Molecular and Phenotypic Characterization of Escherichia coli Associated with Granulomatous Colitis of Boxer Dogs
Abstract
:1. Introduction
2. Results
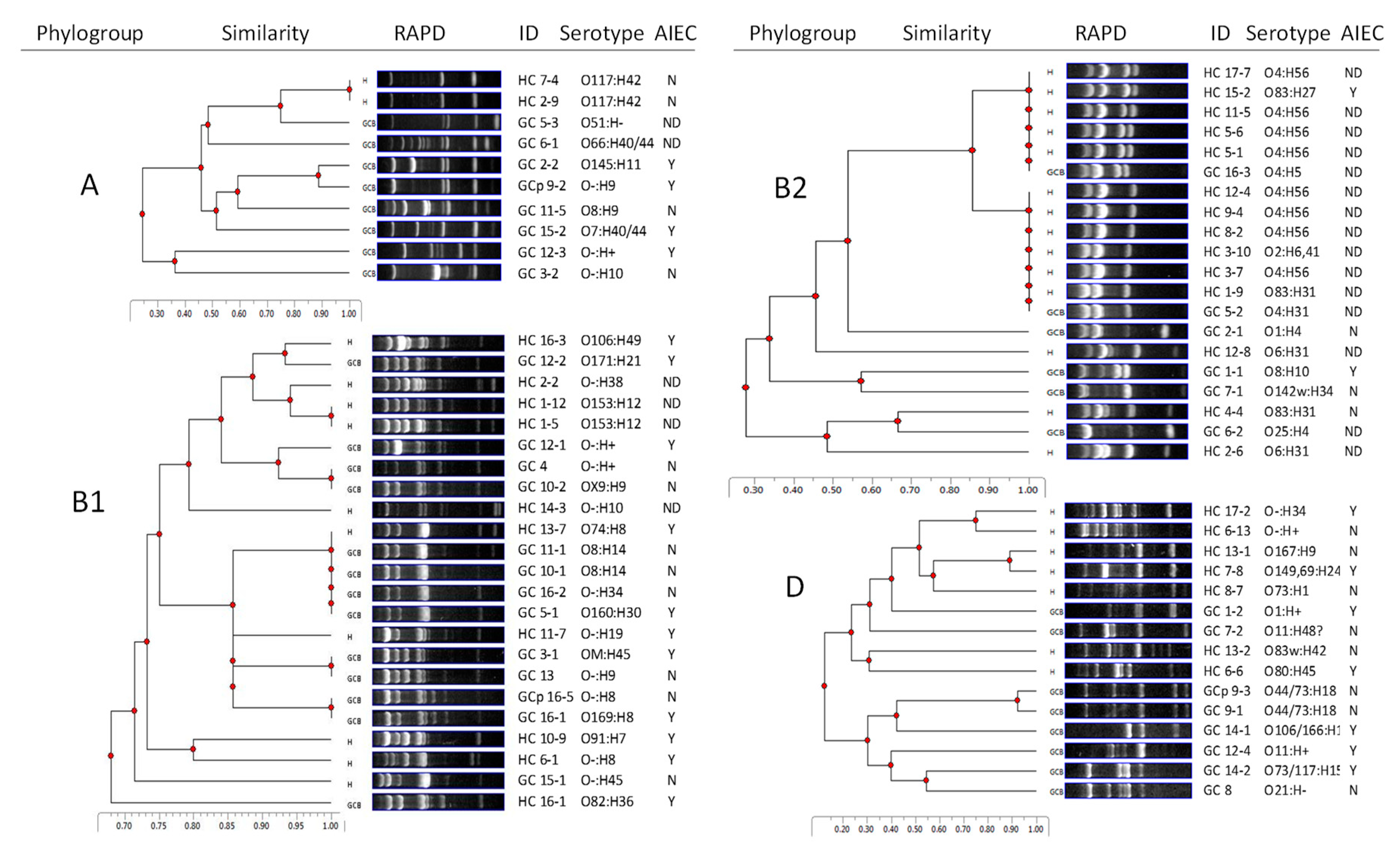
2.1. GC E. coli are Diverse in Genotype, Phylogroup, and Serotype
2.2. HC E. coli Cluster with UPEC
2.3. E. coli from GC and HC Resemble ExPEC and AIEC in Gene Content
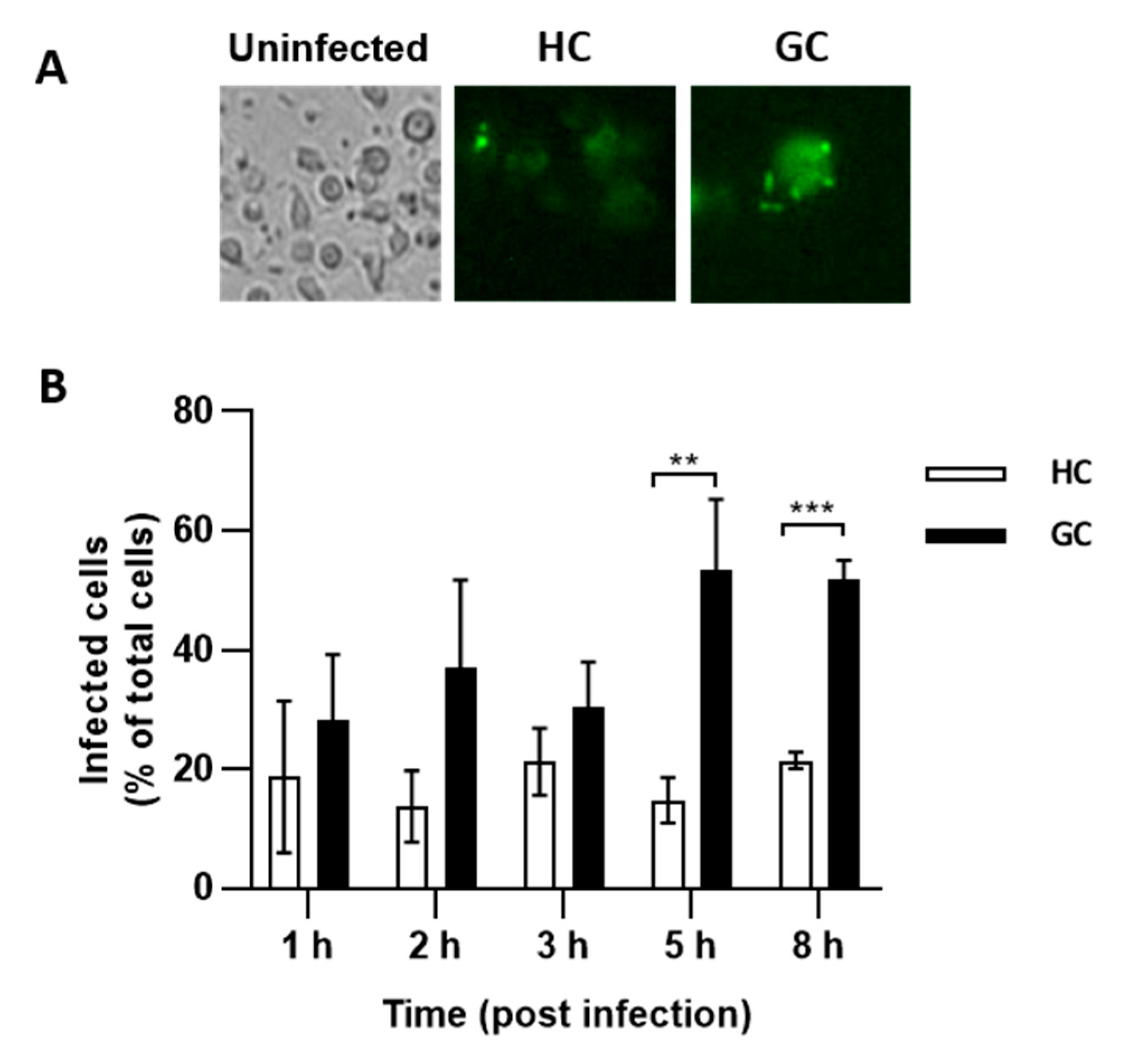
2.4. E. coli from GC and HC Invade Caco-2 Epithelial Cells and Survive in J774 Macrophages
2.5. AIEC are Prevalent in GC and HC
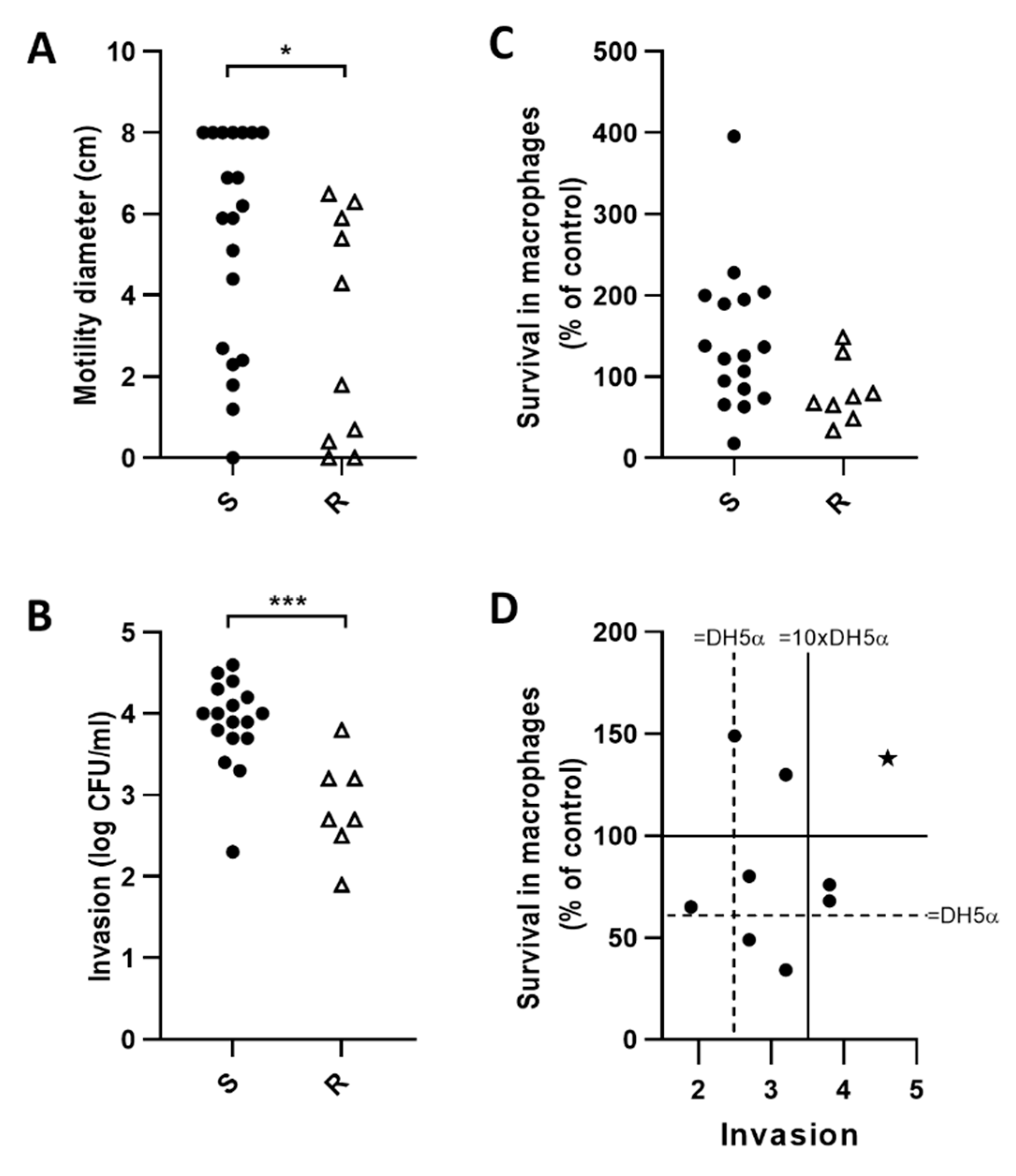
2.6. E. coli Motility Correlates with Invasion of Caco-2 Epithelial Cells but Not Survival in J774 Macrophages
2.7. GC AIEC KD2 Can Replicate in MDMs from GC but Not HC
2.8. Fluoroquinolone Resistant E. coli (FQ-R) are Less Motile and Invasive than FQ-S
3. Discussion
4. Materials and Methods
4.1. Animals and Isolation of Bacterial Strains
4.2. Molecular Characterization of E. coli
4.3. Multilocus Sequence Typing
4.4. Invasion and Persistence in Cultured Cells
4.5. Ability of AIEC to Replicate in MDMs from GC and HC
4.6. Motility
4.7. Antimicrobial Susceptibility Testing
4.8. Fluorescence in Situ Hybridization (FISH)
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Simpson, K.W.; Dogan, B.; Rishniw, M.; Goldstein, R.E.; Klaessig, S.; McDonough, P.L.; German, A.J.; Yates, R.M.; Russell, D.G.; Johnson, S.E.; et al. Adherent and invasive Escherichia coli is associated with granulomatous colitis in Boxer dogs. Infect. Immun. 2006, 74, 4778–4792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manchester, A.C.; Hill, S.; Sabatino, B.; Armentano, R.; Carroll, M.; Kessler, B.; Miller, M.; Dogan, B.; McDonough, S.P.; Simpson, K.W. Association between Granulomatous Colitis in French Bulldogs and Invasive Escherichia coli and Response to Fluoroquinolone Antimicrobials. J. Vet. Intern. Med. 2013, 27, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Hayward, J.J.; Castelhano, M.G.; Oliveira, K.C.; Corey, E.; Balkman, C.; Baxter, T.L.; Casal, M.L.; Center, S.A.; Fang, M.; Garrison, S.J.; et al. Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 2016, 7, 10460. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, C.S.; James, F.E.; Craven, M.; Davies, D.R.; O’Hara, A.J.; Nicholls, P.K.; Dogan, B.; MacDonough, S.P.; Simpson, K.W. Remission of histiocytic ulcerative colitis in Boxer dogs correlates with eradication of invasive intramucosal Escherichia coli. HUC 2009, 23, 964–969. [Google Scholar] [CrossRef] [Green Version]
