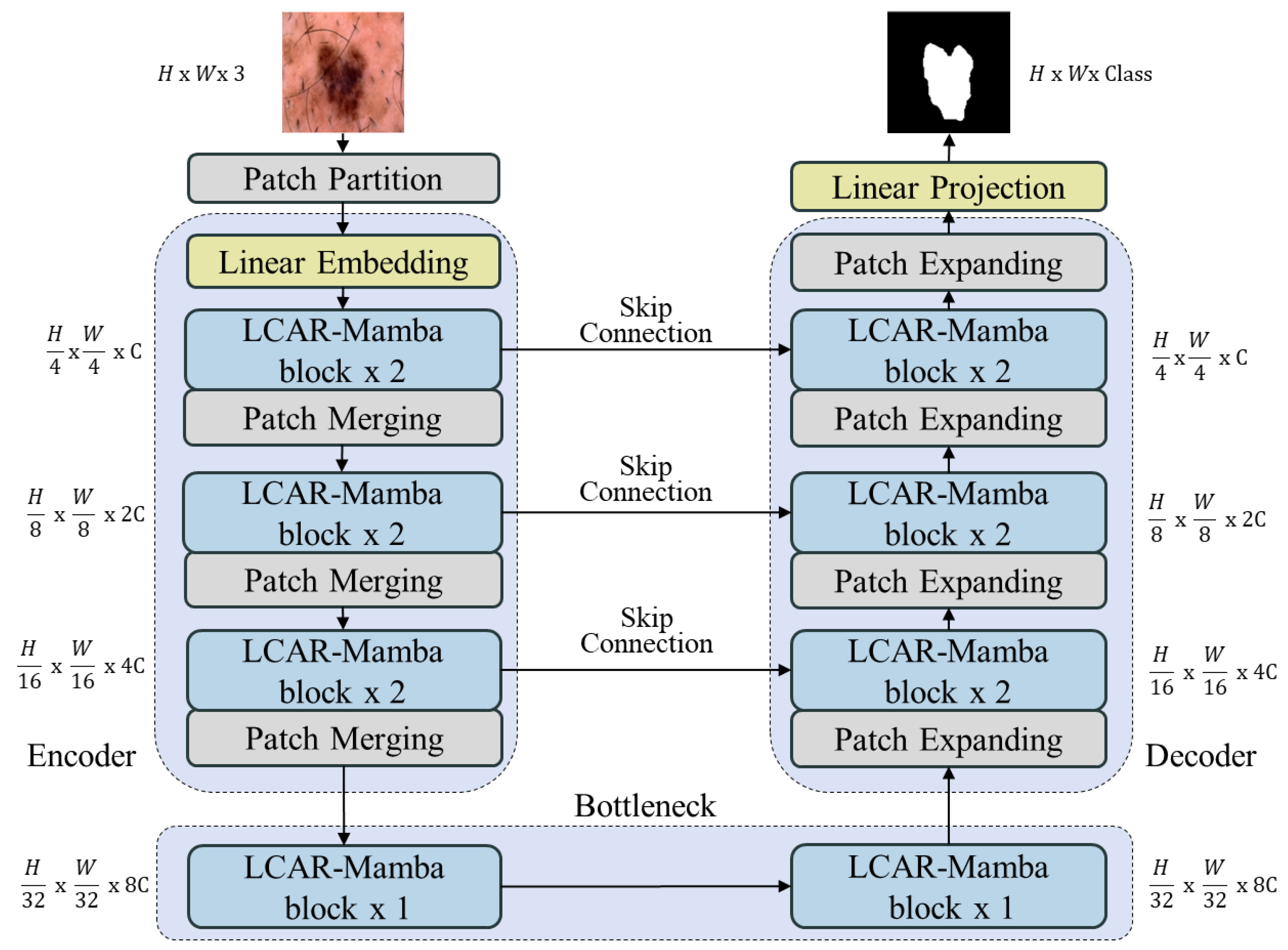
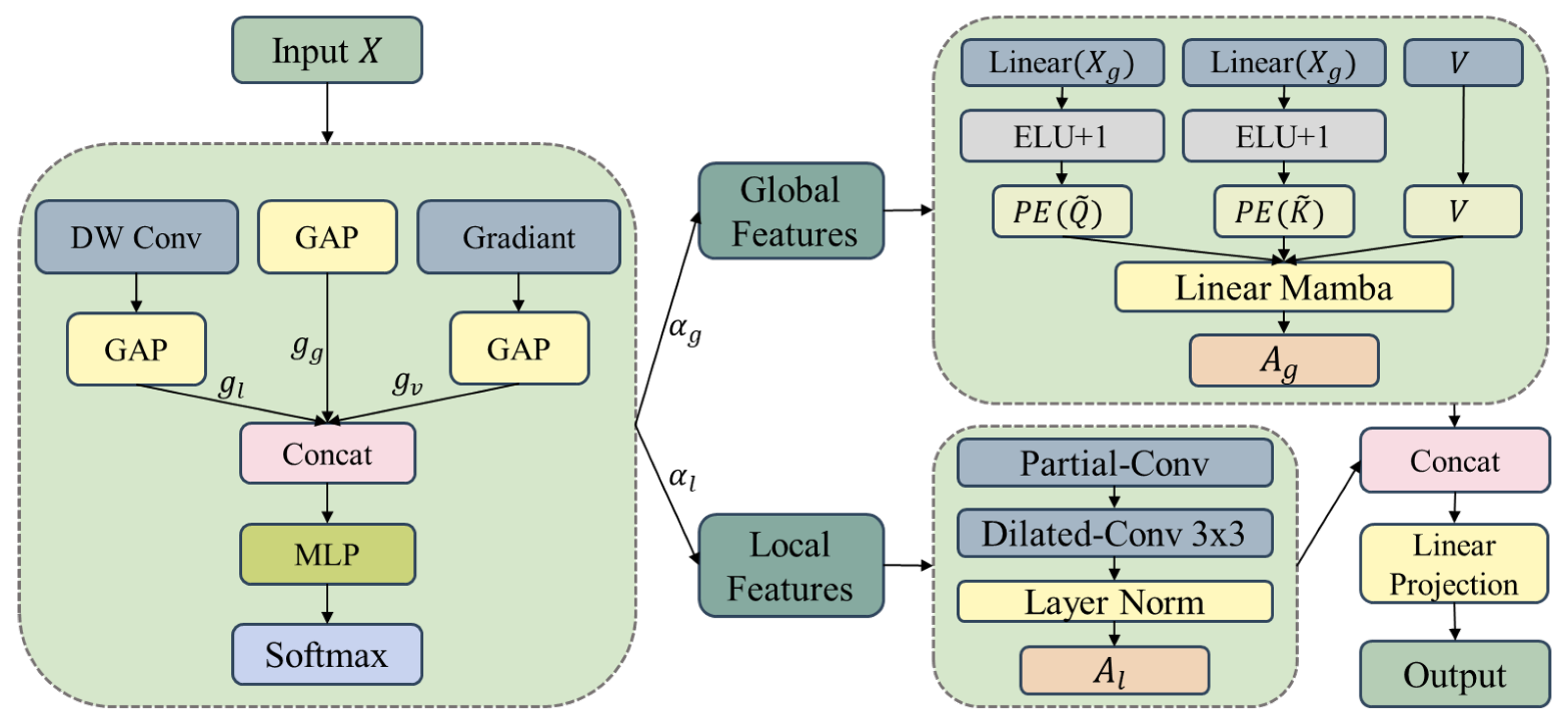
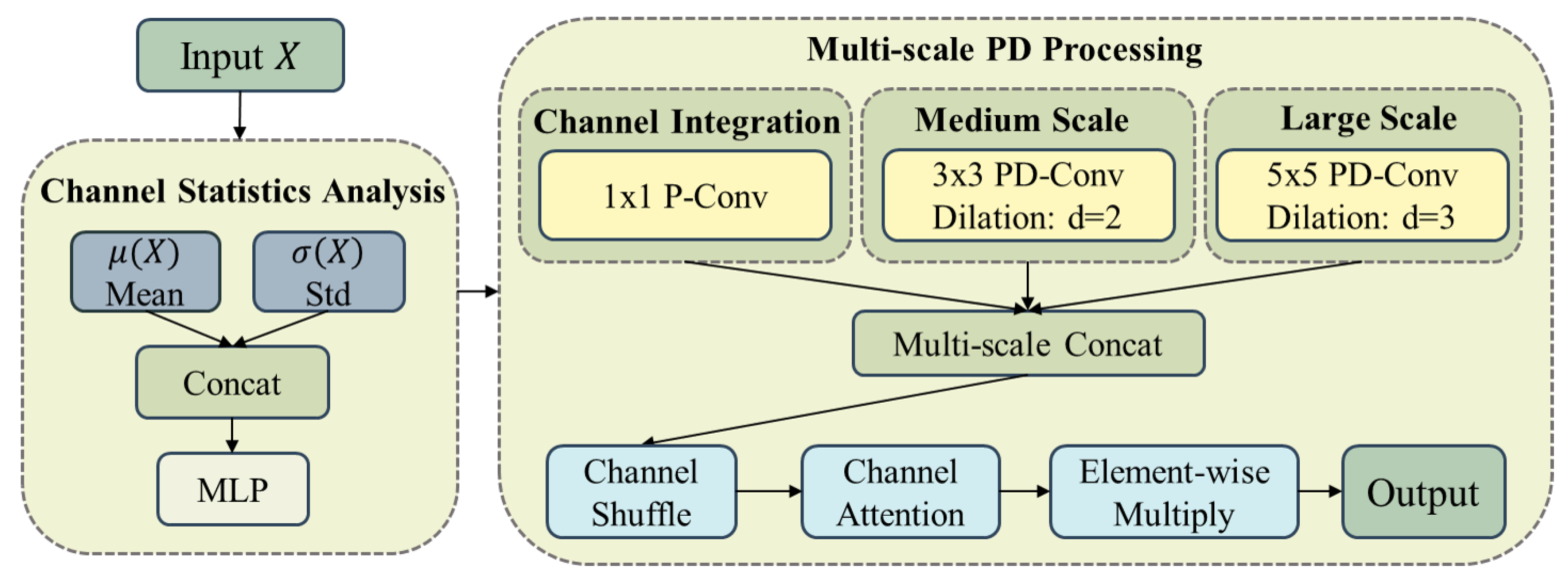
1. Introduction
With the rapid advancement of deep learning techniques, significant progress has been achieved in medical image segmentation [
1,
2,
3,
4]. Despite these advances, current segmentation methods face substantial challenges when dealing with complex medical images. Medical images frequently exhibit intricate multi-scale structural characteristics, ranging from pixel-level textural details to organ-level morphological contours. This requires that segmentation models be capable of capturing fine-grained local information while simultaneously modeling global semantic relationships. In addiction, practical clinical applications require segmentation models to robustly handle highly heterogeneous and complex anatomical structures, which requires adaptive modeling capabilities that can effectively address diverse tissue types, lesion characteristics, and contextual variations across different medical images and imaging modalities.
The main approaches to medical image segmentation are based primarily on two technical paradigms: convolutional neural networks (CNNs) [
5] and vision Transformers (ViTs) [
6]. CNN-based methods, exemplified by U–Net [
1] and its variants, have achieved excellent segmentation performance through encoder–decoder architectures. Building on this foundation, Attention U–Net [
7] introduced attention gates to focus on target structures while suppressing irrelevant regions, and U–Net++ [
