol. 2017, 26, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef] [Green Version]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef]
- Horn, S.; Leonardelli, S.; Sucker, A.; Schadendorf, D.; Griewank, K.G.; Paschen, A. Tumor CDKN2A-Associated JAK2 Loss and Susceptibility to Immunotherapy Resistance. J. Natl. Cancer Inst. 2018, 110, 677–681. [Google Scholar] [CrossRef]
- DeLeon, T.T.; Almquist, D.R.; Kipp, B.R.; Langlais, B.T.; Mangold, A.; Winters, J.L.; Kosiorek, H.E.; Joseph, R.W.; Dronca, R.S.; Block, M.S.; et al. Assessment of clinical outcomes with immune checkpoint inhibitor therapy in melanoma patients with CDKN2A and TP53 pathogenic mutations. PLoS ONE 2020, 15, e0230306. [Google Scholar] [CrossRef] [Green Version]
- Helgadottir, H.; Ghiorzo, P.; van Doorn, R.; Puig, S.; Levin, M.; Kefford, R.; Lauss, M.; Queirolo, P.; Pastorino, L.; Kapiteijn, E.; et al. Efficacy of novel immunotherapy regimens in patients with metastatic melanoma with germline CDKN2A mutations. J. Med. Genet. 2020, 57, 316–321. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.A.; Schuchter, L.M. Chemotherapy for Melanoma. Cancer Treat. Res. 2016, 167, 209–229. [Google Scholar]
- Middleton, M.R.; Grob, J.J.; Aaronson, N.; Fierlbeck, G.; Tilgen, W.; Seiter, S.; Gore, M.; Aamdal, S.; Cebon, J.; Coates, A.; et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J. Clin. Oncol. 2000, 18, 158–166. [Google Scholar] [CrossRef]