- Craven, M.; Dogan, B.; Schukken, A.; Volkman, M.; Chandler, A.; McDonough, P.L.; Simpson, K.W. Antimicrobial resistance impacts clinical outcome of granulomatous colitis in Boxer dogs. J. Vet. Intern. Med. 2010, 24, 819–824. [Google Scholar] [CrossRef]
- Cassmann, E.; White, R.; Atherly, T.; Wang, C.; Sun, Y.; Khoda, S.; Mosher, C.; Ackermann, M.; Jergens, A. Alterations of the Ileal and Colonic Mucosal Microbiota in Canine Chronic Enteropathies. PLoS ONE 2016, 11, e0147321. [Google Scholar] [CrossRef] [Green Version]
- Cochran, L.; Hill, S.; Lotti, U.; Allanspach, K.; Palma, D.; Forman, M.; Gary, A.; Meads, Z.; Dogan, B.; McDonough, S.P.; et al. E. coli Associated Granulomatous Ileo-Colitis in 5 dogs: Clinical Characteristics and Outcome. J. Small Anim. Pract. (under review).
- Ben-Ari, J.; Wolach, O.; Gavrieli, R.; Wolach, B. Infections associated with chronic granulomatous disease: Linking genetics to phenotypic expression. Expert Rev. Anti-Infect. Ther. 2012, 10, 881–894. [Google Scholar] [CrossRef]
- Guérin, A.; Kerner, G.; Marr, N.; Markle, J.G.; Fenollar, F.; Wong, N.; Boughorbel, S.; Avery, D.T.; Ma, C.S.; Bougarn, S.; et al. IRF4 haploinsufficiency in a family with Whipple’s disease. Elife 2018, 7, e32340. [Google Scholar] [CrossRef]
- Magnani, A.; Brosselin, P.; Beauté, J.; de Vergnes, N.; Mouy, R.; Debré, M.; Suarez, F.; Hermine, O.; Lortholary, O.; Blanche, S.; et al. Inflammatory manifestations in a single-center cohort of patients with chronic granulomatous disease. J. Allergy Clin. Immunol. 2014, 134, 655–662. [Google Scholar] [CrossRef]
- Angelino, G.; De Angelis, P.; Faraci, S.; Rea, F.; Romeo, E.F.; Torroni, F.; Tambucci, R.; Claps, A.; Francalanci, P.; Chiriaco, M.; et al. Inflammatory bowel disease in chronic granulomatous disease: An emerging problem over a twenty years’ experience. Pediatr. Allergy Immunol. 2017, 28, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Alimchandani, M.; Lai, J.P.; Aung, P.P.; Khangura, S.; Kamal, N.; Gallin, J.I.; Holland, S.M.; Malech, H.L.; Heller, T.; Miettinen, M.; et al. Gastrointestinal histopathology in chronic granulomatous disease a study of 87 patients. Am. J. Surg. Pathol. 2013, 37, 1365–1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snapper, S.B. Very-Early-Onset inflammatory bowel disease. Gastroenterol. Hepatol. 2015, 11, 554–556. [Google Scholar]