8] redesigned skip connections with nested dense skip pathways for improved feature aggregation across multiple scales. To address multi-scale feature extraction, the DeepLab series [
9] introduced Atrous Spatial Pyramid Pooling (ASPP) to capture multi-scale contextual information, while PSPNet [
10] employed pyramid pooling modules for similar purposes. Efficient convolution strategies have emerged to reduce computational overhead while maintaining local feature extraction capabilities. Partial convolutions [
11] reduce computational cost through selective channel processing, while Ghost convolutions [
12] generate feature maps through efficient transformations. Similarly, depthwise separable convolutions in MobileNets [
13] and mixed-scale convolutions in MixNet [
14] provide efficient alternatives for local feature processing. However, because of the inherently local nature of convolution operations, these models face intrinsic limitations in modeling long-range dependencies. Although researchers have proposed various mitigation strategies such as dilated convolutions and attention mechanisms [
7,
15], these approaches often incur a substantial increase in computational complexity.
ViT-based methods leverage self-attention mechanisms to enable global feature modeling, partially alleviating the challenge of modeling long-range dependencies. Vision Transformers [
6] demonstrated that pure attention mechanisms could achieve competitive performance without convolution operations, addressing CNN limitations in global context modeling. TransUNet [
16] pioneered the integration of Transformers with U–Net architectures, combining global context with fine-grained localization. Building on this foundation, TransFuse [
17] proposed parallel CNN–Transformer processing with attention-based fusion, while UTNetV2 [
18] further explored hybrid strategies to leverage the complementary strengths of both paradigms. Although effective in capturing global dependencies, standard ViTs incur substantial computational costs due to the quadratic complexity of self-attention. To address this challenge, advanced Transformer architectures have been developed specifically for medical imaging. Swin–UNet [
2] employed hierarchical Swin Transformer blocks with windowed attention to reduce complexity, while MedT [
19] proposed gated axial attention optimized for medical image characteristics. Similarly, MISSFormer [
20] introduced an efficient architecture tailored for 2D medical image segmentation through enhanced feature aggregation. To fundamentally overcome the quadratic complexity bottleneck, researchers have explored linear attention mechanisms that approximate self-attention more efficiently. Linear Transformers [
21] achieve linear complexity through kernel-based attention approximations, while Linformer [
22] reduces complexity by projecting attention matrices to lower-dimensional spaces. Nevertheless, these approximations often sacrifice modeling capacity, and processing high-resolution medical images remains challenging because of memory constraints even with linear methods.
