The Cytochrome CYP4 in Breast and Other Cancers
Simple Summary
Abstract
1. Introduction
2. Database Analysis
3. The Effect of CYPs on Several Types of Cancer
3.1. Breast Cancer
3.2. Bioinformatic Analysis of CYPs in Breast Cancer
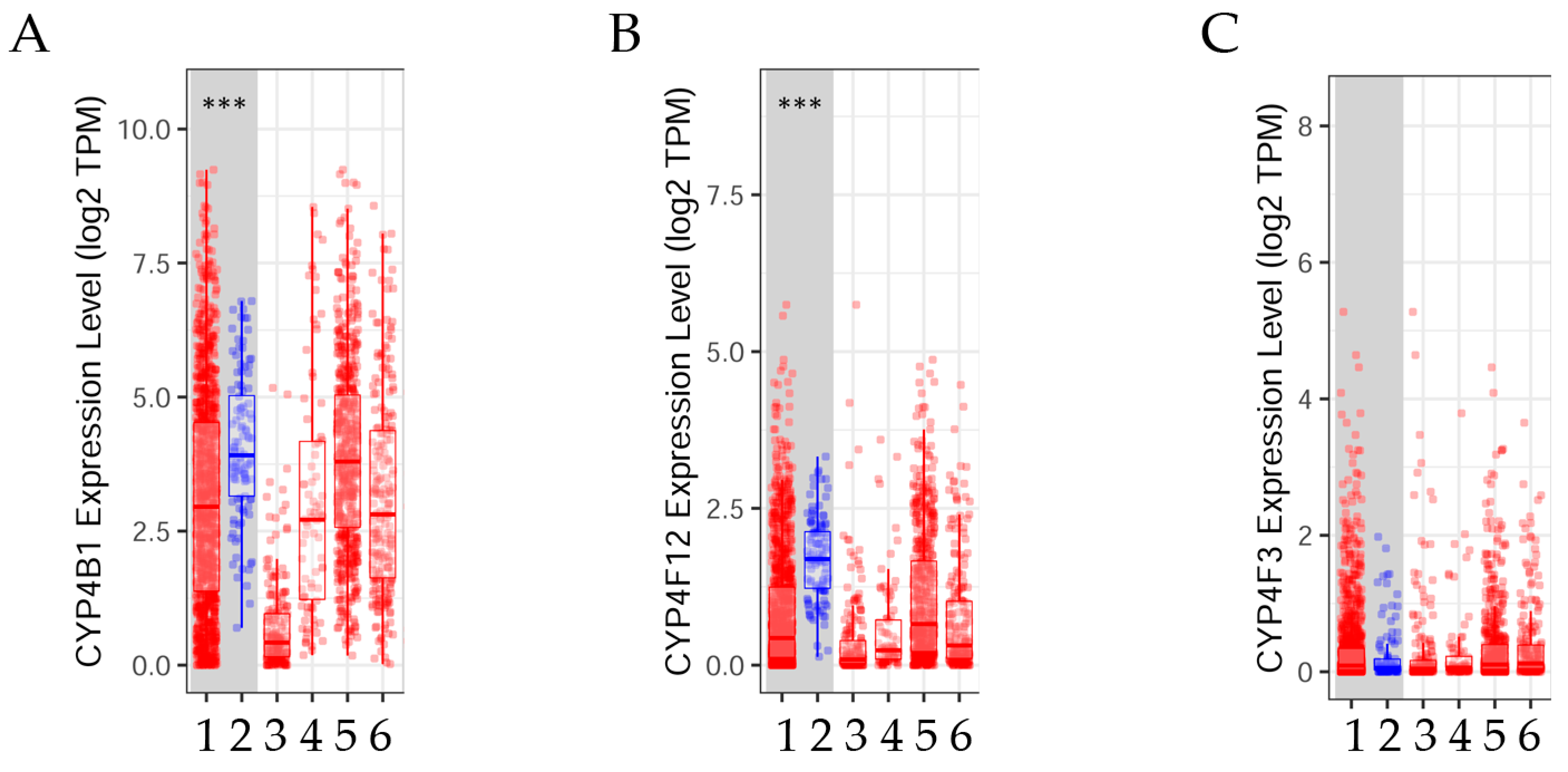
3.2.1. Gene Expression Levels in Normal Versus Tumor Tissues
3.2.2. CYP4B1, CYP4F12, and CYP4F3 Gene Expression Levels Adjusted by Stages
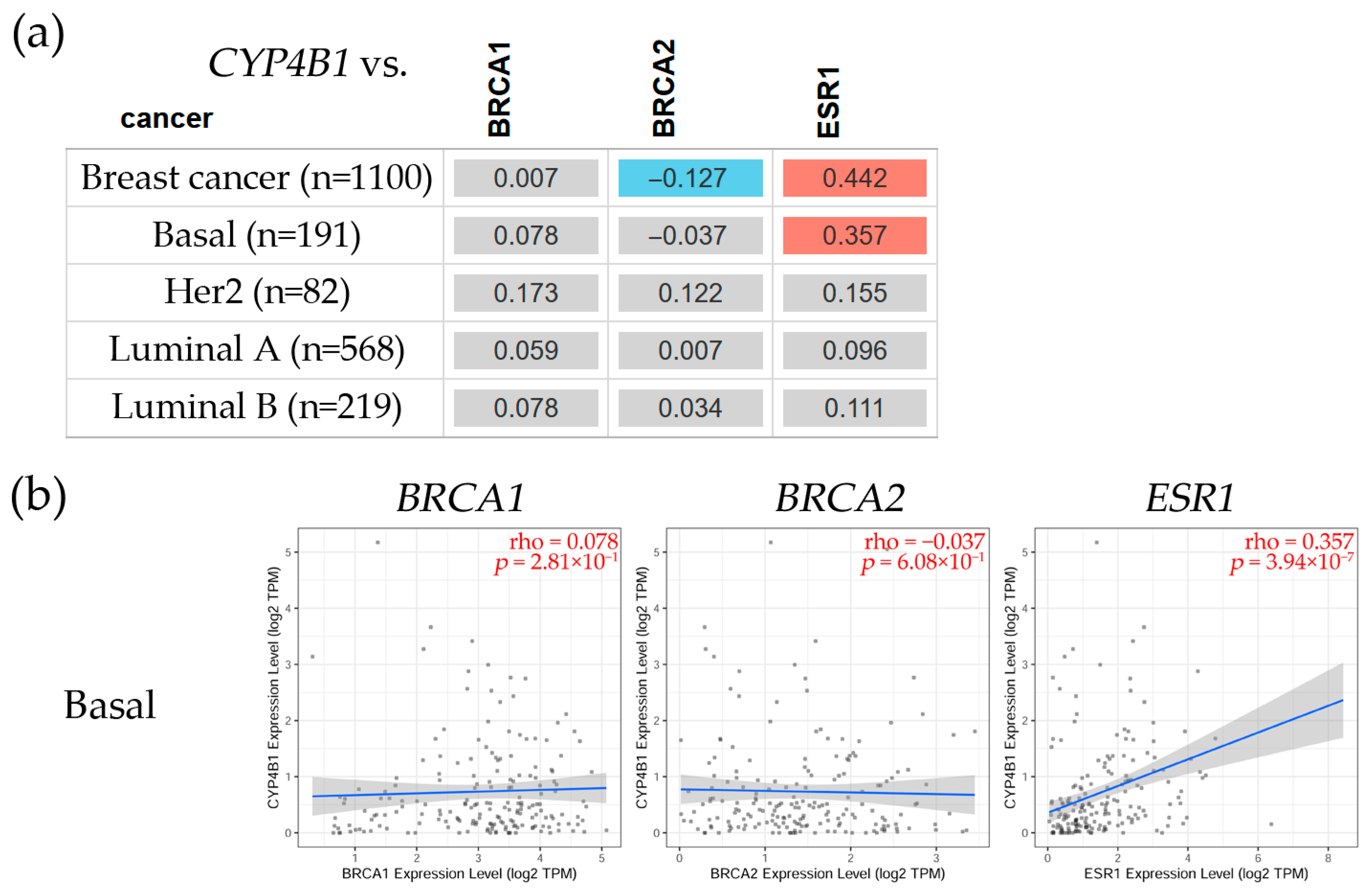
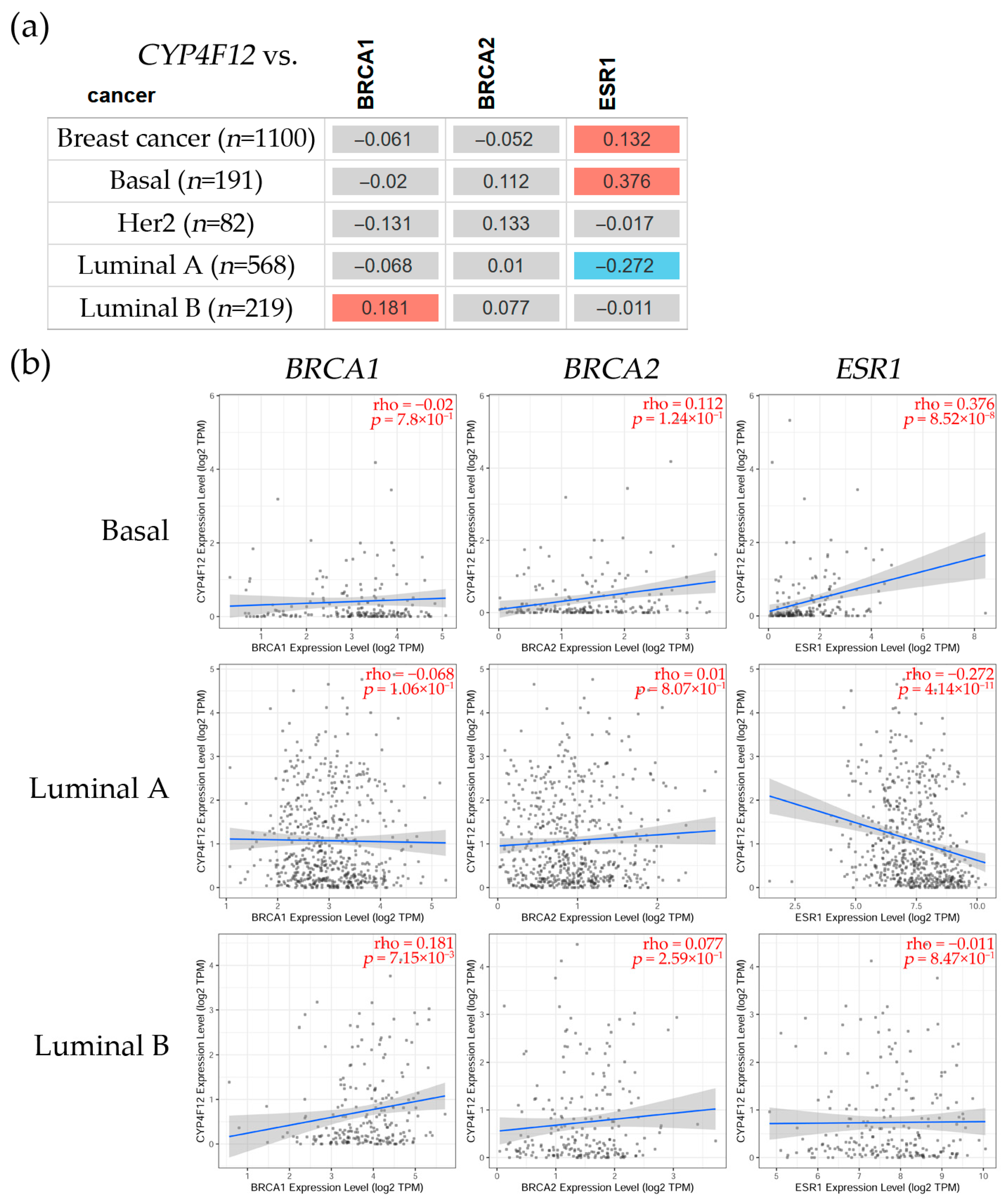
3.2.3. Correlation Between CYP4B1, CYP4F12, and CYP4F3 and BRCA1, BRCA2, and ESR1 Gene Expression Levels in Breast Cancer
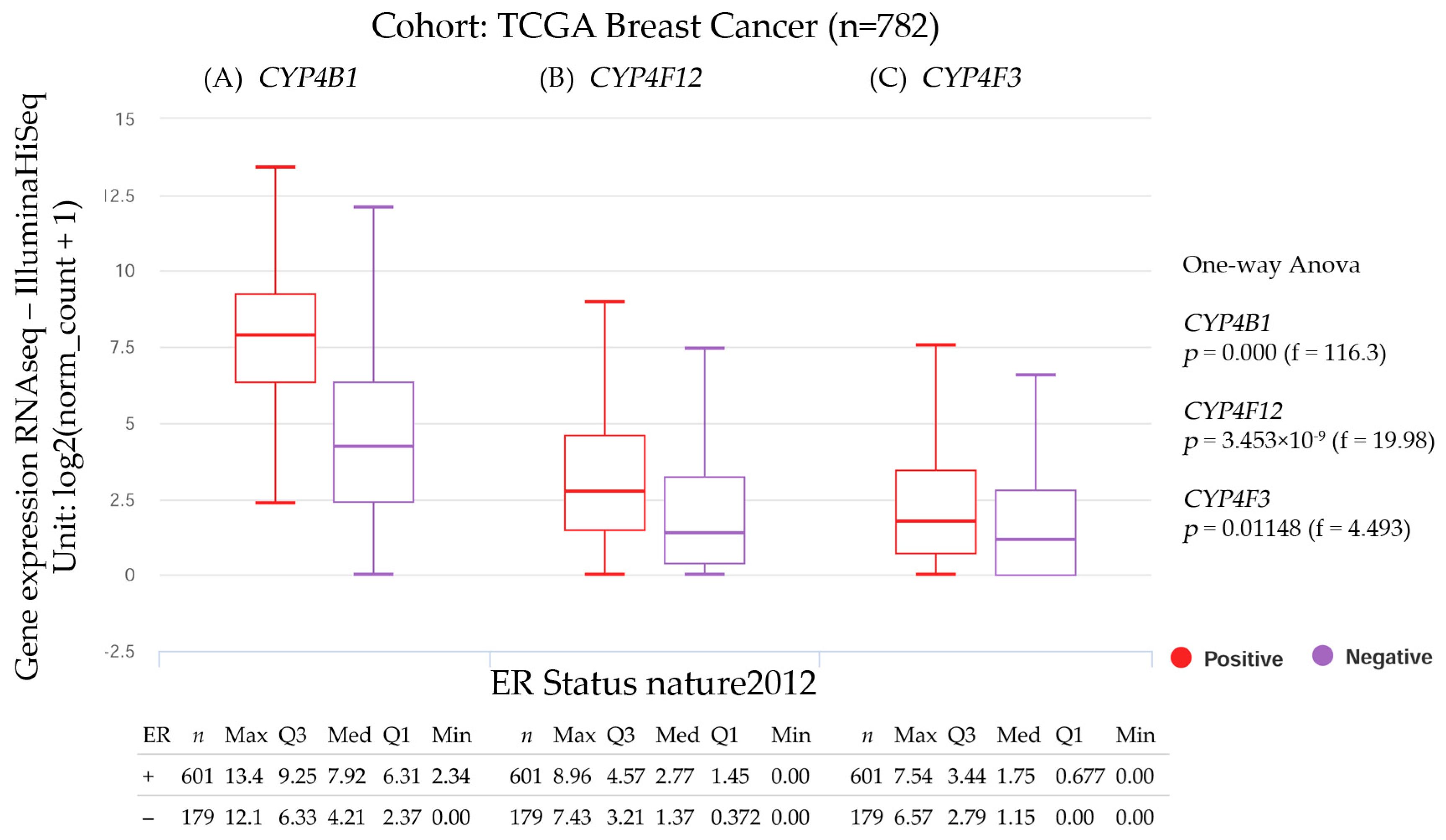
3.2.4. CYP4B1, CYP4F12, and CYP4F3 Gene Expression and the Estrogen Receptor Status
3.2.5. Overall Survival Analysis of Breast Cancer Patients
3.3. Adrenocortical Carcinoma
3.4. Bladder Urothelial Carcinoma
3.5. Cervical and Endocervical Cancer
3.6. Colon Adenocarcinoma
3.7. Esophageal Carcinoma
3.8. Lung Adenocarcinoma
3.9. Lung Squamous Cell
3.10. Ovarian Serous
3.11. Prostate Adenocarcinoma
3.12. Rectum Adenocarcinoma
3.13. Stomach Adenocarcinoma
3.14. Uterine Corpus
3.15. Bioinformatic Analysis of CYPs in Pan-Cancer
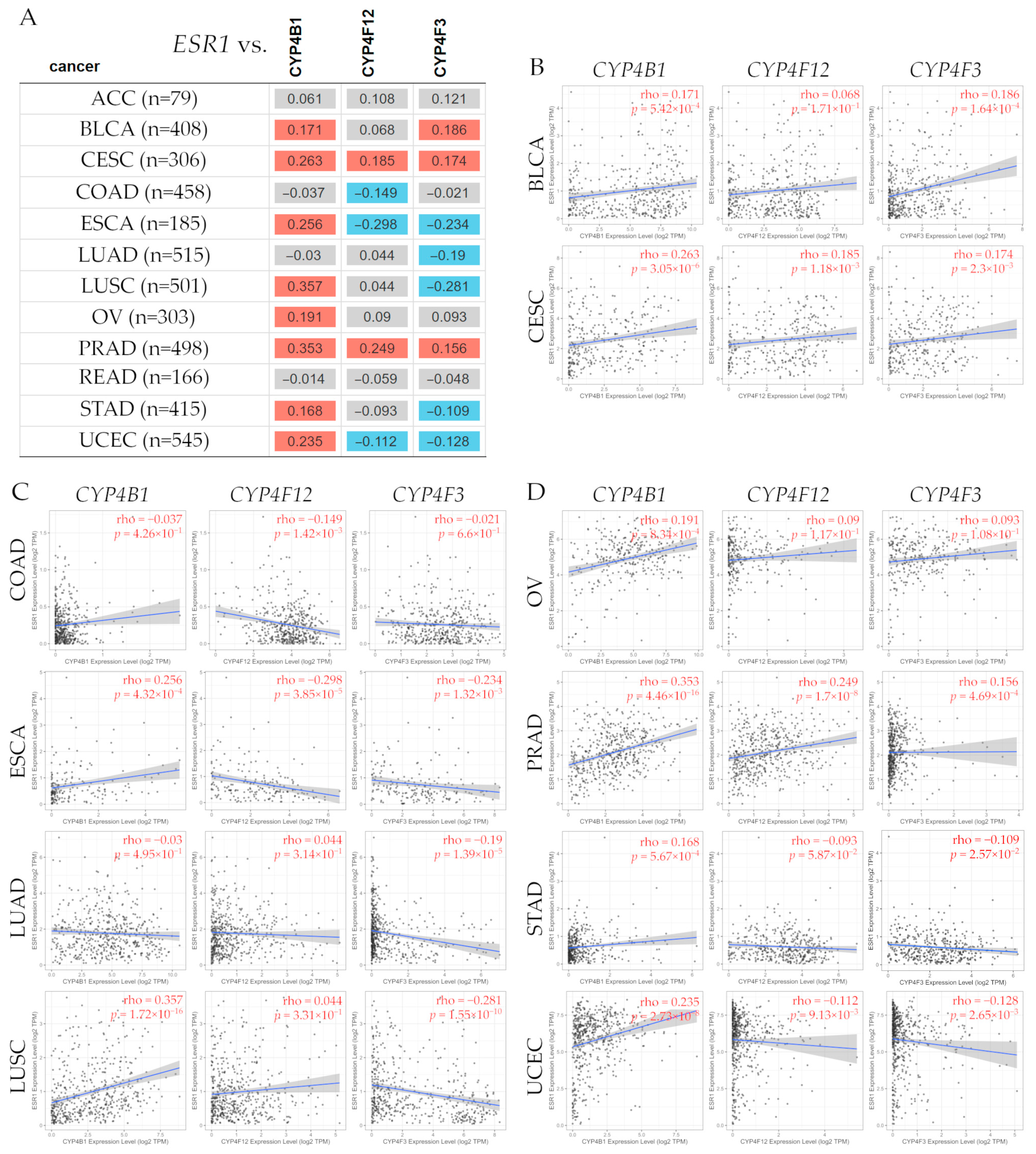
3.15.1. Correlation Between ESR1 and CYP4B1, CYP4F12, and CYP4F3 Gene Expression Levels in Several Types of Cancer
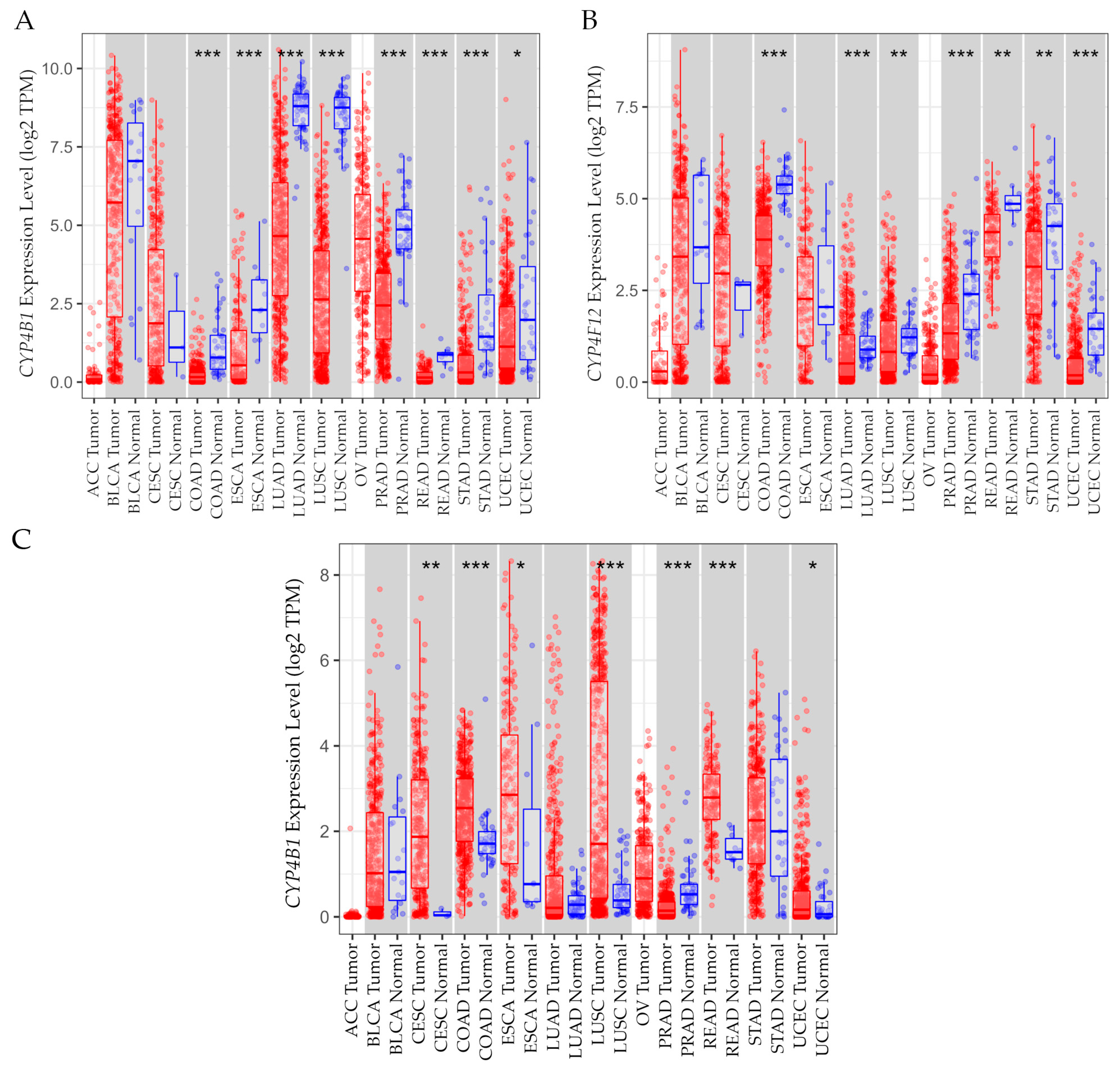
3.15.2. Gene Expression Levels in Tumor Versus Normal Tissues
3.15.3. Overall Survival Analysis of CYPs in Pan-Cancer
3.16. Advantages of Using CYP Enzymes as Biomarkers in Breast Cancer Compared to Other Solid Cancers
3.17. Clinical Trials Based on CYP Family-Based Drugs for Therapeutic Targets in Specific Cancer Settings
3.18. CYP1B1 in Tumor-Selective Activation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ER- | Estrogen receptor-negative |
| ER+ | Estrogen receptor-positive |
| CYP | Cytochrome P450 |
| CYP4B1 | Cytochrome P450 family 4 subfamily B member 1 |
| CYP4F12 | Cytochrome P450 family 4 subfamily F member 12 |
| CYP4F3 | Cytochrome P450 family 4 subfamily F member 3 |
| LTB4 | Leukotriene B4 |
| 20-HETE | 20-hydroxyeicosatetraenoic acid |
| TCGA | The Cancer Genome Atlas |
| GTEx | Genotype-tissue expression |
| GEPIA2 | Gene Expression Profiling Interactive Analysis |
| UALCAN | University of Alabama at Birmingham Cancer Data Analysis Portal |
| HPA | Human Protein Atlas |
| BRCA1 | BRCA1 DNA repair-associated |
| BRCA2 | BRCA2 DNA repair-associated |
| ESR1 | Estrogen receptor alpha |
| TIMER2.0 | Tumor Immune Estimation Resource v2.0 |
| ρ | Correlation coefficients |
| UCSC Xena | University of California, Santa Cruz, Xena explorer |
| ICGC | International Cancer Genome Consortium |
| GDC | Genomic Data Commons |
| OS | Overall survival |
| KM | Kaplan–Meier |
References
- Obeagu, E.I.; Obeagu, G.U. Breast cancer: A review of risk factors and diagnosis. Medicine 2024, 103, e36905. [Google Scholar] [CrossRef]
- WHO. Breast Cancer. World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 9 April 2025).
- Thun, M.J.; DeLancey, J.O.; Center, M.M.; Jemal, A.; Ward, E.M. The global burden of cancer: Priorities for prevention. Carcinogenesis 2010, 31, 100–110. [Google Scholar] [CrossRef]
- Barrios, C.; de Lima Lopes, G.; Yusof, M.M.; Rubagumya, F.; Rutkowski, P.; Sengar, M. Barriers in access to oncology drugs—A global crisis. Nat. Rev. Clin. Oncol. 2023, 20, 7–15. [Google Scholar] [CrossRef]
- Wang, B.; He, F.; Hu, Y.; Wang, Q.; Wang, D.; Sha, Y.; Wu, J. Cancer incidence and mortality and risk factors in member countries of the “ Belt and Road “ initiative. BMC Cancer 2022, 22, 582. [Google Scholar] [CrossRef]
- Antoni, M.H.; Lutgendorf, S.K.; Cole, S.W.; Dhabhar, F.S.; Sephton, S.E.; McDonald, P.G.; Stefanek, M.; Sood, A.K. The influence of bio-behavioural factors on tumour biology: Pathways and mechanisms. Nat. Rev. Cancer 2006, 6, 240–248. [Google Scholar] [CrossRef]