- Ensari, A.; Kelsen, J.; Russo, P. Newcomers in paediatric GI pathology: Childhood enteropathies including very early onset monogenic IBD. Virchows Arch. 2018, 472, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.E.; Azar, A.E.; Chong, H.J.; Jongco, A.M.; Prince, B.T. Considerations in the diagnosis of chronic granulomatous disease. J. Pediatr. Infect. Dis. Soc. 2018, 7, S6–S11. [Google Scholar] [CrossRef] [Green Version]
- Marciano, B.E.; Rosenzweig, S.D.; Kleiner, D.E.; Anderson, V.L.; Darnell, D.N.; Anaya-O’Brien, S.; Hilligoss, D.M.; Malech, H.L.; Gallin, J.I.; Holland, S.M. Gastrointestinal involvement in chronic granulomatous disease. Pediatrics 2004, 114, 462–468. [Google Scholar] [CrossRef] [Green Version]
- Berger, S.B.; Romero, X.; Ma, C.; Wang, G.; Faubion, W.A.; Liao, G.; Compeer, E.; Keszei, M.; Rameh, L.; Wang, N.; et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 2010, 11, 920–927. [Google Scholar] [CrossRef] [Green Version]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Philip Schumm, L.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [Green Version]
- McArdel, S.L.; Terhorst, C.; Sharpe, A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016, 164, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Van Driel, B.J.; Liao, G.; Engel, P.; Terhorst, C. Responses to microbial challenges by SLAMF receptors. Front. Immunol. 2016, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Dogan, B.; Suzuki, H.; Herlekar, D.; Sartor, B.R.B.; Campbell, B.J.; Roberts, C.L.; Stewart, K.; Scherl, E.J.; Araz, Y.; Bitar, P.P.; et al. Inflammation-associated adherent-invasive escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm. Bowel Dis. 2014, 20, 1919–1932. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.L.; Barnich, N.; Bringer, M.A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Lapaquette, P.; Glasser, A.L.; Huett, A.; Xavier, R.J.; Darfeuille-Michaud, A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell. Microbiol. 2010, 12, 99–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hampe, J.; Franke, A.; Rosenstiel, P.; Till, A.; Teuber, M.; Huse, K.; Albrecht, M.; Mayr, G.; De La Vega, F.M.; Briggs, J.; et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007, 39, 207–211. [Google Scholar] [CrossRef]