Recently, state space models (SSMs), such as Mamba [
23], have attracted considerable attention due to their strong long-sequence modeling capabilities and linear computational complexity. Mamba achieves efficient and expressive feature modeling through an innovative selection mechanism and a hardware-friendly design. Vision Mamba [
24] extends this architecture to computer vision tasks, demonstrating substantial potential in image analysis. Building on Vision Mamba, numerous architectures have been developed for medical image segmentation. VM–UNet [
25] integrates Vision Mamba blocks into U–Net frameworks, and H–VMUNet [
26] proposes hierarchical Vision Mamba architectures with multi-scale processing capabilities. UltraLight VM–UNet [
27] focuses on parameter efficiency for resource-constrained deployment, Mamba–UNet [
28] investigates pure Mamba architectures without convolutional components, MambaMIR [
29] addresses medical image reconstruction and uncertainty estimation using arbitrarily masked Mamba architectures, LocalMamba [
30] combines local and global Mamba processing to tackle fine-grained structure extraction, and MSVM–UNet [
31] integrates multi-scale Vision Mamba with U–Net architectures for medical image segmentation.
Despite these advancements, existing advanced architectures, including current Mamba-based methods, still exhibit critical limitations in medical image segmentation. They lack adaptive mechanisms to accommodate varying content complexity and therefore fail to achieve context-aware resource allocation for heterogeneous medical images, in which pathological regions require intensive processing, whereas normal tissues can be handled more efficiently. Moreover, although these methods achieve linear complexity for global modeling, they provide insufficient local feature extraction for fine structures such as organ boundaries and small lesions. This mismatch between existing architectures and the requirements of medical image segmentation—particularly the need for adaptive context-aware processing and enhanced local feature extraction—motivates our LCAR–Mamba architecture, which incorporates novel context-aware linear Mamba modules and multi-scale partial dilated convolution strategies. Recent studies have attempted to mitigate these limitations. For example, Rahman and Marculescu [
32] proposed a medical image segmentation method with cascaded attention decoding, Azad et al. [
33] investigated deformable large-kernel attention, and subsequent works such as G–Cascade [
34] and EMCAD [
35] further introduced efficient cascaded graph convolutional decoding and multi-scale convolutional attention decoding architectures. Ruan et al. proposed MALUNet [
36] and EGE–UNet [
37], which explore lightweight designs with multi-attention and efficient group enhancement strategies for skin lesion segmentation, respectively. However, conventional partial convolution strategies employed in these approaches may overlook important information and thereby adversely affect the segmentation accuracy of fine structures such as boundaries.
To address these issues, this paper proposes a Linear Context-Aware Robust Mamba architecture that integrates two synergistic modules designed to directly tackle the identified limitations. The design philosophy of LCAR-Mamba builds upon recent Vision Mamba architectures for medical imaging [
25] and draws inspiration from efficient multi-stage feature extraction strategies in medical image super-resolution [
38], where combining attention mechanisms with lightweight convolutional operations has proven effective for balancing performance and computational cost. LCAR-Mamba comprises two key modules: the Context-Aware Linear Mamba (CALM) module and the Multi-scale Partial Dilated Convolution (MSPD) module. CALM implements a four-stage adaptive processing framework that performs context complexity analysis, adaptive resource allocation, dual-path parallel processing, and intelligent feature fusion, enabling context-aware feature representation and substantially improving model performance. MSPD employs channel statistics analysis, multi-scale parallel processing, and adaptive feature integration to achieve data-driven channel utilization and multi-scale feature fusion while maintaining feature integrity and significantly reducing computational complexity.