- Nebert, D.W.; Russell, D.W. Clinical importance of the cytochromes P450. Lancet 2002, 360, 1155–1162. [Google Scholar] [CrossRef]
- Nelson, D.R.; Zeldin, D.C.; Hoffman, S.M.; Maltais, L.J.; Wain, H.M.; Nebert, D.W. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 2004, 14, 1–18. [Google Scholar] [CrossRef]
- Ma, M.K.; Woo, M.H.; McLeod, H.L. Genetic basis of drug metabolism. Am. J. Health Syst. Pharm. 2002, 59, 2061–2069. [Google Scholar] [CrossRef]
- Luthra, A.; Denisov, I.G.; Sligar, S.G. Spectroscopic features of cytochrome P450 reaction intermediates. Arch. Biochem. Biophys. 2011, 507, 26–35. [Google Scholar] [CrossRef]
- Hodges, N.J.; Green, R.M.; Chipman, J.K.; Graham, M. Induction of DNA strand breaks and oxidative stress in HeLa cells by ethanol is dependent on CYP2E1 expression. Mutagenesis 2007, 22, 189–194. [Google Scholar] [CrossRef]
- Hu, Z.; Fan, C.; Oh, D.S.; Marron, J.S.; He, X.; Qaqish, B.F.; Livasy, C.; Carey, L.A.; Reynolds, E.; Dressler, L.; et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006, 7, 96. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Rakha, E.A.; Reis-Filho, J.S.; Ellis, I.O. Basal-like breast cancer: A critical review. J. Clin. Oncol. 2008, 26, 2568–2581. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Sorlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef]
- Gusterson, B. Do ‘basal-like’ breast cancers really exist? Nat. Rev. Cancer 2009, 9, 128–134. [Google Scholar] [CrossRef]
- Rakha, E.A.; El-Sayed, M.E.; Reis-Filho, J.; Ellis, I.O. Patho-biological aspects of basal-like breast cancer. Breast Cancer Res. Treat. 2009, 113, 411–422. [Google Scholar] [CrossRef]
- Rakha, E.A.; Reis-Filho, J.S.; Ellis, I.O. Impact of basal-like breast carcinoma determination for a more specific therapy. Pathobiology 2008, 75, 95–103. [Google Scholar] [CrossRef]
- Livasy, C.A.; Karaca, G.; Nanda, R.; Tretiakova, M.S.; Olopade, O.I.; Moore, D.T.; Perou, C.M. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod. Pathol. 2006, 19, 264–271. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef]
- Cheang, M.C.; Voduc, D.; Bajdik, C.; Leung, S.; McKinney, S.; Chia, S.K.; Perou, C.M.; Nielsen, T.O. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin. Cancer Res. 2008, 14, 1368–1376. [Google Scholar] [CrossRef]
- Fulford, L.G.; Easton, D.F.; Reis-Filho, J.S.; Sofronis, A.; Gillett, C.E.; Lakhani, S.R.; Hanby, A. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology 2006, 49, 22–34. [Google Scholar] [CrossRef]
- Banerjee, S.; Reis-Filho, J.S.; Ashley, S.; Steele, D.; Ashworth, A.; Lakhani, S.R.; Smith, I.E. Basal-like breast carcinomas: Clinical outcome and response to chemotherapy. J. Clin. Pathol. 2006, 59, 729–735. [Google Scholar] [CrossRef]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef]
- Haffty, B.G.; Yang, Q.; Reiss, M.; Kearney, T.; Higgins, S.A.; Weidhaas, J.; Harris, L.; Hait, W.; Toppmeyer, D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. 2006, 24, 5652–5657. [Google Scholar] [CrossRef]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Rakha, E.A.; Ellis, I.O.; Reis-Filho, J.S. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2/neu (basal-like breast carcinomas). Mod. Pathol. 2008, 21, 1060–1061; author reply 1061–1062. [Google Scholar] [CrossRef]
- Thike, A.A.; Cheok, P.Y.; Jara-Lazaro, A.R.; Tan, B.; Tan, P.; Tan, P.H. Triple-negative breast cancer: Clinicopathological characteristics and relationship with basal-like breast cancer. Mod. Pathol. 2010, 23, 123–133. [Google Scholar] [CrossRef]