- Massey, D.C.; Parkes, M. Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn’s disease. Autophagy 2007, 3, 649–651. [Google Scholar] [CrossRef] [Green Version]
- Buisson, A.; Douadi, C.; Ouchchane, L.; Goutte, M.; Hugot, J.P.; Dubois, A.; Minet-Quinard, R.; Bouvier, D.; Bommelaer, G.; Vazeille, E.; et al. Macrophages Inability to Mediate Adherent-Invasive E. coli Replication is Linked to Autophagy in Crohn’s Disease Patients. Cells 2019, 8, 1394. [Google Scholar] [CrossRef] [Green Version]
- Lapaquette, P.; Bringer, M.A.; Darfeuille-Michaud, A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell. Microbiol. 2012, 14, 791–807. [Google Scholar] [CrossRef] [Green Version]
- Boudeau, J.; Glasser, A.L.; Masseret, E.; Joly, B.; Darfeuille-Michaud, A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect. Immun. 1999, 67, 4499–4509. [Google Scholar] [CrossRef] [Green Version]
- Elhenawy, W.; Tsai, C.N.; Coombes, B.K. Host-Specific Adaptive Diversification of Crohn’s Disease-Associated Adherent-Invasive Escherichia coli. Cell Host Microbe 2019, 25, 301–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.R.; Stell, A.L.; Delavari, P. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect. Immun. 2001, 69, 1306–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Medina, M.; Garcia-Gil, J.; Barnich, N.; Wieler, L.H.; Ewers, C. Adherent-invasive Escherichia coli phenotype displayed by intestinal pathogenic E. coli strains from cats, dogs, and swine. Appl. Environ. Microbiol. 2011, 77, 5813–5817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nash, J.H.; Villegas, A.; Kropinski, A.M.; Aguilar-Valenzuela, R.; Konczy, P.; Mascarenhas, M.; Ziebell, K.; Torres, A.G.; Karmali, M.A.; Coombes, B.K. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genom. 2010, 11, 667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumgart, M.; Dogan, B.; Rishniw, M.; Weitzman, G.; Bosworth, B.; Yantiss, R.; Orsi, R.H.; Wiedmann, M.; McDonough, P.; Kim, S.G.; et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007, 1, 403–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, T.R.D.