The main contributions of this work are summarized as follows:
We propose the CALM module, which enables context-aware feature modeling through a four-stage adaptive processing framework, significantly enhancing the feature representation capability and robustness of linear Mamba;
We design the MSPD module, which utilizes channel statistics analysis and multi-scale feature fusion strategies to substantially reduce computational complexity while maintaining segmentation accuracy;
We construct the LCAR–Mamba architecture, which seamlessly integrates global and local feature modeling through the collaborative optimization of CALM and MSPD, achieving superior performance in medical image segmentation by combining context-aware adaptive processing with intelligent multi-scale feature extraction;
We conduct comprehensive experiments on three public medical image segmentation datasets [
39,
40,
41], and the results demonstrate that our method achieves state-of-the-art performance on various segmentation tasks.
3. Experiments and Results
3.1. Datasets
Our method was evaluated on three public datasets: ISIC2017, ISIC2018 and Synapse.
3.1.1. ISIC2017 and ISIC2018
The ISIC2017 [
40] and ISIC2018 [
39] datasets are publicly available skin lesion segmentation benchmarks, containing 2150 and 2694 images, respectively. Following the previous work [
36] on ISIC2017 and ISIC2018, we used a 7:3 train–test split. For ISIC2017, this resulted in 1500 training images and 650 test images. For ISIC2018, we obtained 1886 training images and 808 test images. All images were resized to
. The experiments used fixed random seed = 42. Segmentation performance was evaluated using five metrics: mean Intersection over Union (mIoU), Dice Similarity Coefficient (DSC), Sensitivity (SE), Specificity (SP) and Accuracy (ACC).
3.1.2. Synapse
The Synapse dataset [
41] is a publicly available multi-organ segmentation dataset comprising 30 abdominal CT cases with 3779 axial images across eight organs. Following [
16], 18 cases were used for training and 12 for testing. Performance was evaluated using the Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95).
We employ a 2D slice-wise approach. Each 3D CT volume is decomposed into axial slices and resized from to pixels using bicubic interpolation for images and nearest-neighbor interpolation for masks. The dataset contains volumes with voxel spacing ranging from ([0.54 0.54] × [0.98 0.98] × [2.5 5.0]) mm3. Following standard evaluation protocols, Hausdorff Distance 95 (HD95) was computed using a normalized spacing of 1.0 mm for fair comparison with baseline methods.
3.2. Implementation Details
All experiments were conducted using the PyTorch 2.2.2 framework on an NVIDIA RTX 3090 GPU. We adopted the AdamW optimizer with an initial learning rate of , weight decay of , and cosine annealing learning rate scheduler. The batch size was set to 48 and the network was trained for up to 300 epochs.
Inference and Post-processing. For ISIC datasets, predictions were obtained by applying a threshold of 0.5 to the sigmoid output. For Synapse, we used argmax over the softmax outputs to generate per-pixel class predictions. No additional post-processing techniques (e.g., conditional random fields, test-time augmentation, or largest connected component filtering) were employed in our experiments, ensuring that all reported results reflect the direct model output.
Baseline Implementation. To ensure fair comparison, all baseline methods were trained under identical experimental settings to LCAR-Mamba, including the same input resolution (256 × 256 for ISIC, 224 × 224 for Synapse), training configuration (300 epochs, AdamW optimizer with learning rate , cosine annealing scheduler), and data augmentation strategies.
Data Augmentation. During training, the following augmentations were applied:
ISIC datasets: Random horizontal flip (), random vertical flip (), and random rotation (0°–360°)
Synapse dataset: Random rotation by 90° increments (0°, 90°, 180°, and 270°) with and random flip along vertical or horizontal axis with
3.3. Comparisons with State of the Art
To demonstrate the effectiveness of the proposed LCAR–Mamba, we conducted extensive comparisons with state-of-the-art approaches on three publicly available medical image segmentation datasets.
Skin Lesion Segmentation.