- Thike, A.A.; Iqbal, J.; Cheok, P.Y.; Chong, A.P.; Tse, G.M.; Tan, B.; Tan, P.; Wong, N.S.; Tan, P.H. Triple negative breast cancer: Outcome correlation with immunohistochemical detection of basal markers. Am. J. Surg. Pathol. 2010, 34, 956–964. [Google Scholar] [CrossRef]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Tse, C.K.; Edmiston, S.; et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef]
- Putti, T.C.; El-Rehim, D.M.; Rakha, E.A.; Paish, C.E.; Lee, A.H.; Pinder, S.E.; Ellis, I.O. Estrogen receptor-negative breast carcinomas: A review of morphology and immunophenotypical analysis. Mod. Pathol. 2005, 18, 26–35. [Google Scholar] [CrossRef]
- Guedj, M.; Marisa, L.; de Reynies, A.; Orsetti, B.; Schiappa, R.; Bibeau, F.; MacGrogan, G.; Lerebours, F.; Finetti, P.; Longy, M.; et al. A refined molecular taxonomy of breast cancer. Oncogene 2012, 31, 1196–1206. [Google Scholar] [CrossRef]
- Farmer, P.; Bonnefoi, H.; Becette, V.; Tubiana-Hulin, M.; Fumoleau, P.; Larsimont, D.; Macgrogan, G.; Bergh, J.; Cameron, D.; Goldstein, D.; et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 2005, 24, 4660–4671. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Naderi, A.; Barbosa-Morais, N.L.; Caldas, C. PACK: Profile Analysis using Clustering and Kurtosis to find molecular classifiers in cancer. Bioinformatics 2006, 22, 2269–2275. [Google Scholar] [CrossRef]
- Doane, A.S.; Danso, M.; Lal, P.; Donaton, M.; Zhang, L.; Hudis, C.; Gerald, W.L. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 2006, 25, 3994–4008. [Google Scholar] [CrossRef]
- Guengerich, F.P. Characterization of human microsomal cytochrome P-450 enzymes. Annu. Rev. Pharmacol. Toxicol. 1989, 29, 241–264. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome P-450 3A4: Regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 1–17. [Google Scholar] [CrossRef]
- Niwa, T.; Murayama, N.; Yamazaki, H. Oxidation of endobiotics mediated by xenobiotic-metabolizing forms of human cytochrome. Curr. Drug Metab. 2009, 10, 700–712. [Google Scholar] [CrossRef]
- Al-Saraireh, Y.M.; Alshammari, F.; Abu-Azzam, O.H.; Al-Dalain, S.M.; Al-Sarayra, Y.M.; Haddad, M.; Makeen, H.; Al-Qtaitat, A.; Almermesh, M.; Al-Sarayreh, S.A. Targeting Cytochrome P450 Enzymes in Ovarian Cancers: New Approaches to Tumor-Selective Intervention. Biomedicines 2023, 11, 2898. [Google Scholar] [CrossRef]
- Porter, T.D.; Coon, M.J. Cytochrome P-450. Multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J. Biol. Chem. 1991, 266, 13469–13472. [Google Scholar] [CrossRef]
- Chang, G.W.; Kam, P.C. The physiological and pharmacological roles of cytochrome P450 isoenzymes. Anaesthesia 1999, 54, 42–50. [Google Scholar] [CrossRef]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef]
- Thelen, K.; Dressman, J.B. Cytochrome P450-mediated metabolism in the human gut wall. J. Pharm. Pharmacol. 2009, 61, 541–558. [Google Scholar] [CrossRef]
- Renaud, H.J.; Cui, J.Y.; Khan, M.; Klaassen, C.D. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol. Sci. 2011, 124, 261–277. [Google Scholar] [CrossRef]
- Guengerich, F.P. Mechanisms of cytochrome P450 substrate oxidation: MiniReview. J. Biochem. Mol. Toxicol. 2007, 21, 163–168. [Google Scholar] [CrossRef]
- Simpson, A.E. The cytochrome P450 4 (CYP4) family. Gen. Pharmacol. 1997, 28, 351–359. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, Y.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Global impact of somatic structural variation on the cancer proteome. Nat. Commun. 2023, 14, 5637. [Google Scholar] [CrossRef]
- Goldman, M.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv 2019, 1–16. [Google Scholar] [CrossRef]
- Leung, T.; Rajendran, R.; Singh, S.; Garva, R.; Krstic-Demonacos, M.; Demonacos, C. Cytochrome P450 2E1 (CYP2E1) regulates the response to oxidative stress and migration of breast cancer cells. Breast Cancer Res. 2013, 15, R107. [Google Scholar] [CrossRef]