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Rolhion, N.; de Vallee, A.; Salim, S.Y.; Prorok-Hamon, M.; Neut, C.; Campbell, B.J.; Soderholm, J.D.; Hugot, J.P.; Colombel, J.F.; et al. Crohn disease—Associated adherent-invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. J. Clin. Investig. 2011, 121, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Galan, C.; Hill, A.A.; Wu, W.-J.; Fehlner-Peach, H.; Song, H.W.; Schady, D.; Bettini, M.L.; Simpson, K.W.; Longman, R.S.; et al. Critical Role for the Microbiota in CX3CR1+ Intestinal Mononuclear Phagocyte Regulation of Intestinal T Cell Responses. Immunity 2018, 49, 151–163.e5. [Google Scholar] [CrossRef]
- Viladomiu, M.; Kivolowitz, C.; Abdulhamid, A.; Dogan, B.; Victorio, D.; Castellanos, J.G.; Woo, V.; Teng, F.; Tran, N.L.; Sczesnak, A.; et al. IgA-coated E. Coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci. Transl. Med. 2017, 9, eaaf9655. [Google Scholar] [CrossRef]
- Camprubí-Font, C.; Ewers, C.; Lopez-Siles, M.; Martinez-Medina, M. Genetic and phenotypic features to screen for putative adherent-invasive Escherichia coli. Front. Microbiol. 2019, 10, 108. [Google Scholar] [CrossRef] [Green Version]
- Kingsley, R.A.; Humphries, A.D.; Weening, E.H.; De Zoete, M.R.; Winter, S.; Papaconstantinopoulou, A.; Dougan, G.; Baumler, A.J. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: Identification of intestinal colonization and persistence determinants. Infect. Immun. 2003, 71, 629–640. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.S. Strategy of Escherichia coli for crossing the blood-brain barrier. J. Infect. Dis. 2002, 186 (Suppl. 2), S220–S224. [Google Scholar] [CrossRef] [Green Version]
- Camprubí-Font, C.; Martinez-Medina, M. Why the discovery of adherent-invasive Escherichia coli molecular markers is so challenging? World J. Biol. Chem. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Dogan, B.; Belcher-Timme, H.F.; Dogan, E.I.; Jiang, Z.D.; DuPont, H.L.; Snyder, N.; Yang, S.; Chandler, B.; Scherl, E.J.; Simpson, K.W. Evaluation of Escherichia coli pathotypes associated with irritable bowel syndrome. FEMS Microbiol. Lett. 2018, 365, fny249. [Google Scholar] [CrossRef]
