Strain-Specific Liver Metabolite Profiles in Medaka
Abstract
:1. Introduction
2. Results
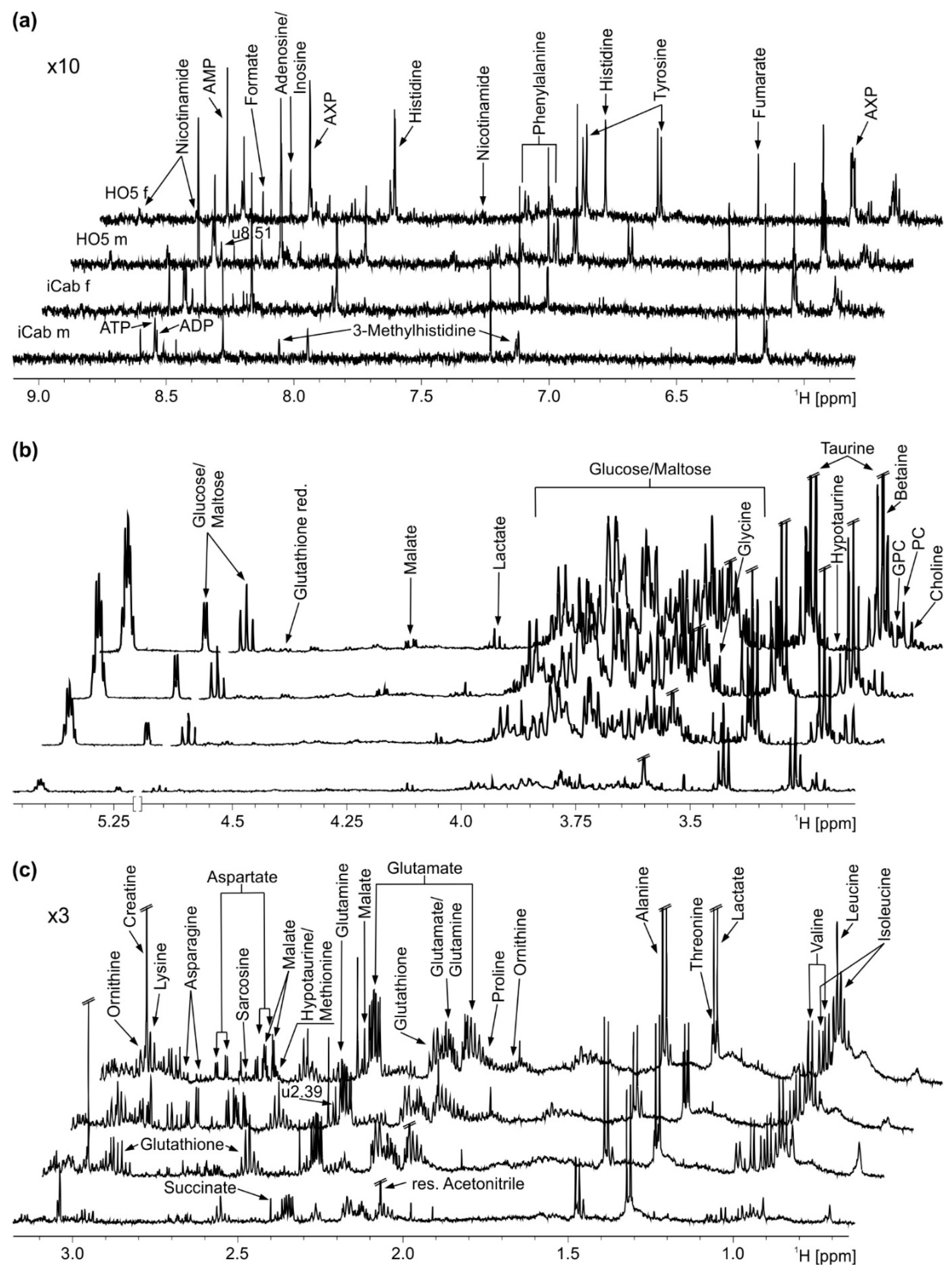
2.1. NMR Spectroscopy of Liver Extracts
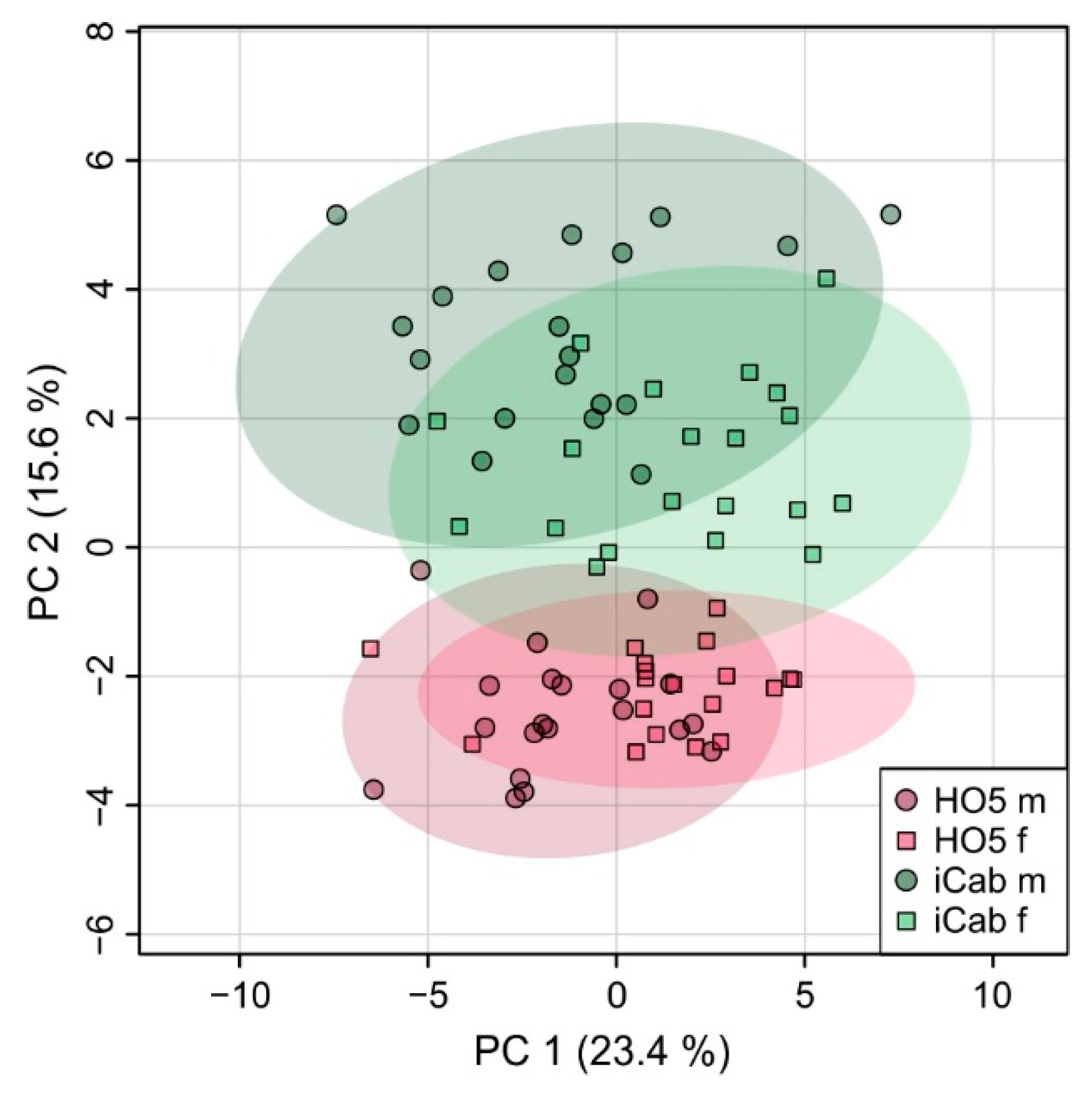
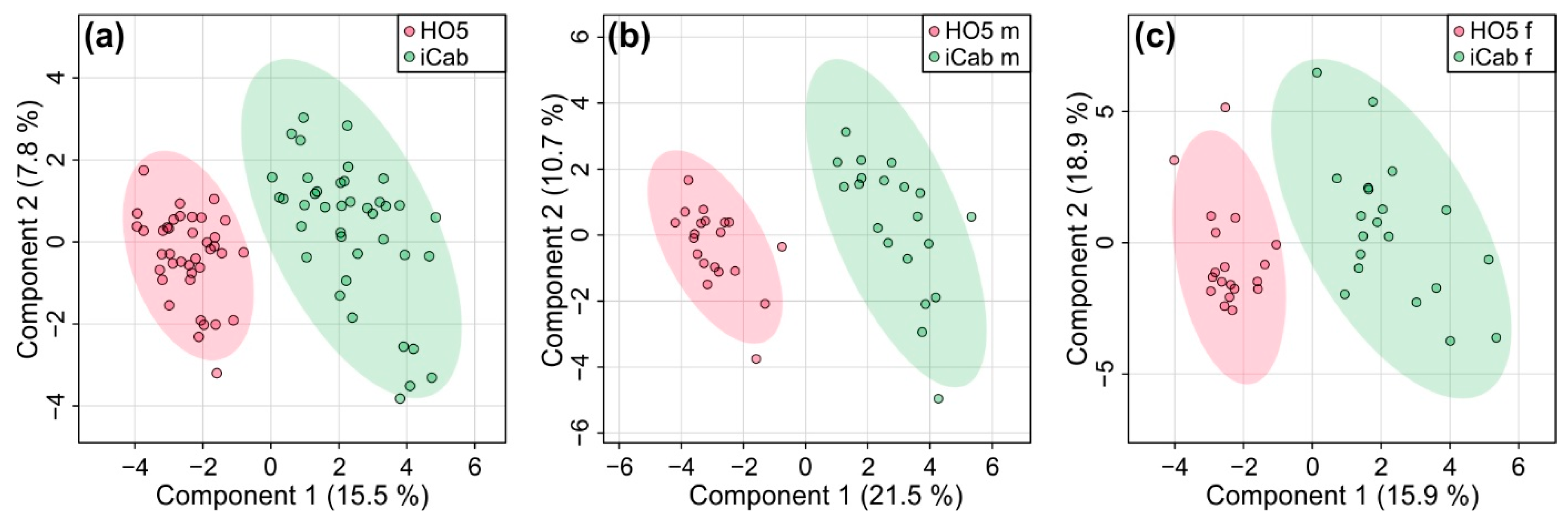
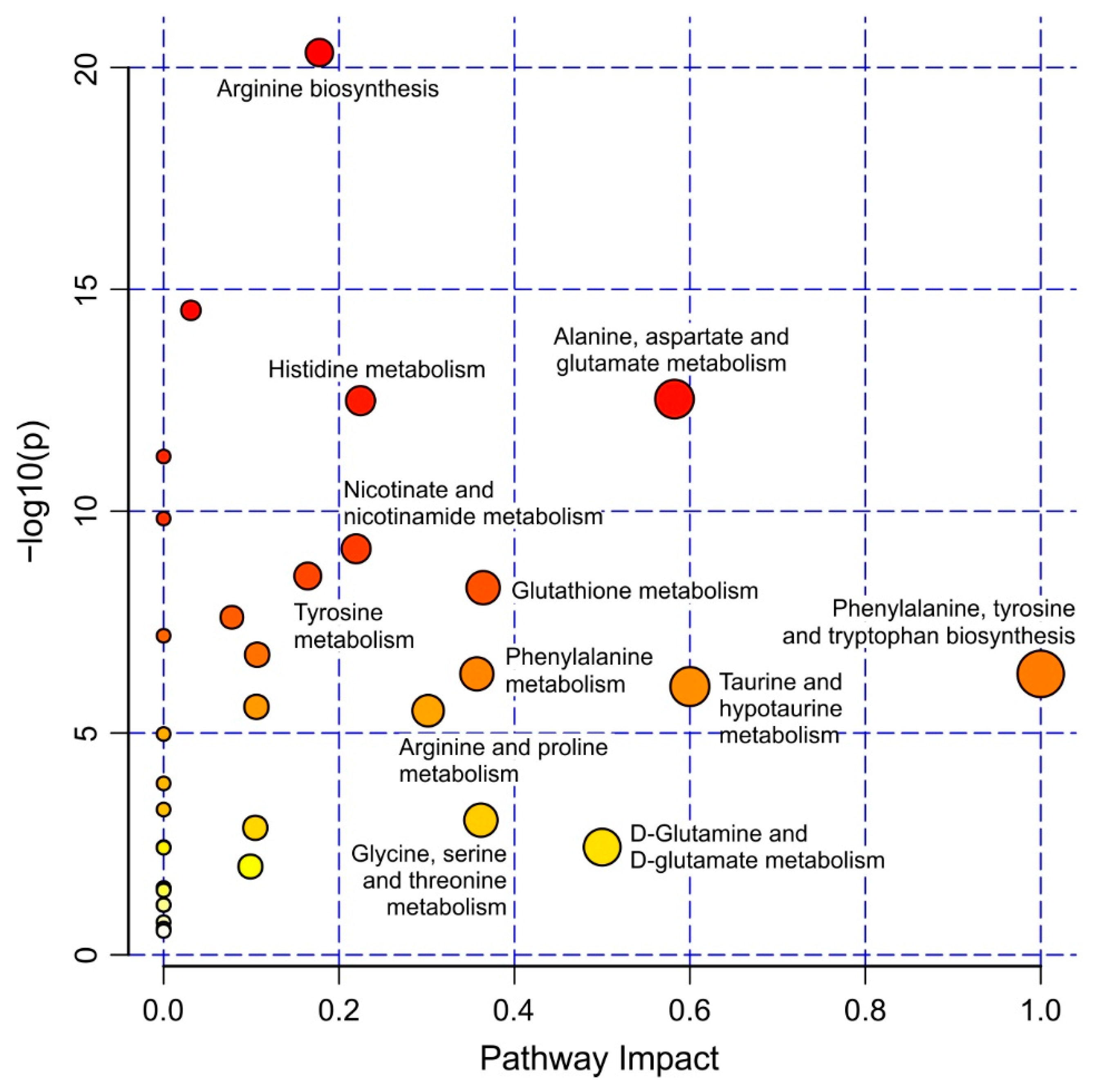
2.2. Strain-Specific Metabolic Profile of Liver Extracts
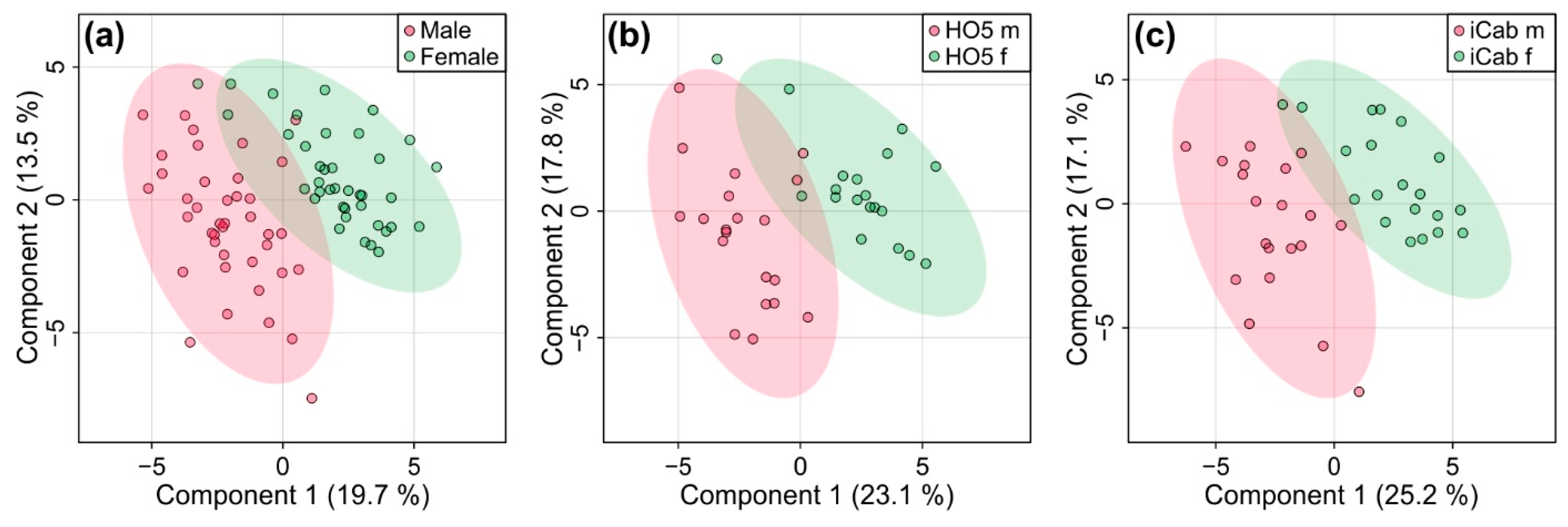
2.3. Metabolic Differences between Male and Female Fish Are Not as Pronounced as between the Inbred Strains
3. Discussion
4. Materials and Methods
4.1. Animal Handling and Tissue Collection
4.2. Sample Preparation
4.3. 1H-NMR Spectroscopy of Liver Samples
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228, . [Google Scholar] [CrossRef]
- Hoffman, E.P.; Brown, R.H.; Kunkel, L.M. Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell 1987, 51, 919–928. [Google Scholar] [CrossRef]
- Claussnitzer, M.; Cho, J.H.; Collins, R.; Cox, N.J.; Dermitzakis, E.T.; Hurles, M.E.; Kathiresan, S.; Kenny, E.E.; Lindgren, C.M.; MacArthur, D.G.; et al. A brief history of human disease genetics. Nature 2020, 577, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Sinnott-Armstrong, N.; Naqvi, S.; Rivas, M.; Pritchard, J.K. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. eLife 2021, 10, e58615. [Google Scholar] [CrossRef]