Table 1 and
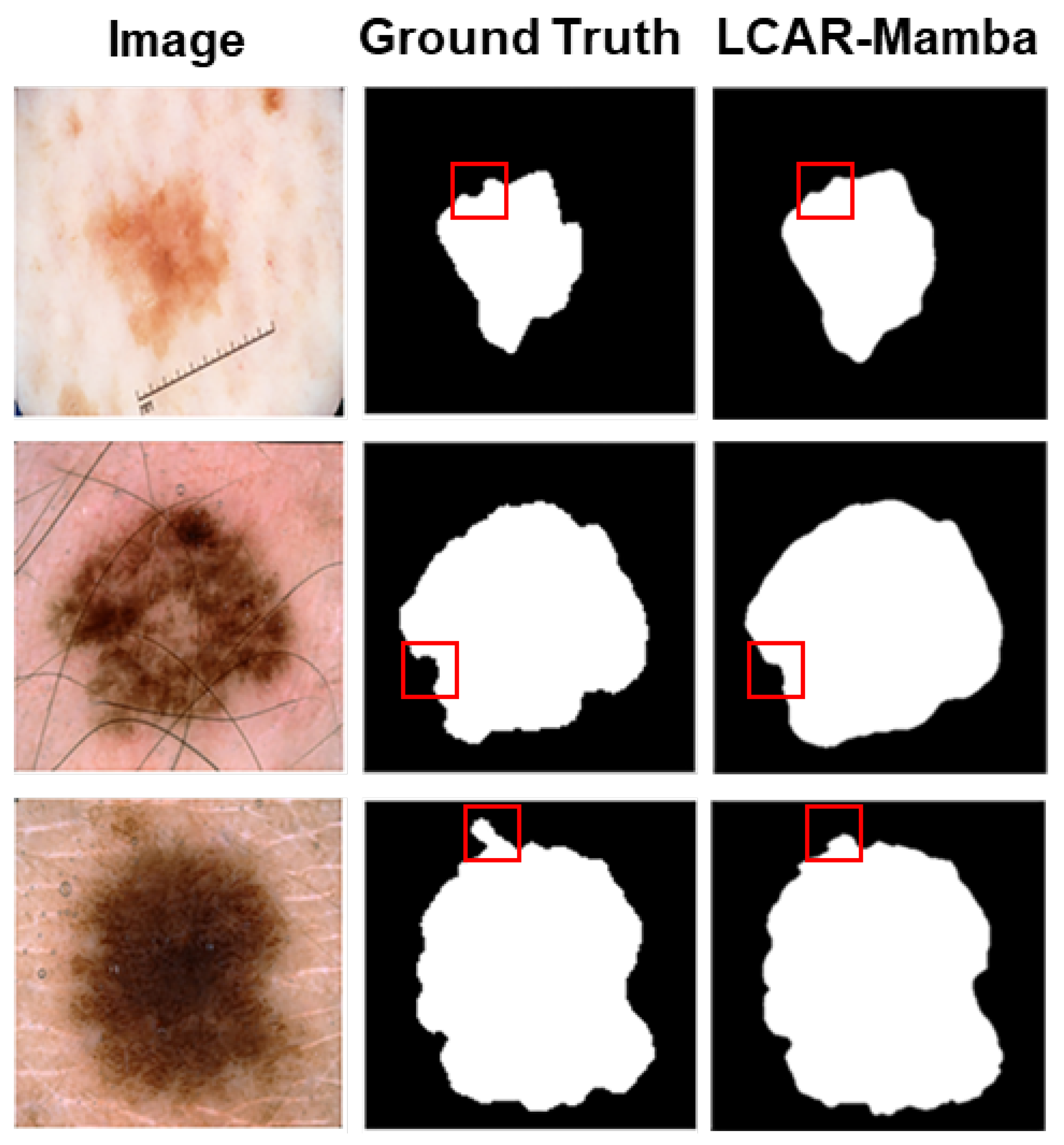
Table 2 report the quantitative results on the ISIC2017 and ISIC2018 datasets, respectively. On ISIC2017, LCAR–Mamba achieves the highest mIoU of 80.79% and DSC of 89.42%, outperforming the Mamba-based model VM–UNet by 0.62% in mIoU and 0.52% in DSC. Notably, our method yields substantial gains in sensitivity, reaching 89.54%, which is 1.95% higher than VM–UNet (87.59%). On ISIC2018, LCAR–Mamba achieving an mIoU of 81.68% and a DSC of 89.93%, surpassing VM–UNet by 1.25% and 0.89%, respectively. Moreover, LCAR–Mamba attains the highest specificity of 97.29%, representing a 1.10% improvement over VM–UNet (96.19%). These consistent improvements on both skin lesion datasets highlight the effectiveness of combining context-aware adaptive processing through the CALM module with enhanced local feature extraction capabilities through the MSPD module. The qualitative comparisons shown in
Figure 4 further demonstrate that LCAR–Mamba produces more precise boundary delineations and better captures fine-grained lesion details compared to existing methods.