- Sevrin, G.; Massier, S.; Chassaing, B.; Agus, A.; Delmas, J.; Denizot, J.; Billard, E.; Barnich, N. Adaptation of adherent-invasive E. coli to gut environment: Impact on flagellum expression and bacterial colonization ability. Gut Microbes 2018, 11, 364–380. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Neut, C.; Barnich, N.; Lederman, E.; Di, M.P.; Desreumaux, P.; Gambiez, L.; Joly, B.; Cortot, A.; Colombel, J.F. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 1998, 115, 1405–1413. [Google Scholar] [CrossRef]
- Lacroix, M. Persistent use of “false” cell lines. Int. J. Cancer 2008, 122, 1–4. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, C.L.; Bringer, M.A.; Holt, K.E.; Gordon, D.M.; Dubois, A.L.; Barnich, N.; Darfeuille-Michaud, A.; Pavli, P. Comparative genomics of Crohn’s disease-Associated adherent-invasive Escherichia coli. Gut 2017, 66, 1382–1389. [Google Scholar] [CrossRef] [Green Version]
- Abadía-Molina, A.C.; Ji, H.; Faubion, W.A.; Julien, A.; Latchman, Y.; Yagita, H.; Sharpe, A.; Bhan, A.K.; Terhorst, C. CD48 Controls T-Cell and Antigen-Presenting Cell Functions in Experimental Colitis. Gastroenterology 2006, 130, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Ralph, P.; Prichard, J.; Cohn, M. Reticulum cell sarcoma: An effector cell in antibody-dependent cell-mediated immunity. J. Immunol. 1975, 114, 898–905. [Google Scholar] [PubMed]
- Craven, M.; Mansfield, C.S.; Simpson, K.W. Granulomatous Colitis of Boxer Dogs. Vet. Clin. N. Am.-Small Anim. Pract. 2011, 41, 433–445. [Google Scholar] [CrossRef]
- Ryan, P.; Kelly, R.G.; Lee, G.; Collins, J.K.; O’Sullivan, G.C.; O’Connell, J.; Shanahan, F. Bacterial DNA within granulomas of patients with Crohn’s disease—Detection by laser capture microdissection and PCR. Am. J. Gastroenterol. 2004, 99, 1539–1543. [Google Scholar] [CrossRef]
- Cattin, R.P.; Hardcastle, M.R.; Simpson, K.W. Successful treatment of vaginal malakoplakia in a young cat. J. Feline Med. Surg. Open Rep. 2016, 2, 2055116916674871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuesta-Garcia, N.; Marchesi, F.; Ramsey, I.; Simpson, K.W. Concurrent granulomatous nephritis and colitis associated with invasive Escherichia coli in a boxer dog. J. Comp. Pathol. 2015, 152, 80–81. [Google Scholar] [CrossRef]
- Manchester, A.C. Antimicrobial Susceptibility Testing Informs Treatment of E. coli-Associated Granulomatous Colitis. J. Vet. Intern. Med. (under review).