- Elfaki, I.; Mir, R.; Almutairi, F.M.; Duhier, F.M.A. Cytochrome P450: Polymorphisms and Roles in Cancer, Diabetes and Atherosclerosis. Asian Pac. J. Cancer Prev. 2018, 19, 2057–2070. [Google Scholar] [CrossRef]
- Murray, G.I.; Patimalla, S.; Stewart, K.N.; Miller, I.D.; Heys, S.D. Profiling the expression of cytochrome P450 in breast cancer. Histopathology 2010, 57, 202–211. [Google Scholar] [CrossRef]
- Roder, A.; Husken, S.; Hutter, M.C.; Rettie, A.E.; Hanenberg, H.; Wiek, C.; Girhard, M. Spotlight on CYP4B1. Int. J. Mol. Sci. 2023, 24, 2038. [Google Scholar] [CrossRef]
- Iscan, M.; Klaavuniemi, T.; Coban, T.; Kapucuoglu, N.; Pelkonen, O.; Raunio, H. The expression of cytochrome P450 enzymes in human breast tumours and normal breast tissue. Breast Cancer Res. Treat. 2001, 70, 47–54. [Google Scholar] [CrossRef]
- Hashizume, T.; Imaoka, S.; Mise, M.; Terauchi, Y.; Fujii, T.; Miyazaki, H.; Kamataki, T.; Funae, Y. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J. Pharmacol. Exp. Ther. 2002, 300, 298–304. [Google Scholar] [CrossRef]
- Johnson, A.L.; Edson, K.Z.; Totah, R.A.; Rettie, A.E. Cytochrome P450 omega-Hydroxylases in Inflammation and Cancer. Adv. Pharmacol. 2015, 74, 223–262. [Google Scholar] [CrossRef]
- Corcos, L.; Lucas, D.; Le Jossic-Corcos, C.; Dreano, Y.; Simon, B.; Plee-Gautier, E.; Amet, Y.; Salaun, J.P. Human cytochrome P450 4F3: Structure, functions, and prospects. Drug Metabol. Drug Interact. 2012, 27, 63–71. [Google Scholar] [CrossRef]
- Calaf, G.M.; Roy, D. Human drug metabolism genes in parathion-and estrogen-treated breast cells. Int. J. Mol. Med. 2007, 20, 875–881. [Google Scholar] [CrossRef]
- Edson, K.Z.; Rettie, A.E. CYP4 enzymes as potential drug targets: Focus on enzyme multiplicity, inducers and inhibitors, and therapeutic modulation of 20-hydroxyeicosatetraenoic acid (20-HETE) synthase and fatty acid omega-hydroxylase activities. Curr. Top. Med. Chem. 2013, 13, 1429–1440. [Google Scholar] [CrossRef]
- McDonald, M.G.; Ray, S.; Amorosi, C.J.; Sitko, K.A.; Kowalski, J.P.; Paco, L.; Nath, A.; Gallis, B.; Totah, R.A.; Dunham, M.J.; et al. Expression and Functional Characterization of Breast Cancer-Associated Cytochrome P450 4Z1 in Saccharomyces cerevisiae. Drug Metab. Dispos. 2017, 45, 1364–1371. [Google Scholar] [CrossRef]
- Borin, T.F.; Zuccari, D.A.; Jardim-Perassi, B.V.; Ferreira, L.C.; Iskander, A.S.; Varma, N.R.; Shankar, A.; Guo, A.M.; Scicli, G.; Arbab, A.S. HET0016, a selective inhibitor of 20-HETE synthesis, decreases pro-angiogenic factors and inhibits growth of triple negative breast cancer in mice. PLoS ONE 2014, 9, e116247. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Han, X.; Li, T.; Li, D.; Liu, Q.; Yan, L.; Li, F.; Pei, X.; Feng, Y.; et al. Targeting estrogen mediated CYP4F2/CYP4F11-20-HETE metabolic disorder decelerates tumorigenesis in ER+ breast cancer. Biochem. Biophys. Rep. 2024, 38, 101706. [Google Scholar] [CrossRef]
- Nunna, V.; Jalal, N.; Bureik, M. Anti-CYP4Z1 autoantibodies detected in breast cancer patients. Cell. Mol. Immunol. 2017, 14, 572–574. [Google Scholar] [CrossRef]
- Li, C.; Zheng, L.; Xin, Y.; Tan, Z.; Zhang, Y.; Meng, X.; Wang, Z.; Xi, T. The competing endogenous RNA network of CYP4Z1 and pseudogene CYP4Z2P exerts an anti-apoptotic function in breast cancer. FEBS Lett. 2017, 591, 991–1000. [Google Scholar] [CrossRef]
- Hossam Abdelmonem, B.; Abdelaal, N.M.; Anwer, E.K.E.; Rashwan, A.A.; Hussein, M.A.; Ahmed, Y.F.; Khashana, R.; Hanna, M.M.; Abdelnaser, A. Decoding the Role of CYP450 Enzymes in Metabolism and Disease: A Comprehensive Review. Biomedicines 2024, 12, 1467. [Google Scholar] [CrossRef]
- Kartikasari, A.E.R.; Huertas, C.S.; Mitchell, A.; Plebanski, M. Tumor-Induced Inflammatory Cytokines and the Emerging Diagnostic Devices for Cancer Detection and Prognosis. Front. Oncol. 2021, 11, 692142. [Google Scholar] [CrossRef]