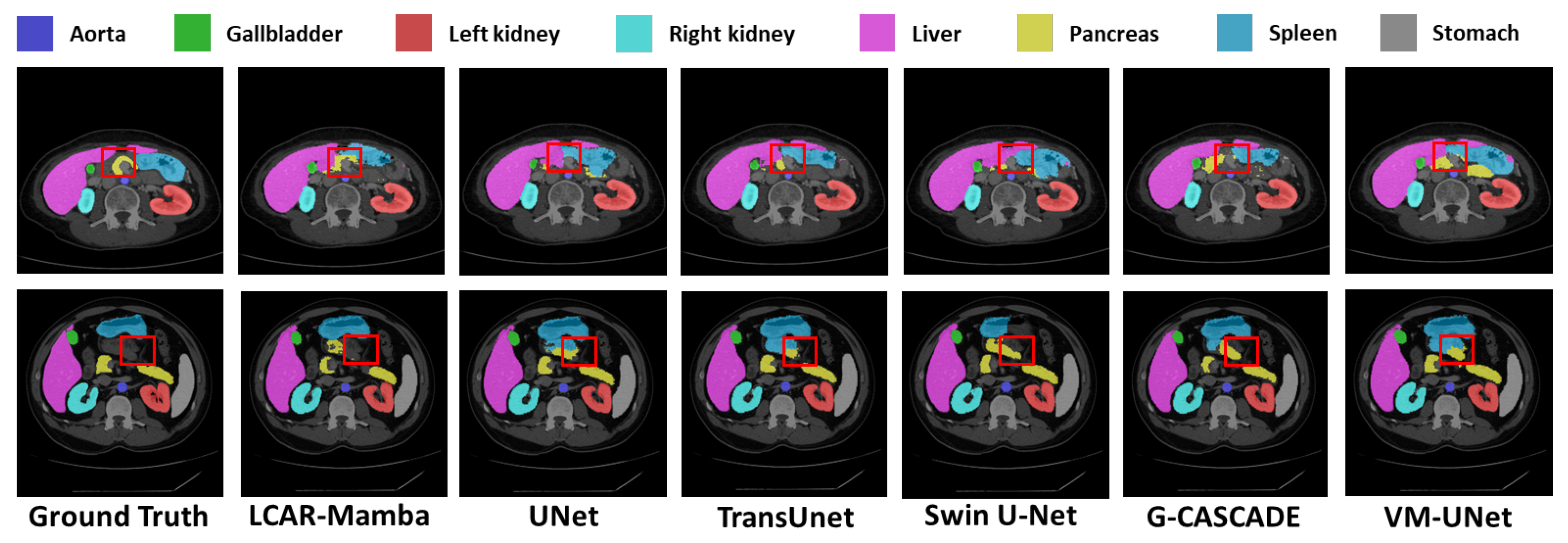
Multi-Organ Segmentation.
Table 3 demonstrates the performance of LCAR–Mamba on the Synapse multi-organ segmentation dataset. LCAR–Mamba achieves the highest overall DSC score of 85.56% and an HD95 value of 13.15 mm, reflecting strong segmentation accuracy and boundary precision. Compared to its closest competitor MSVM–UNet, LCAR–Mamba improves DSC by 0.48% and HD95 by 1.28 mm. Compared to the Mamba-based baseline VM–UNet, LCAR–Mamba yields substantial gains of 2.90% in DSC and 2.75 mm in HD95, highlighting the advantages of the CALM and MSPD modules over conventional Mamba architectures. Compared to pure Transformer-based methods such as Swin–UNet and TransUNet, LCAR–Mamba outperforms them by 6.15% and 8.48% in DSC, along with substantial HD95 improvements of 8.13 mm and 30.74 mm, respectively, demonstrating the effectiveness of the proposed hybrid architecture. LCAR–Mamba also achieves competitive performance across most anatomical structures. In particular, it achieves the highest DSC scores for challenging organs, including the aorta (89.52%), gallbladder (76.43%), left kidney (88.75%), liver (96.21%) and stomach (88.15%). Even for small and irregularly shaped organs such as the pancreas, LCAR–Mamba maintains competitive performance with a DSC of 67.87%, comparable to state-of-the-art methods. Qualitative comparisons in
Figure 5 further show that LCAR–Mamba produces more precise organ boundaries and better preserves fine anatomical structures.
3.4. Statistical Significance Analysis
To rigorously assess the reliability of our results, we conducted statistical significance testing using paired
t-tests across 3 independent runs with different random seeds (42, 123, and 456).
Table 4 summarizes the statistical analysis comparing LCAR–Mamba with VM-UNet on ISIC2017 and ISIC2018 datasets.
All improvements achieved by LCAR-Mamba are statistically significant at the level (p < 0.05), confirming that the observed performance gains are not due to random variation. The relatively small standard deviations (0.12–0.21%) across different seeds indicate stable and reproducible training dynamics.