- Fàbrega, A.; Soto, S.M.; Ballesté-Delpierre, C.; Fernández-Orth, D.; Jiménez de Anta, M.T.; Vila, J. Impact of quinolone-resistance acquisition on biofilm production and fitness in Salmonella enterica. J. Antimicrob. Chemother. 2014, 69, 1815–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Céspedes, J.; Sáez-López, E.; Frimodt-Møller, N.; Vila, J.; Soto, S.M. Effects of a mutation in the gyrA gene on the virulence of uropathogenic Escherichia coli. Antimicrob. Agents Chemother. 2015, 59, 4662–4668. [Google Scholar] [CrossRef] [Green Version]
- Ormsby, M.J.; Johnson, S.A.; Wall, D.M. Draft genome sequence of the Commensal Escherichia coli strain F-18. Genome Announc. 2016, 4, e01416-16. [Google Scholar] [CrossRef] [Green Version]
- Craven, M.; Egan, C.E.; Dowd, S.E.; McDonough, S.P.; Dogan, B.; Denkers, E.Y.; Bowman, D.; Scherl, E.J.; Simpson, K.W. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn’s Disease. PLoS ONE 2012, 7, e41594. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Whittam, T.S.; Berg, C.M.; Berg, D.E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993, 21, 5930–5933. [Google Scholar] [CrossRef] [Green Version]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- DebRoy, C.; Maddox, C.W. Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev. 2001, 2, 129–140. [Google Scholar] [CrossRef]
- Dogan, B.; Rishniw, M.; Bruant, G.; Harel, J.; Schukken, Y.H.; Simpson, K.W. Phylogroup and lpfA influence epithelial invasion by mastitis associated Escherichia coli. Vet. Microbiol. 2012, 159, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, I.M.; Rycroft, A.N.; Dogan, B.; Craven, M.; Bromfield, J.J.; Chandler, A.; Roberts, M.H.; Price, S.B.; Gilbert, R.O.; Simpson, K.W. Specific Strains of Escherichia coli Are Pathogenic for the Endometrium of Cattle and Cause Pelvic Inflammatory Disease in Cattle and Mice. PLoS ONE 2010, 5, e9192. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Function | Genes | Disease | Pathotype # | Fluoroquinolone Resistance | Phylogroup | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GC (n = 36) | Healthy (n = 33) | AIEC (n = 24) | non-AIEC (n = 23) | S (n = 25) | R (n = 11) | A (n = 10) | B1 (n = 23) | B2 (n = 20) | D (n = 16) | ||
| Adhesins | lpfA141 | 17 | 15 | 25 | 13 | 24 | 0 | 0 | 26 | 25 | 0 |
| lpfA154 | 44 | 45 | 58 | 52 | 36 | 64 | 0 | 100 | 5 | 44 | |
| lpfA | 50 | 55 | 67 | 57 | 44 | 64 | 0 | 100 | 30 | 44 | |