- Geindreau, M.; Bruchard, M.; Vegran, F. Role of Cytokines and Chemokines in Angiogenesis in a Tumor Context. Cancers 2022, 14, 2446. [Google Scholar] [CrossRef]
- Rocic, P.; Schwartzman, M.L. 20-HETE in the regulation of vascular and cardiac function. Pharmacol. Ther. 2018, 192, 74–87. [Google Scholar] [CrossRef]
- Alzahrani, A.M.; Rajendran, P. The Multifarious Link between Cytochrome P450s and Cancer. Oxid. Med. Cell. Longev. 2020, 2020, 3028387. [Google Scholar] [CrossRef]
- Carrera-Pacheco, S.E.; Mueller, A.; Puente-Pineda, J.A.; Zuniga-Miranda, J.; Guaman, L.P. Designing cytochrome P450 enzymes for use in cancer gene therapy. Front. Bioeng. Biotechnol. 2024, 12, 1405466. [Google Scholar] [CrossRef]
- Murtha, T.D.; Korah, R.; Carling, T. Suppression of cytochrome P450 4B1: An early event in adrenocortical tumorigenesis. Surgery 2017, 161, 257–263. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Mear, L.; Zhong, W.; Digre, A.; Katona, B.; Sjostedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Ahmed, K.M.; Veeramachaneni, R.; Deng, D.; Putluri, N.; Putluri, V.; Cardenas, M.F.; Wheeler, D.A.; Decker, W.K.; Frederick, A.I.; Kazi, S.; et al. Glutathione peroxidase 2 is a metabolic driver of the tumor immune microenvironment and immune checkpoint inhibitor response. J. Immunother. Cancer 2022, 10, e004752. [Google Scholar] [CrossRef]
- Alshammari, F.; Al-Saraireh, Y.M.; Youssef, A.M.M.; Al-Sarayra, Y.M.; Alrawashdeh, H.M. Cytochrome P450 1B1 Overexpression in Cervical Cancers: Cross-sectional Study. Interact. J. Med. Res. 2021, 10, e31150. [Google Scholar] [CrossRef]
- Dai, G.; Ou, J.; Wu, B. A predictive study of metabolism reprogramming in cervical carcinoma. Ann. Transl. Med. 2022, 10, 414. [Google Scholar] [CrossRef]
- Yin, J.; Liu, H.; Liu, Z.; Owzar, K.; Han, Y.; Su, L.; Wei, Y.; Hung, R.J.; Brhane, Y.; McLaughlin, J.; et al. Pathway-analysis of published genome-wide association studies of lung cancer: A potential role for the CYP4F3 locus. Mol. Carcinog. 2017, 56, 1663–1672. [Google Scholar] [CrossRef]
- Tang, X.L.; Xu, Z.Y.; Guan, J.; Yao, J.; Tang, X.L.; Zhou, Z.Q.; Zhang, Z.Y. Establishment of a neutrophil extracellular trap-related prognostic signature for colorectal cancer liver metastasis and expression validation of CYP4F3. Clin. Exp. Med. 2024, 24, 112. [Google Scholar] [CrossRef]
- Lim, S.; Alshagga, M.; Ong, C.E.; Chieng, J.Y.; Pan, Y. Cytochrome P450 4B1 (CYP4B1) as a target in cancer treatment. Hum. Exp. Toxicol. 2020, 39, 785–796. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, L.; Wang, Y.; Fang, Y.; Shen, W.; Si, Y.; Zheng, X.; Zeng, S. Integrative Analysis of Histone Acetylation Regulated CYP4F12 in Esophageal Cancer Development. Drug Metab. Dispos. 2024, 52, 813–823. [Google Scholar] [CrossRef]
- Smith, J.D.A.; Johnson, M. Clinical outcomes with perioperative chemotherapy in sarcomatoid carcinomas of the lung. J. Thorac. Oncol. 2022, 7, 123–130. [Google Scholar] [CrossRef]
- Smith, J.; Doe, A.; Johnson, M. Obinutuzumab pretreatment as a novel approach to mitigate formation of anti-drug antibodies against cergutuzumab amunaleukin in patients with solid tumors. Ann. Oncol. 2022, 33, 1422–1424. [Google Scholar] [CrossRef]
- Barlin, J.N.; Jelinic, P.; Olvera, N.; Bogomolniy, F.; Bisogna, M.; Dao, F.; Barakat, R.R.; Chi, D.S.; Levine, D.A. Validated gene targets associated with curatively treated advanced serous ovarian carcinoma. Gynecol. Oncol. 2013, 128, 512–517. [Google Scholar] [CrossRef]
- Marchini, S.; Mariani, P.; Chiorino, G.; Marrazzo, E.; Bonomi, R.; Fruscio, R.; Clivio, L.; Garbi, A.; Torri, V.; Cinquini, M.; et al. Analysis of gene expression in early-stage ovarian cancer. Clin. Cancer Res. 2008, 14, 7850–7860. [Google Scholar] [CrossRef]