3.5. Failure Case Analysis
Figure 6 shows representative failure cases on ISIC2017. LCAR–Mamba struggles with fuzzy boundaries in low-contrast regions (Row 1), hair occlusion artifacts (Row 2), and pigmentation heterogeneity (Row 3). These cases reveal ongoing challenges in boundary refinement and noise robustness.
3.6. Ablation Studies
To validate the effectiveness of each component in the LCAR–Mamba architecture, we conducted comprehensive ablation studies on the Synapse dataset. The experiments are organized into two levels: module-level ablation and component-level ablation.
3.6.1. Module-Level Ablation
We first conduct module-level ablation experiments to assess the contribution of the CALM and MSPD modules.
Table 5 summarizes the effect of each module on model performance. Incorporating the CALM module improves DSC from 82.66% to 84.12% while reducing the number of parameters from 44.75 M to 38.21 M, demonstrating the effectiveness of the proposed context–aware linear Mamba. Introducing the MSPD module substantially reduces the computational cost while maintaining high accuracy (83.87%), with the parameter count further reduced to 31.45 M. The complete LCAR–Mamba model achieves the best overall performance, reaching a DSC of 85.56% with only 29.83 M parameters, corresponding to an approximate 33% reduction in parameters compared to the baseline.
3.6.2. MSPD Component-Level Ablation
We further analyze the contribution of each component in the MSPD module.
Table 6 highlights several key observations regarding the MSPD design. The statistics-based channel selection strategy is more effective than random selection, improving DSC from 82.91% to 83.34%. Multi-scale processing further enhances feature representation, with the three-scale configuration achieving the best trade-off between performance and complexity, reaching 83.87% DSC and yielding a 1.21% improvement over the baseline model. However, extending to four scales introduces parameter redundancy with only marginal performance gains, indicating that three scales constitute the optimal configuration for this architecture.
3.6.3. CALM Component-Level Ablation
We systematically analyze the contribution of each functional component in the CALM module.
Table 7 presents controlled ablation experiments isolating the complexity analysis mechanism and the adaptive fusion strategy.
The complexity analysis mechanism proves essential for content-aware processing. Without complexity–driven resource allocation (using uniform weights throughout), performance improves to 83.45% DSC, representing a 0.79% gain over the baseline but 0.67% below full CALM. This confirms that analyzing spatial heterogeneity via the Context-Aware Feature Analyzer is crucial for optimal resource distribution.
The adaptive fusion strategy also contributes independently. When complexity descriptors guide feature partitioning but fusion uses fixed weights, the model achieves 83.68% DSC—-a 1.02% improvement over the baseline yet 0.44% below full CALM. This demonstrates that while complexity-aware partitioning provides benefits, adaptive fusion in the Dual-Path Processor is necessary for optimal feature integration.
4. Conclusions
This paper introduces LCAR–Mamba, a hybrid architecture that addresses key limitations of current medical image segmentation methods through two synergistic components: the CALM module and the MSPD module. Extensive experiments on three public datasets show consistent and substantial performance gains across diverse imaging modalities and anatomical structures. The CALM module addresses the fundamental challenge of adaptive resource allocation in heterogeneous medical images, enabling context-aware feature modeling with linear computational complexity, as evidenced by a 1.46% DSC increase and a 14.6% reduction in parameters on the Synapse dataset. The MSPD module further demonstrates that intelligent channel selection and multi-scale processing can significantly enhance feature representations while reducing computational overhead, with the statistics-based selection strategy improving DSC by 0.43% compared with random selection.
Compared with pure CNN architectures such as U–Net [
1], LCAR–Mamba achieves a 10.51% DSC improvement on Synapse by more effectively modeling long-range dependencies, while surpassing Transformer-based methods [
2,
16] with approximately 33% fewer parameters. In addition, the 2.90% DSC improvement over existing Mamba-based methods [
25] validates the effectiveness of the proposed context-aware adaptive processing and multi-scale feature extraction mechanisms. These results underscore both the superior performance and parameter efficiency of LCAR–Mamba across different architectural paradigms.
Despite its strong empirical performance, several promising research directions remain. Future work includes extending the framework to 3D volumetric segmentation, enhancing small lesion detection (e.g., pancreas segmentation at 67.87% DSC), exploring semi-supervised or weakly supervised learning settings, and investigating dynamic network adaptation strategies for real-time clinical deployment. In summary, LCAR–Mamba integrates global context modeling with local feature extraction through adaptive processing mechanisms, achieving state-of-the-art performance across diverse medical segmentation tasks, with notable gains in accuracy (85.56% DSC on Synapse) and boundary precision (13.15 mm HD95), and offering a robust solution for practical clinical applications.