- Aygen, S.; Dürr, U.; Hegele, P.; Kunig, J.; Spraul, M.; Schäfer, H.; Krings, D.; Cannet, C.; Fang, F.; Schütz, B.; et al. NMR-Based Screening for Inborn Errors of Metabolism: Initial Results from a Study on Turkish Neonates. JIMD Rep. 2014, 16, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Edghill, E.L.; Bingham, C.; Ellard, S.; Hattersley, A.T. Mutations in hepatocyte nuclear factor-1beta and their related phenotypes. J. Med. Genet. 2006, 43, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Torell, F.; Bennett, K.; Cereghini, S.; Fabre, M.; Rännar, S.; Lundstedt-Enkel, K.; Moritz, T.; Haumaitre, C.; Trygg, J.; Lundstedt, T. Metabolic Profiling of Multiorgan Samples: Evaluation of MODY5/RCAD Mutant Mice. J. Proteome Res. 2018, 17, 2293–2306. [Google Scholar] [CrossRef] [PubMed]
- Burlikowska, K.; Stryjak, I.; Bogusiewicz, J.; Kupcewicz, B.; Jaroch, K.; Bojko, B. Comparison of Metabolomic Profiles of Organs in Mice of Different Strains Based on SPME-LC-HRMS. Metabolites 2020, 10, 255. [Google Scholar] [CrossRef]
- Pann, P.; de Angelis, M.H.; Prehn, C.; Adamski, J. Mouse Age Matters: How Age Affects the Murine Plasma Metabolome. Metabolites 2020, 10, 472. [Google Scholar] [CrossRef]