- Yang, A.S.; Chapman, P.B. The history and future of chemotherapy for melanoma. Hematol. Oncol. Clin. N. Am. 2009, 23, 583–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallagher, S.; Kefford, R.F.; Rizos, H. Enforced expression of p14ARF induces p53-dependent cell cycle arrest but not apoptosis. Cell Cycle 2005, 4, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.W.; Zhu, X.F.; Huang, X.F.; Sheng, P.Y.; He, A.S.; Yang, Z.B.; Deng, R.; Feng, G.K.; Liao, W.M. P14ARF sensitizes human osteosarcoma cells to cisplatin-induced apoptosis in a p53-independent manner. Cancer Biol. Ther. 2007, 6, 1074–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, M.; Voss, D.; Park-Simon, T.W.; Mahlberg, R.; Koster, G. Role of p16 and p14ARF in radio- and chemosensitivity of malignant gliomas. Oncol. Rep. 2006, 16, 127–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallagher, S.J.; Thompson, J.F.; Indsto, J.; Scurr, L.L.; Lett, M.; Gao, B.F.; Dunleavey, R.; Mann, G.J.; Kefford, R.F.; Rizos, H. p16INK4a expression and absence of activated B-RAF are independent predictors of chemosensitivity in melanoma tumors. Neoplasia 2008, 10, 1231–1239. [Google Scholar] [CrossRef] [Green Version]
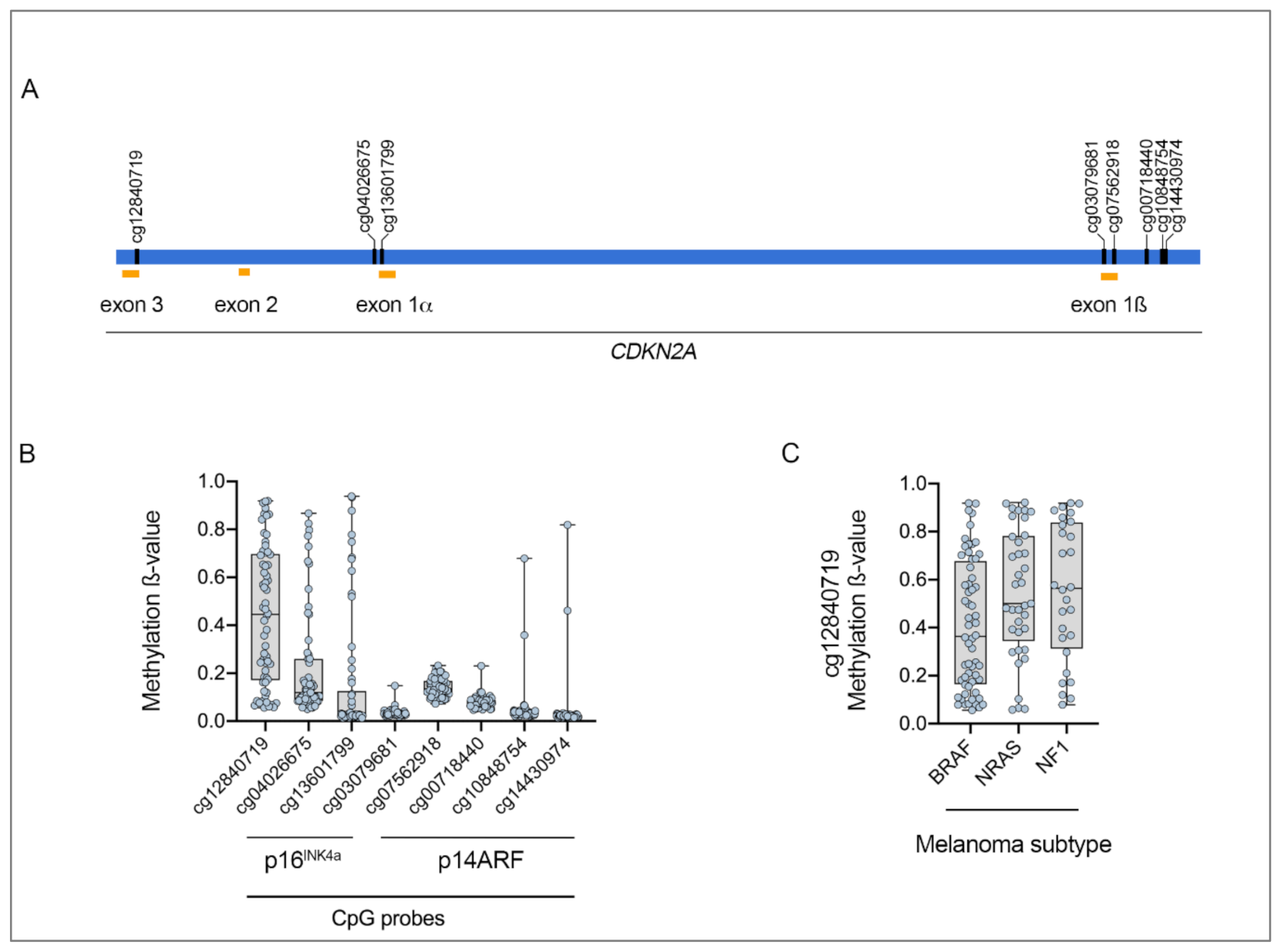
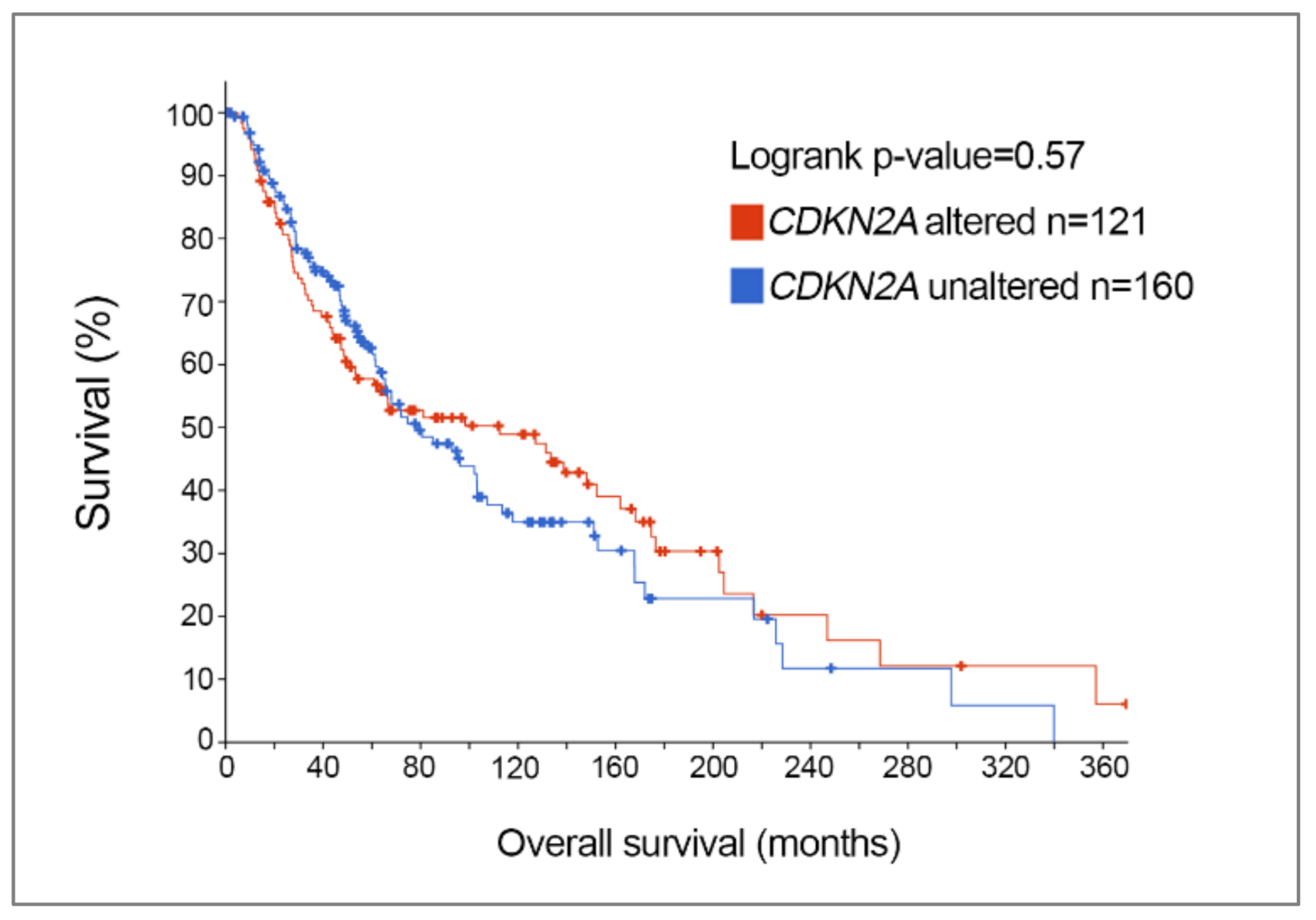
| Molecular Subtype | BRAF (n = 188) | NRAS (n = 114) | NF1 (n = 66) | Triple Wild-Type (n = 45) |