- Zencir, S.; Alptekin, D.; Celiktas, M.; Canturk, P.; Colak, D.; Caner, V.; Luleyap, U.H.; Topcu, Z. Detection of cytochrome P450-2A6, -3A5 and -4B1 with real-time polymerase chain reaction in prostate tissue. Z. Naturforsch. C J. Biosci. 2008, 63, 780–784. [Google Scholar] [CrossRef]
- Abdul Aziz, A.A.; Md Salleh, M.S.; Yahya, M.M.; Zakaria, A.D.; Ankathil, R. Genetic Association of CYP1B1 4326 C>G Polymorphism with Disease-Free Survival in TNBC Patients Undergoing TAC Chemotherapy Regimen. Asian Pac. J. Cancer Prev. 2021, 22, 1319–1324. [Google Scholar] [CrossRef]
- Aiyappa-Maudsley, R.; Storr, S.J.; Rakha, E.A.; Green, A.R.; Ellis, I.O.; Martin, S.G. CYP2S1 and CYP2W1 expression is associated with patient survival in breast cancer. J. Pathol. Clin. Res. 2022, 8, 550–566. [Google Scholar] [CrossRef]
- Manthalkar, L.; Ajazuddin; Bhattacharya, S. Evidence-based capacity of natural cytochrome enzyme inhibitors to increase the effectivity of antineoplastic drugs. Discov. Oncol. 2022, 13, 142. [Google Scholar] [CrossRef]
- Blackburn, H.L.; Ellsworth, D.L.; Shriver, C.D.; Ellsworth, R.E. Role of cytochrome P450 genes in breast cancer etiology and treatment: Effects on estrogen biosynthesis, metabolism, and response to endocrine therapy. Cancer Causes Control 2015, 26, 319–332. [Google Scholar] [CrossRef]
- Fortin, S.; Charest-Morin, X.; Turcotte, V.; Lauvaux, C.; Lacroix, J.; Cote, M.F.; Gobeil, S.; René, C.G. Activation of Phenyl 4-(2-Oxo-3-alkylimidazolidin-1-yl)benzenesulfonates Prodrugs by CYP1A1 as New Antimitotics Targeting Breast Cancer Cells. J. Med. Chem. 2017, 60, 4963–4982. [Google Scholar] [CrossRef]
- Alaei, M.; Nazari, S.E.; Pourali, G.; Asadnia, A.; Moetamani-Ahmadi, M.; Fiuji, H.; Tanzadehpanah, H.; Asgharzadeh, F.; Babaei, F.; Khojasteh-Leylakoohi, F.; et al. Therapeutic Potential of Targeting the Cytochrome P450 Enzymes Using Lopinavir/Ritonavir in Colorectal Cancer: A Study in Monolayers, Spheroids and In Vivo Models. Cancers 2023, 15, 3939. [Google Scholar] [CrossRef]
- Watkins, P.B.; Wrighton, S.A.; Schuetz, E.G.; Molowa, D.T.; Guzelian, P.S. Identification of glucocorticoid-inducible cytochromes P-450 in the intestinal mucosa of rats and man. J. Clin. Investig. 1987, 80, 1029–1036. [Google Scholar] [CrossRef]
- Maksimovic-Ivanic, D.; Fagone, P.; McCubrey, J.; Bendtzen, K.; Mijatovic, S.; Nicoletti, F. HIV-protease inhibitors for the treatment of cancer: Repositioning HIV protease inhibitors while developing more potent NO-hybridized derivatives? Int. J. Cancer 2017, 140, 1713–1726. [Google Scholar] [CrossRef]
- Molenaar-Kuijsten, L.; Braal, C.L.; Groenland, S.L.; de Vries, N.; Rosing, H.; Beijnen, J.H.; Koolen, S.L.W.; Vulink, A.J.E.; van Dongen, M.G.J.; Mathijssen, R.H.J.; et al. Effects of the Moderate CYP3A4 Inhibitor Erythromycin on the Pharmacokinetics of Palbociclib: A Randomized Crossover Trial in Patients With Breast Cancer. Clin. Pharmacol. Ther. 2022, 111, 477–484. [Google Scholar] [CrossRef]
- Rochat, B.; Morsman, J.M.; Murray, G.I.; Figg, W.D.; McLeod, H.L. Human CYP1B1 and anticancer agent metabolism: Mechanism for tumor-specific drug inactivation? J. Pharmacol. Exp. Ther. 2001, 296, 537–541. [Google Scholar] [CrossRef]
- Sneha, S.; Baker, S.C.; Green, A.; Storr, S.; Aiyappa, R.; Martin, S.; Pors, K. Intratumoural Cytochrome P450 Expression in Breast Cancer: Impact on Standard of Care Treatment and New Efforts to Develop Tumour-Selective Therapies. Biomedicines 2021, 9, 290. [Google Scholar] [CrossRef]
- Brodie, A.; Njar, V.; Macedo, L.F.; Vasaitis, T.S.; Sabnis, G. The Coffey Lecture: Steroidogenic enzyme inhibitors and hormone dependent cancer. Urol. Oncol. 2009, 27, 53–63. [Google Scholar] [CrossRef]