- Takeda, H.; Shimada, A. The art of medaka genetics and genomics: What makes them so unique? Annu. Rev. Genet. 2010, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Gao, N.; Wang, R.; Zhang, L. Proteomic and metabolomic analysis of marine medaka (Oryzias melastigma) after acute ammonia exposure. Ecotoxicology 2018, 27, 267–277. [Google Scholar] [CrossRef]
- Le Manach, S.; Sotton, B.; Huet, H.; Duval, C.; Paris, A.; Marie, A.; Yépremian, C.; Catherine, A.; Mathéron, L.; Vinh, J.; et al. Physiological effects caused by microcystin-producing and non-microcystin producing Microcystis aeruginosa on medaka fish: A proteomic and metabolomic study on liver. Environ. Pollut. 2018, 234, 523–537. [Google Scholar] [CrossRef]
- Lai, K.P.; Gong, Z.; Tse, W.K.F. Zebrafish as the toxicant screening model: Transgenic and omics approaches. Aquat. Toxicol. 2021, 234, 105813. [Google Scholar] [CrossRef]
- Fujisawa, K.; Takami, T.; Kimoto, Y.; Matsumoto, T.; Yamamoto, N.; Terai, S.; Sakaida, I. Circadian variations in the liver metabolites of medaka (Oryzias latipes). Sci. Rep. 2016, 6, 20916. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, T.; Okimura, K.; Shen, J.; Guh, Y.-J.; Tamai, T.K.; Shimada, A.; Minou, S.; Okushi, Y.; Shimmura, T.; Furukawa, Y.; et al. Seasonal changes in NRF2 antioxidant pathway regulates winter depression-like behavior. Proc. Natl. Acad. Sci. USA 2020, 117, 9594–9603. [Google Scholar] [CrossRef] [Green Version]
