DNA Sequencing by Hexagonal Boron Nitride Nanopore: A Computational Study
Abstract
:1. Introduction
2. Results and Discussions
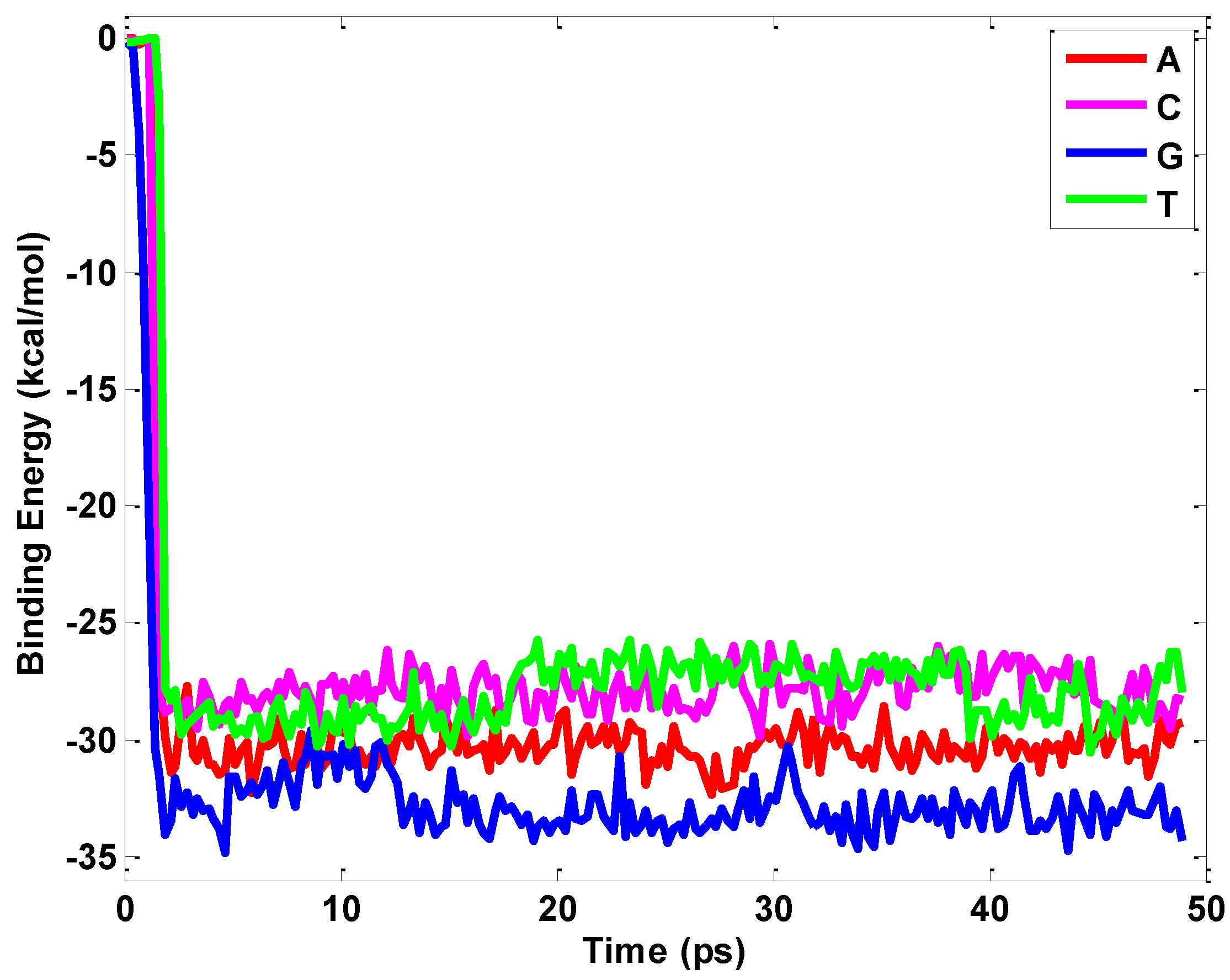
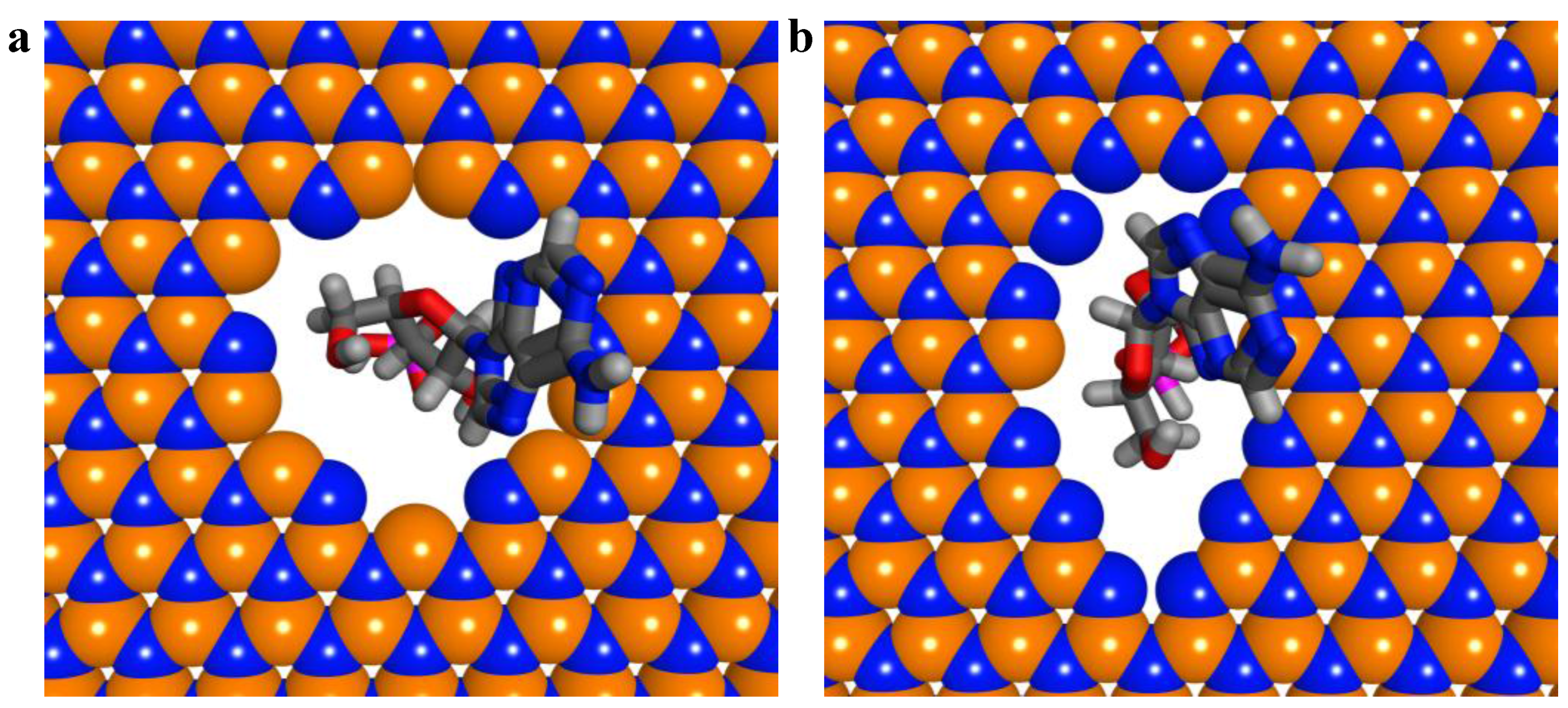
2.1. Interaction between Nucleobases and hBN Sheet
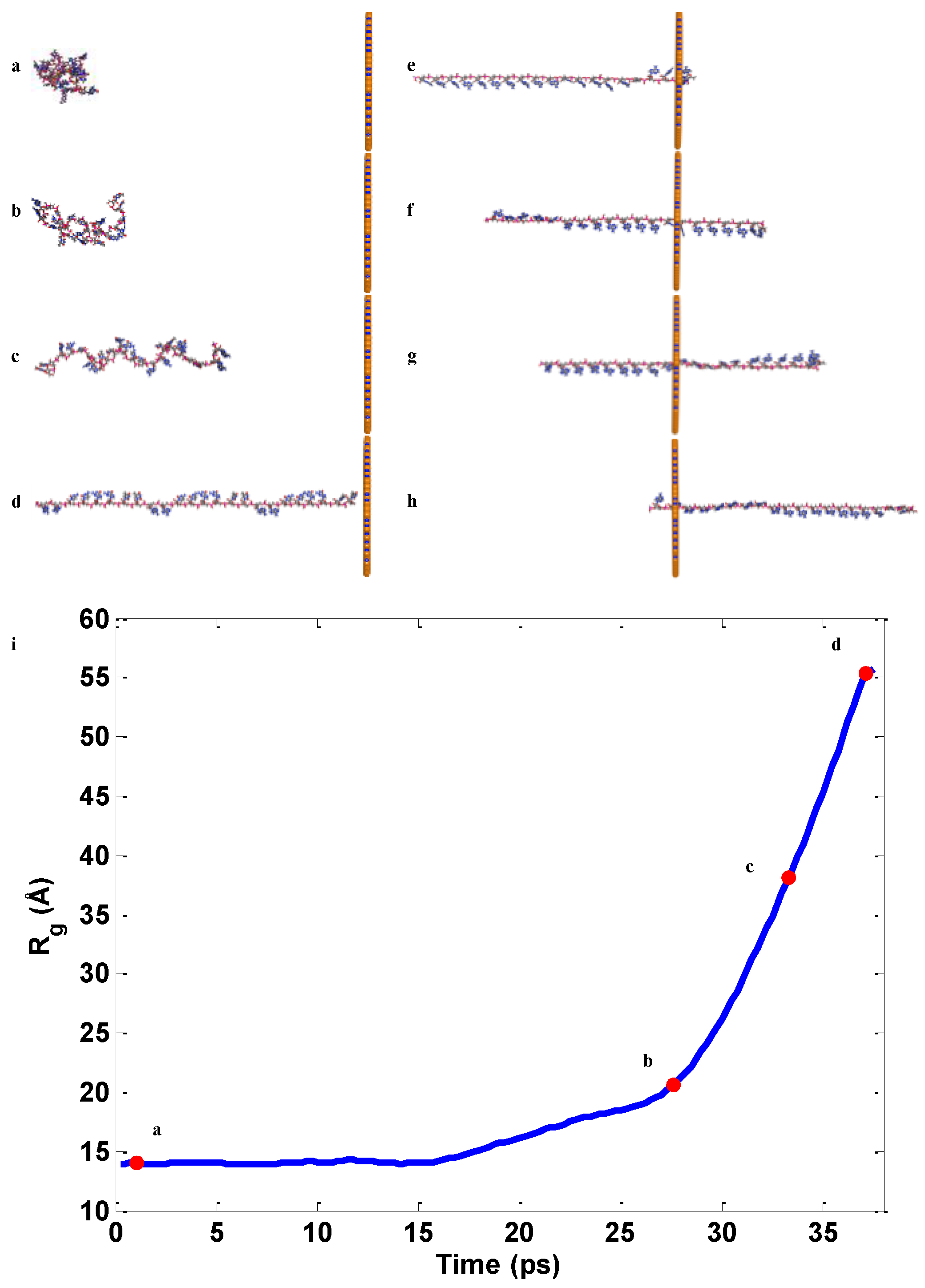
2.2. ssDNA Aligning
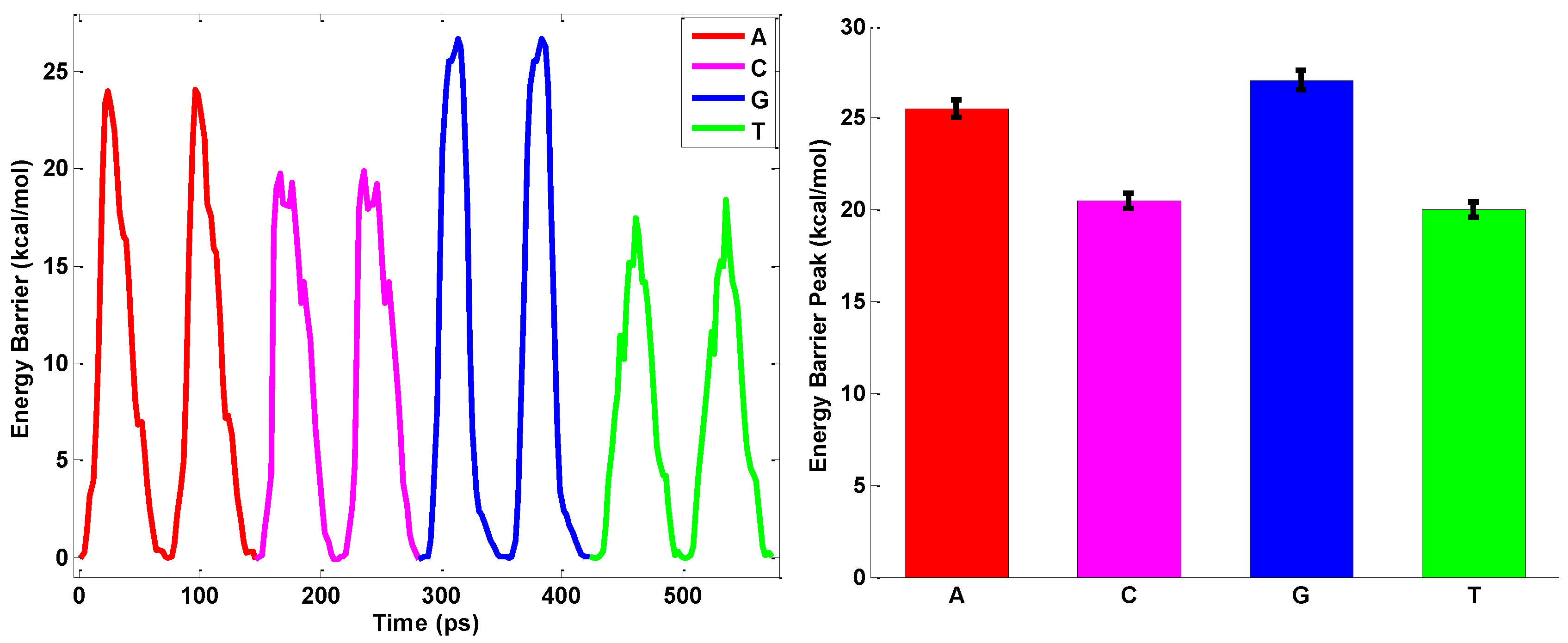
2.3. ssDNA Sequencing
3. Methods
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Benner, S.; Chen, R.J.; Wilson, N.A.; Abu-Shumays, R.; Hurt, N.; Lieberman, K.R.; Deamer, D.W.; Dunbar, W.B.; Akeson, M. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat. Nanotechnol. 2007, 2, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.C.; Rezapour, M.R.; Yun, J.; Cho, Y.; Cho, W.J.; Min, S.K.; Lee, G.; Kim, K.S. Two Dimensional Molecular Electronics Spectroscopy for Molecular Fingerprinting, DNA Sequencing, and Cancerous DNA Recognition. ACS Nano 2014, 8, 1827–1833. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.-Y.; Zeng, T.; Wu, H.-C. Recent advances of DNA sequencing via nanopore-based technologies. Sci. Bull. 2015, 60, 287–295. [Google Scholar] [CrossRef]
- Min, S.K.; Kim, W.Y.; Cho, Y.; Kim, K.S. Fast DNA sequencing with a graphene-based nanochannel device. Nat. Nano 2011, 6, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Laszlo, A.H.; Derrington, I.M.; Ross, B.C.; Brinkerhoff, H.; Adey, A.; Nova, I.C.; Craig, J.M.; Langford, K.W.; Samson, J.M.; Daza, R.; et al. Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 2014, 32, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X. Atomistic Insights into the Nanohelix of Hydrogenated Graphene: Formation, Characterization and Application. Phys. Chem. Chem. Phys. 2014, 16, 2981–2988. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X. Computational Insights of Water Droplet Transport on Graphene Sheet with Chemical Density. J. Appl. Phys. 2014, 115. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Zhao, Y.; Wang, X.; Ke, C. Graphene folding on flat substrates. J. Appl. Phys. 2014, 116. [Google Scholar] [CrossRef]
- Zhang, L.; Becton, M.; Wang, X. Mechanical Analysis of Graphene-Based Woven Nano-Fabric. Mater. Sci. Eng. A 2015, 620, 367–374. [Google Scholar] [CrossRef]
- Becton, M.; Zhang, L.; Wang, X. Molecular Dynamics Study of Programmable Nanoporous Graphene. J. Nanomech. Micromech. 2014, 4, 2153–5477. [Google Scholar] [CrossRef]
- Zhang, L.; Zeng, X.; Wang, X. Programmable Hydrogenation of Graphene for Novel Nanocages. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Merchant, C.A.; Healy, K.; Wanunu, M.; Ray, V.; Peterman, N.; Bartel, J.; Fischbein, M.D.; Venta, K.; Luo, Z.; Johnson, A.T.C.; et al. DNA Translocation through Graphene Nanopores. Nano Lett. 2010, 10, 2915–2921. [Google Scholar] [CrossRef] [PubMed]
- Sathe, C.; Zou, X.; Leburton, J.-P.; Schulten, K. Computational Investigation of DNA Detection Using Graphene Nanopores. ACS Nano 2011, 5, 8842–8851. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.B.; Belkin, M.; Comer, J.; Aksimentiev, A. Assessing Graphene Nanopores for Sequencing DNA. Nano Lett. 2012, 12, 4117–4123. [Google Scholar] [CrossRef] [PubMed]
- Prasongkit, J.; Grigoriev, A.; Pathak, B.; Ahuja, R.; Scheicher, R.H. Transverse Conductance of DNA Nucleotides in a Graphene Nanogap from First Principles. Nano Lett. 2011, 11, 1941–1945. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.K.; Drndić, M.; Nikolić, B.K. DNA Base-Specific Modulation of Microampere Transverse Edge Currents through a Metallic Graphene Nanoribbon with a Nanopore. Nano Lett. 2012, 12, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Heerema, S.J.; Schneider, G.F.; Rozemuller, M.; Vicarelli, L.; Zandbergen, H.W.; Dekker, C. 1/f noise in graphene nanopores. Nanotechnology 2015, 26. [Google Scholar] [CrossRef] [PubMed]