- Yap, T.A.; Carden, C.P.; Attard, G.; de Bono, J.S. Targeting CYP17: Established and novel approaches in prostate cancer. Curr. Opin. Pharmacol. 2008, 8, 449–457. [Google Scholar] [CrossRef]
- Bhutani, K.; Vishwakarma, S.; Yadav, P.; Yadav, M.K. The current landscape of aromatase inhibitors for the treatment of estrogen receptor-positive breast carcinoma. J. Steroid Biochem. Mol. Biol. 2025, 250, 106729. [Google Scholar] [CrossRef]
- Burstein, H.J.; Prestrud, A.A.; Seidenfeld, J.; Anderson, H.; Buchholz, T.A.; Davidson, N.E.; Gelmon, K.E.; Giordano, S.H.; Hudis, C.A.; Malin, J.; et al. American Society of Clinical Oncology clinical practice guideline: Update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. J. Clin. Oncol. 2010, 28, 3784–3796. [Google Scholar] [CrossRef]
- Yang, Y.; Pan, W.; Tang, X.; Wu, S.; Sun, X. A meta-analysis of randomized controlled trials comparing the efficacy and safety of anastrozole versus tamoxifen for breast cancer. Oncotarget 2017, 8, 48362–48374. [Google Scholar] [CrossRef]
- Lum, D.W.; Perel, P.; Hingorani, A.D.; Holmes, M.V. CYP2D6 genotype and tamoxifen response for breast cancer: A systematic review and meta-analysis. PLoS ONE 2013, 8, e76648. [Google Scholar] [CrossRef]
- Tamura, K.; Imamura, C.K.; Takano, T.; Saji, S.; Yamanaka, T.; Yonemori, K.; Takahashi, M.; Tsurutani, J.; Nishimura, R.; Sato, K.; et al. CYP2D6 Genotype-Guided Tamoxifen Dosing in Hormone Receptor-Positive Metastatic Breast Cancer (TARGET-1): A Randomized, Open-Label, Phase II Study. J. Clin. Oncol. 2020, 38, 558–566. [Google Scholar] [CrossRef]
- Schroth, W.; Goetz, M.P.; Hamann, U.; Fasching, P.A.; Schmidt, M.; Winter, S.; Fritz, P.; Simon, W.; Suman, V.J.; Ames, M.M.; et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 2009, 302, 1429–1436. [Google Scholar] [CrossRef]
- Eisen, A.; Trudeau, M.; Shelley, W.; Messersmith, H.; Pritchard, K.I. Aromatase inhibitors in adjuvant therapy for hormone receptor positive breast cancer: A systematic review. Cancer Treat. Rev. 2008, 34, 157–174. [Google Scholar] [CrossRef]
- Mokbel, R.; Karat, I.; Mokbel, K. Adjuvant endocrine therapy for postmenopausal breast cancer in the era of aromatase inhibitors: An update. Int. Semin. Surg. Oncol. 2006, 3, 31. [Google Scholar] [CrossRef]
- Goss, P.E.; Smith, R.E. Letrozole for the management of breast cancer. Expert Rev. Anticancer Ther. 2002, 2, 249–260. [Google Scholar] [CrossRef]
- Smith, I.; Yardley, D.; Burris, H.; De Boer, R.; Amadori, D.; McIntyre, K.; Ejlertsen, B.; Gnant, M.; Jonat, W.; Pritchard, K.I.; et al. Comparative Efficacy and Safety of Adjuvant Letrozole Versus Anastrozole in Postmenopausal Patients With Hormone Receptor-Positive, Node-Positive Early Breast Cancer: Final Results of the Randomized Phase III Femara Versus Anastrozole Clinical Evaluation (FACE) Trial. J. Clin. Oncol. 2017, 35, 1041–1048. [Google Scholar] [CrossRef]
- Meinhardt, U.; Mullis, P.E. The aromatase cytochrome P-450 and its clinical impact. Horm. Res. 2002, 57, 145–152. [Google Scholar] [CrossRef]
- Santen, R.J.; Harvey, H.A. Use of aromatase inhibitors in breast carcinoma. Endocr. Relat. Cancer 1999, 6, 75–92. [Google Scholar] [CrossRef]
- Bhatnagar, A.S. The discovery and mechanism of action of letrozole. Breast Cancer Res. Treat. 2007, 105 (Suppl. S1), 7–17. [Google Scholar] [CrossRef]
- Yang, W.H.; Hammes, S.R. Xenopus laevis CYP17 regulates androgen biosynthesis independent of the cofactor cytochrome b5. J. Biol. Chem. 2005, 280, 10196–10201. [Google Scholar] [CrossRef]
- Harshman, L.C.; Taplin, M.E. Abiraterone acetate: Targeting persistent androgen dependence in castration-resistant prostate cancer. Adv. Ther. 2013, 30, 727–747. [Google Scholar] [CrossRef]
- Chan, C.W.H.; Law, B.M.H.; So, W.K.W.; Chow, K.M.; Waye, M.M.Y. Pharmacogenomics of breast cancer: Highlighting CYP2D6 and tamoxifen. J. Cancer Res. Clin. Oncol. 2020, 146, 1395–1404. [Google Scholar] [CrossRef]