- Weger, B.D.; Weger, M.; Görling, B.; Schink, A.; Gobet, C.; Keime, C.; Poschet, G.; Jost, B.; Krone, N.; Hell, R.; et al. Extensive Regulation of Diurnal Transcription and Metabolism by Glucocorticoids. PLoS Genet. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Spivakov, M.; Auer, T.O.; Peravali, R.; Dunham, I.; Dolle, D.; Fujiyama, A.; Toyoda, A.; Aizu, T.; Minakuchi, Y.; Loosli, F.; et al. Genomic and phenotypic characterization of a wild medaka population: Towards the establishment of an isogenic population genetic resource in fish. G3 Genes Genomes Genet. 2014, 4, 433–445. [Google Scholar] [CrossRef] [Green Version]
- Qiao, Q.; Le Manach, S.; Sotton, B.; Huet, H.; Duvernois-Berthet, E.; Paris, A.; Duval, C.; Ponger, L.; Marie, A.; Blond, A.; et al. Deep sexual dimorphism in adult medaka fish liver highlighted by multi-omic approach. Sci. Rep. 2016, 6, 32459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry, 6th ed.; W.H. Freeman and Company: New York, NY, USA, 2013; ISBN 978-1-4641-0962-1. [Google Scholar]
- Ruoppolo, M.; Caterino, M.; Albano, L.; Pecce, R.; Di Girolamo, M.G.; Crisci, D.; Costanzo, M.; Milella, L.; Franconi, F.; Campesi, I. Targeted metabolomic profiling in rat tissues reveals sex differences. Sci. Rep. 2018, 8, 4663. [Google Scholar] [CrossRef] [PubMed]
- Leskanicova, A.; Chovancova, O.; Babincak, M.; Verboova, L.; Benetinova, Z.; Macekova, D.; Kostolny, J.; Smajda, B.; Kiskova, T. Sexual Dimorphism in Energy Metabolism of Wistar Rats Using Data Analysis. Molecules 2020, 25, 2353. [Google Scholar] [CrossRef] [PubMed]
- Leskanicova, A.; Chovancova, O.; Babincak, M.; Blicharova, A.; Kolesarova, M.; Macekova, D.; Kostolny, J.; Smajda, B.; Kiskova, T. Defining sex differences in selected lipid metabolites of blood plasma in Wistar rats. J. Physiol. Pharmacol. 2019, 70. [Google Scholar] [CrossRef]
- Vignoli, A.; Tenori, L.; Luchinat, C.; Saccenti, E. Age and Sex Effects on Plasma Metabolite Association Networks in Healthy Subjects. J. Proteome Res. 2018, 17, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loosli, F.; Köster, R.W.; Carl, M.; Kühnlein, R.; Henrich, T.; Mücke, M.; Krone, A.; Wittbrodt, J. A genetic screen for mutations affecting embryonic development in medaka fish (Oryzias latipes). Mech. Dev. 2000, 97, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.F.; Kanehisa, M. Using the KEGG database resource. Curr. Protoc. Bioinform. 2005. Chapter 1, Unit 1.12. Available online: https://www.genome.jp/kegg/ (accessed on 13 October 2021). [CrossRef]