|---|---|---|---|---|
| Mutations in p16INK4a only | 7.4% (14/188) | 6.1% (7/114) | 9.1% (6/66) | 0% (0/45) |
| Mutations Affecting p16INK4a and p14ARF | 6.9% (13/188) | 7.9% (9/114) | 12.1% (8/66) | 4.4% (2/45) |
| CDKN2A Deletions | 34.6% (65/188) | 30.7% (35/114) | 28.8% (19/66) | 17.8% (8/45) |
| CDKN2A Hyper-Methylation | 67.0% (126/188) | 87.7% (100/114) | 77.3% (51/66) | 66.7% (30/45) |
| Germline Mutations | CDKN2 ANM_000077.4 (p16INK4a) | CDKN2A NM_058195.3 (p14ARF) |
|---|---|---|
| Total Number of Coding Variants | 435 | 179 |
| Number of Coding Variants | ||
| Exon 1a | 140/435 (32%) | 0/477 (0%) |
| Exon 1b | 0/435 (0% | 2/477 (0%) |
| Exon 2 | 295/435 (68%) | 177/477 (37%) |
| Exon 3 | 0/435 (0%) | 0/477 (0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ming, Z.; Lim, S.Y.; Rizos, H. Genetic Alterations in the INK4a/ARF Locus: Effects on Melanoma Development and Progression. Biomolecules 2020, 10, 1447. https://doi.org/10.3390/biom10101447
Ming Z, Lim SY, Rizos H. Genetic Alterations in the INK4a/ARF Locus: Effects on Melanoma Development and Progression. Biomolecules. 2020; 10(10):1447. https://doi.org/10.3390/biom10101447
Chicago/Turabian StyleMing, Zizhen, Su Yin Lim, and Helen Rizos. 2020. "Genetic Alterations in the INK4a/ARF Locus: Effects on Melanoma Development and Progression" Biomolecules 10, no. 10: 1447. https://doi.org/10.3390/biom10101447