- NGR-Oncology. Recently Activated! NRG-BR007: The DEBRA Trial. Available online: https://www.nrgoncology.org/Home/News/Post/NRG-BR007-The-DEBRA-Trial (accessed on 24 June 2025).
- NIH. De-Escalation of Breast Radiation Trial for Hormone Sensitive, HER-2 Negative, Oncotype Recurrence Score Less Than or Equal to 18 Breast Cancer (DEBRA) (DEBRA). National Library of Medicine: National Center for Biotechnology Information. Available online: https://clinicaltrials.gov/study/NCT04852887?utm_source= (accessed on 24 June 2025).
- Vanderbilt-Ingram. Clinical Trials Search at Vanderbilt-Ingram Cancer Center. Vanderbilt University Medical Center. Available online: https://vicc.org/clinical-trials/protocol-nrgbrebr007?utm_source= (accessed on 24 June 2025).
- SWOG. LoTam: A Randomized, Phase III Clinical Trial of Low-Dose Tamoxifen for Selected Patients with Molecular Low-Risk Early-Stage Breast Cancer. SWOG Cancer Research Network. Available online: https://www.swog.org/clinical-trials/ctsu-a012301?utm_source= (accessed on 24 June 2025).
- MedPath. Tamoxifen: Clinical Trials. Available online: https://trial.medpath.com/drug/26419669936bcbf8?utm_source= (accessed on 24 June 2025).
- Hamilton, E.P.; Loibl, S.; Bachelot, T.; Gnant, M.; Niikura, N.; Park, Y.H.; Tolaney, S.M.; Pistilli, B.; Rastogi, P.; Saini, K.S.; et al. CAMBRIA-1 & CAMBRIA-2 phase III trials: Camizestrant versus standard endocrine therapy in ER+/HER2- early breast cancer. Future Oncol. 2025, 21, 795–806. [Google Scholar] [CrossRef]
- AstraZeneca. An Adjuvant Endocrine-Based Therapy Study of Camizestrant (AZD9833) in ER+/HER2- Early Breast Cancer (CAMBRIA-2)—CAMBRIA-2. Available online: https://www.astrazenecaclinicaltrials.com/study/D8535C00001/?utm_source= (accessed on 24 June 2025).
- Ahmed, J.H.; Makonnen, E.; Fotoohi, A.; Aseffa, A.; Howe, R.; Aklillu, E. CYP2D6 Genotype Predicts Plasma Concentrations of Tamoxifen Metabolites in Ethiopian Breast Cancer Patients. Cancers 2019, 11, 1353. [Google Scholar] [CrossRef]
- Ferraldeschi, R.; Newman, W.G. The Impact of CYP2D6 Genotyping on Tamoxifen Treatment. Pharmaceuticals 2010, 3, 1122–1138. [Google Scholar] [CrossRef]
- Mulder, T.A.M.; de With, M.; Del Re, M.; Danesi, R.; Mathijssen, R.H.J.; van Schaik, R.H.N. Clinical CYP2D6 Genotyping to Personalize Adjuvant Tamoxifen Treatment in ER-Positive Breast Cancer Patients: Current Status of a Controversy. Cancers 2021, 13, 771. [Google Scholar] [CrossRef]
- Campos, S.M. Aromatase inhibitors for breast cancer in postmenopausal women. Oncologist 2004, 9, 126–136. [Google Scholar] [CrossRef]
- Riemsma, R.; Forbes, C.A.; Kessels, A.; Lykopoulos, K.; Amonkar, M.M.; Rea, D.W.; Kleijnen, J. Systematic review of aromatase inhibitors in the first-line treatment for hormone sensitive advanced or metastatic breast cancer. Breast Cancer Res. Treat. 2010, 123, 9–24. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calaf, G.M.; Crispin, L.A.; Ossandon-Acosta, F.; Perez-Tapia, S.; Ardiles, L.N. The Cytochrome CYP4 in Breast and Other Cancers. Biology 2025, 14, 812. https://doi.org/10.3390/biology14070812
Calaf GM, Crispin LA, Ossandon-Acosta F, Perez-Tapia S, Ardiles LN. The Cytochrome CYP4 in Breast and Other Cancers. Biology. 2025; 14(7):812. https://doi.org/10.3390/biology14070812
Chicago/Turabian StyleCalaf, Gloria M., Leodan A. Crispin, Felipe Ossandon-Acosta, Summer Perez-Tapia, and Luis N. Ardiles. 2025. "The Cytochrome CYP4 in Breast and Other Cancers" Biology 14, no. 7: 812. https://doi.org/10.3390/biology14070812
APA StyleCalaf, G. M., Crispin, L. A., Ossandon-Acosta, F., Perez-Tapia, S., & Ardiles, L. N. (2025). The Cytochrome CYP4 in Breast and Other Cancers. Biology, 14(7), 812. https://doi.org/10.3390/biology14070812