| Both Sexes | Male | Female | ||||
|---|---|---|---|---|---|---|
| Multivariate | Univariate | Multivariate | Univariate | Multivariate | Univariate | |
| VIP Score | FC (p-Value) | VIP Score | FC (p-Value) | VIP Score | FC (p-Value) | |
| Aspartate | 1.86 | 1.54 (1.39 × 10−9) | 1.79 | 1.70 (1.62 × 10−7) | 1.67 | 1.41 (0.0012) |
| Lactate/Threonine | 1.82 | 0.56 (2.88 × 10−9) | 1.57 | 0.46 (1.51 × 10−5) | 1.87 | 0.67 (1.16 × 10−4) |
| ATP | 1.80 | 0.56 (2.90 × 10−9) | 1.71 | 0.48 (9.37 × 10−7) | 1.65 | 0.67 (0.0012) |
| u8.51 | 1.66 | 0.54 (1.08 × 10−7) | 1.48 | 0.53 (5.01 × 10−5) | 1.91 | 0.53 (1.16 × 10−4) |
| Fumarate | 1.58 | 2.21 (6.94 × 10−7) | 1.52 | 3.09 (2.77 × 10−5) | 1.29 | 1.69 (0.0170) |
| Phenylalanine | 1.35 | 1.65 (5.40 × 10−5) | 1.30 | 2.00 (6.78 × 10−4) | 1.09 | 1.39 (0.0428) |
| Malate | 1.29 | 1.34 (1.42 × 10−4) | 0.96 | 1.32 (0.0154) | 1.44 | 1.37 (0.0066) |
| Ornithine | 1.22 | 1.34 (2.78 × 10−4) | 1.06 | 1.35 (0.0073) | 1.46 | 1.34 (0.0066) |
| Tyrosine | 1.22 | 1.49 (2.78 × 10−4) | 1.23 | 1.61 (0.0013) | 1.35 | 1.43 (0.0118) |