- Sourav, K.; Karmakar, S.N. Detection of base-pair mismatches in DNA using graphene-based nanopore device. Nanotechnology 2016, 27. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Park, C.; Fay, C.C.; Wang, X.; Ke, C. Mechanical strength of boron nitride nanotube-polymer interfaces. Appl. Phys. Lett. 2015, 107. [Google Scholar] [CrossRef]
- Liu, H.; Turner, C.H. Adsorption properties of nitrogen dioxide on hybrid carbon and boron-nitride nanotubes. Phys. Chem. Chem. Phys. 2014, 16, 22853–22860. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, X.; Zeng, X.C.; Yang, J. Band-Gap Engineering via Tailored Line Defects in Boron-Nitride Nanoribbons, Sheets, and Nanotubes. ACS Nano 2012, 6, 4104–4112. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xie, Y.; Liu, Z.; Wang, X.; Chai, Y.; Yan, F. Two-Dimensional Material Membranes: An Emerging Platform for Controllable Mass Transport Applications. Small 2014, 10, 4521–4542. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lu, B.; Zhao, Q.; Li, J.; Gao, T.; Chen, Y.; Zhang, Y.; Liu, Z.; Fan, Z.; Yang, F.; et al. Boron Nitride Nanopores: Highly Sensitive DNA Single-Molecule Detectors. Adv. Mater. 2013, 25, 4549–4554. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zou, X.; Zhou, G.; Liu, R.; Wu, J.; Li, J.; Duan, W. Adsorption of DNA/RNA nucleobases on hexagonal boron nitride sheet: An ab initio study. Phys. Chem. Chem. Phys. 2011, 13, 12225–12230. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Zhang, Y.; Luan, B.; Zhou, R. DNA translocation through single-layer boron nitride nanopores. Soft Matter 2016, 12, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Chen, X.; Wu, C.-M.L.; Li, H. Adsorption of nucleobase pairs on hexagonal boron nitride sheet: hydrogen bonding versus stacking. Phys. Chem. Chem. Phys. 2013, 15, 10767–10776. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Choi, Y.-K.; Kim, H.-J.; Scheicher, R.H.; Cho, J.-H. Physisorption of DNA Nucleobases on h-BN and Graphene: vdW-Corrected DFT Calculations. J. Phys. Chem. C 2013, 117, 13435–13441. [Google Scholar] [CrossRef]
- Johnson, R.R.; Johnson, A.T.C.; Klein, M.L. The Nature of DNA-Base–Carbon-Nanotube Interactions. Small 2010, 6, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.R.; Kohlmeyer, A.; Johnson, A.T.C.; Klein, M.L. Free Energy Landscape of a DNA–Carbon Nanotube Hybrid Using Replica Exchange Molecular Dynamics. Nano Lett. 2009, 9, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Enyashin, A.N.; Gemming, S.; Seifert, G. DNA-wrapped carbon nanotubes. Nanotechnology 2007, 18. [Google Scholar] [CrossRef]
- Gowtham, S.; Scheicher, R.H.; Pandey, R.; Karna, S.P.; Ahuja, R. First-principles study of physisorption of nucleic acid bases on small-diameter carbon nanotubes. Nanotechnology 2008, 19. [Google Scholar] [CrossRef] [PubMed]
- Varghese, N.; Mogera, U.; Govindaraj, A.; Das, A.; Maiti, P.K.; Sood, A.K.; Rao, C.N.R. Binding of DNA Nucleobases and Nucleosides with Graphene. ChemPhysChem 2009, 10, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Gowtham, S.; Scheicher, R.H.; Ahuja, R.; Pandey, R.; Karna, S.P. Physisorption of nucleobases on graphene: Density-functional calculations. Phys. Rev. B 2007, 76. [Google Scholar] [CrossRef]