| afaBC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| sfaDE | 6 | 33 * | 0 | 0 | 8 | 0 | 0 | 0 | 65 | 0 | |
| papC | 6 | 33 * | 0 | 0 | 8 | 0 | 0 | 0 | 65 | 0 | |
| focG | 3 | 15 | 0 | 0 | 4 | 0 | 0 | 0 | 30 | 0 | |
| Toxins | sta | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| stb | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | |
| stx1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| stx2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| cnf1 | 6 | 36 * | 0 | 0 | 8 | 0 | 0 | 4 | 65 | 0 | |
| Iron acquisition | chuA | 42 | 64 | 42 | 43 | 44 | 36 | 0 | 0 | 100 | 100 |
| fyuA | 44 | 48 | 29 | 30 | 36 | 64 | 30 | 13 | 95 | 44 | |
| aer | 17 | 3 | 7 | 19 | 8 | 36 | 20 | 4 | 5 | 19 | |
| Secretion systems (II, IV, VI) | gsp | 72 | 64 | 63 | 78 | 72 | 82 | 40 | 70 | 55 | 75 |
| traC | 56 | 91 * | 63 | 74 | 48 | 73 | 70 | 70 | 75 | 75 | |
| hcp | 58 | 45 | 63 | 57 | 60 | 55 | 80 | 83 | 5 | 50 | |
| Various functions | pduC | 19 | 36 | 33 | 43 | 20 | 18 | 10 | 30 | 30 | 31 |
| kpsMII | 22 | 33 | 21 | 30 | 24 | 18 | 10 | 0 | 50 | 50 | |
| iss | 6 | 3 | 0 | 4 | 4 | 9 | 10 | 4 | 0 | 6 | |
| malX | 28 | 45 | 17 | 22 | 28 | 27 | 10 | 0 | 95 | 31 | |
| eae | 3 | 0 | 0 | 4 | 4 | 0 | 0 | 0 | 5 | 0 | |
| ibeA | 3 | 9 | 0 | 4 | 4 | 0 | 0 | 4 | 15 | 0 | |
| ratA | 22 | 55 * | 21 | 26 | 16 | 36 | 10 | 0 | 85 | 50 | |
| pmt1 | 3 | 9 | 4 | 9 | 4 | 0 | 0 | 9 | 5 | 6 | |
| colV | 22 | 45 | 21 | 17 | 28 | 9 | 10 | 0 | 90 | 25 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dogan, B.; Zhang, S.; Kalla, S.E.; Dogan, E.I.; Guo, C.; Ang, C.R.; Simpson, K.W. Molecular and Phenotypic Characterization of Escherichia coli Associated with Granulomatous Colitis of Boxer Dogs. Antibiotics 2020, 9, 540. https://doi.org/10.3390/antibiotics9090540
Dogan B, Zhang S, Kalla SE, Dogan EI, Guo C, Ang CR, Simpson KW. Molecular and Phenotypic Characterization of Escherichia coli Associated with Granulomatous Colitis of Boxer Dogs. Antibiotics. 2020; 9(9):540. https://doi.org/10.3390/antibiotics9090540
Chicago/Turabian StyleDogan, Belgin, Shiying Zhang, Sarah E. Kalla, Esra I. Dogan, Cindy Guo, Chelston R. Ang, and Kenneth W. Simpson. 2020. "Molecular and Phenotypic Characterization of Escherichia coli Associated with Granulomatous Colitis of Boxer Dogs" Antibiotics 9, no. 9: 540. https://doi.org/10.3390/antibiotics9090540