| Hypotaurine | 1.20 | 5.10 (3.54 × 10−4) | 1.57 | 5.69 (1.51 × 10−5) | 1.10 | 5.09 (0.0428) |
| Alanine | 1.19 | 0.78 (3.70 × 10−4) | 1.52 | 0.72 (2.77 × 10−5) | 0.90 | 0.83 (0.0971) |
| u2.39 | 1.15 | 1.40 (5.39 × 10−4) | 1.30 | 1.88 (6.78 × 10−4) | 0.56 | 1.10 (0.3076) |
| AMP | 1.14 | 1.36 (5.40 × 10−4) | 0.85 | 1.39 (0.0318) | 1.43 | 1.33 (0.0066) |
| Choline | 1.06 | 0.79 (0.0013) | 0.72 | 0.87 (0.0741) | 1.28 | 0.72 (0.0170) |
| Sarcosine | 1.02 | 0.77 (0.0022) | 0.45 | 0.88 (0.3004) | 1.42 | 0.69 (0.0066) |
| Both Strains | HO5 | iCab | ||||
|---|---|---|---|---|---|---|
| Multivariate | Univariate | Multivariate | Univariate | Multivariate | Univariate | |
| VIP Score | FC (p-Value) | VIP Score | FC (p-Value) | VIP Score | FC (p-Value) | |
| 3-Methylhistidine | 2.31 | 12.77 (2.63 × 10−12) | 2.10 | 12.48 (1.31 × 10−6) | 1.84 | 13.00 (1.46 × 10−5) |
| Creatine | 2.28 | 0.42 (3.98 × 10−12) | 1.84 | 0.42 (9.39 × 10−5) | 2.06 | 0.42 (1.64 × 10−7) |
| Glutathione_red | 1.68 | 0.65 (1.11 × 10−5) | 1.33 | 0.76 (0.0114) | 1.55 | 0.55 (0.0006) |
| Glutamine | 1.57 | 0.80 (5.15 × 10−5) | 1.83 | 0.75 (9.39 × 10−5) | 0.94 | 0.85 (0.0579) |
| Tyrosine | 1.47 | 0.66 (0.0002) | 1.49 | 0.69 (0.0037) | 1.32 | 0.61 (0.0053) |
| Glycerophosphocholine | 1.45 | 1.39 (0.0002) | 0.82 | 1.19 (0.1397) | 1.57 | 1.60 (0.0005) |