- Ortmann, F.; Schmidt, W.G.; Bechstedt, F. Attracted by Long-Range Electron Correlation: Adenine on Graphite. Phys. Rev. Lett. 2005, 95. [Google Scholar] [CrossRef] [PubMed]
- Nasrabadi, A.T.; Foroutan, M. Interactions between Polymers and Single-Walled Boron Nitride Nanotubes: A Molecular Dynamics Simulation Approach. J. Phys. Chem. B 2010, 114, 15429–15436. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Zheng, M.; Park, C.; Wang, X.; Ke, C. Quantitative nanomechanical characterization of the van der Waals interfaces between carbon nanotubes and epoxy. Carbon 2015, 82, 214–228. [Google Scholar] [CrossRef]
- Becton, M.; Zhang, L.; Wang, X. Mechanics of Graphyne Crumpling. Phys. Chem. Chem. Phys. 2014, 16, 18233–18240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, B.; Wang, X. Cholesterol Extraction from Cell Membrane by Graphene Nanosheets: A Computational Study. J. Phys. Chem. B 2016, 120, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Gao, J.; Guo, W.; Jiang, L. Mechanical exfoliation of track-etched two-dimensional layered materials for the fabrication of ultrathin nanopores. Chem. Commun. 2014, 50, 14149–14152. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Peng, H.; Polonsky, S.; Rossnagel, S.; Stolovitzky, G.; Martyna, G. Base-By-Base Ratcheting of Single Stranded DNA through a Solid-State Nanopore. Phys. Rev. Lett. 2010, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shen, J.; Wang, H.; Wang, Q.; Zhang, J.; Liang, L.; Ågren, H.; Tu, Y. Effects of Graphene Nanopore Geometry on DNA Sequencing. J. Phys. Chem. Lett. 2014, 5, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Prasongkit, J.; Feliciano, G.T.; Rocha, A.R.; He, Y.; Osotchan, T.; Ahuja, R.; Scheicher, R.H. Theoretical assessment of feasibility to sequence DNA through interlayer electronic tunneling transport at aligned nanopores in bilayer graphene. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Nelson, T.; Zhang, B.; Prezhdo, O.V. Detection of Nucleic Acids with Graphene Nanopores: Ab Initio Characterization of a Novel Sequencing Device. Nano Lett. 2010, 10, 3237–3242. [Google Scholar] [CrossRef] [PubMed]
- Avdoshenko, S.M.; Nozaki, D.; da Rocha, C.G.; González, J.W.; Lee, M.H.; Gutierrez, R.; Cuniberti, G. Dynamic and Electronic Transport Properties of DNA Translocation through Graphene Nanopores. Nano Lett. 2013, 13, 1969–1976. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Baowan, D.; Hill, J.M. Nested boron nitride and carbon-boron nitride nanocones. IET Micro Nano Lett. 2007, 2, 46–49. [Google Scholar] [CrossRef]
- Neek-Amal, M.; Peeters, F.M. Graphene on boron-nitride: Moiré pattern in the van der Waals energy. Appl. Phys. Lett. 2014, 104. [Google Scholar] [CrossRef]
- Won, C.Y.; Aluru, N. Structure and dynamics of water confined in a boron nitride nanotube. J. Phys. Chem. C 2008, 112, 1812–1818. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Bashford, D.; Case, D.A. Generalized Born Models OF Macromolecular Solvation Effects. Ann. Rev. Phys. Chem. 2000, 51, 129–152. [Google Scholar] [CrossRef] [PubMed]




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wang, X. DNA Sequencing by Hexagonal Boron Nitride Nanopore: A Computational Study. Nanomaterials 2016, 6, 111. https://doi.org/10.3390/nano6060111
Zhang L, Wang X. DNA Sequencing by Hexagonal Boron Nitride Nanopore: A Computational Study. Nanomaterials. 2016; 6(6):111. https://doi.org/10.3390/nano6060111
Chicago/Turabian StyleZhang, Liuyang, and Xianqiao Wang. 2016. "DNA Sequencing by Hexagonal Boron Nitride Nanopore: A Computational Study" Nanomaterials 6, no. 6: 111. https://doi.org/10.3390/nano6060111