| Ornithine | 1.36 | 0.75 (0.0006) | 1.23 | 0.75 (0.0145) | 1.61 | 0.75 (0.0003) |
| Hypotaurine | 1.31 | 0.20 (0.0010) | 1.05 | 0.20 (0.0433) | 1.72 | 0.18 (8.43 × 10−5) |
| Formate | 1.29 | 0.16 (0.0010) | 1.07 | 0.20 (0.0394) | 1.27 | 0.16 (0.0061) |
| Nicotinamide | 1.28 | 1.57 (0.0010) | 1.27 | 1.56 (0.0125) | 1.04 | 1.59 (0.0343) |
| Alanine | 1.19 | 0.80 (0.0022) | 1.51 | 0.74 (0.0033) | 0.79 | 0.86 (0.1132) |
| u8.51 | 1.16 | 1.46 (0.0028) | 1.36 | 1.47 (0.0091) | 1.27 | 1.47 (0.0061) |
| Aspartate | 0.75 | 0.86 (0.0743) | 0.52 | 0.92 (0.4048) | 1.30 | 0.76 (0.0058) |
| ATP | 0.74 | 1.23 (0.0749) | 0.03 | 1.01 (0.9656) | 1.33 | 1.39 (0.0053) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soergel, H.; Loosli, F.; Muhle-Goll, C. Strain-Specific Liver Metabolite Profiles in Medaka. Metabolites 2021, 11, 744. https://doi.org/10.3390/metabo11110744
Soergel H, Loosli F, Muhle-Goll C. Strain-Specific Liver Metabolite Profiles in Medaka. Metabolites. 2021; 11(11):744. https://doi.org/10.3390/metabo11110744
Chicago/Turabian StyleSoergel, Hannah, Felix Loosli, and Claudia Muhle-Goll. 2021. "Strain-Specific Liver Metabolite Profiles in Medaka" Metabolites 11, no. 11: 744. https://doi.org/10.3390/metabo11110744






