Predicting Cytotoxicity of Metal Oxide Nanoparticles Using Isalos Analytics Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Toxicological Data from Metal Oxide NPs
2.2. Dataset Enrichment with Computational Descriptors
2.3. NanoPharos Database and Data Management
2.4. Model Development, Validation, Read Across and Domain of Applicability
- Gathering of the required descriptors (physicochemical, molecular and atomistic) for each NP.
- Construction of a data matrix including properties and endpoints.
- Development of an initial grouping hypothesis that correlates an endpoint, to different behaviour and reactivity properties. Assignment of the samples to groups.
- Assessment of the applicability of the approach using computational techniques and data gap filling. If no regular pattern emerged, an alternative grouping hypothesis must be proposed.
- If the grouping hypothesis is robust, but adequate data are not available, additional testing should be considered.
- Justification of the method.
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jeevanandam, J.; Barhoum, A.; Chan, Y.S.; Dufresne, A.; Danquah, M.K. Review on nanoparticles and nanostructured materials: History, sources, toxicity and regulations. Beilstein J. Nanotechnol. 2018, 9, 1050–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buzea, C.; Pacheco, I.I.; Robbie, K. Nanomaterials and nanoparticles: Sources and toxicity. Biointerphases 2007, 2, MR17–MR71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, J.A.; Fernández-García, M. Synthesis, Properties, and Applications of Oxide Nanomaterials; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Djurišić, A.B.; Leung, Y.H.; Ng, A.M.C.; Xu, X.Y.; Lee, P.K.H.; Degger, N.; Wu, R.S.S. Toxicity of Metal Oxide Nanoparticles: Mechanisms, Characterization, and Avoiding Experimental Artefacts. Small 2015, 11, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.L.; Russo, A.E.; Carro, R.V.; Stephens, M.A.; LePage, A.R.; Spalding, T.C.; Petersen, E.L.; Seal, S. Nanoscale Additives Tailor Energetic Materials. Nano Lett. 2007, 7, 2157–2161. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Dave, P.N. Nano-metal oxide: Potential catalyst on thermal decomposition of ammonium perchlorate. J. Exp. Nanosci. 2012, 7, 205–231. [Google Scholar] [CrossRef]
- Laurent, S.; Forge, D.; Port, M.; Roch, A.; Robic, C.; Vander Elst, L.; Muller, R.N. Magnetic Iron Oxide Nanoparticles: Synthesis, Stabilization, Vectorization, Physicochemical Characterizations, and Biological Applications. Chem. Rev. 2008, 108, 2064–2110. [Google Scholar] [CrossRef] [PubMed]
- Hasany, S.; Ahmed, I.; Rajan, J.; Rehman, A. Systematic review of the preparation techniques of iron oxide magnetic nanoparticles. Nanosci. Nanotechnol. 2012, 2, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Falcaro, P.; Ricco, R.; Yazdi, A.; Imaz, I.; Furukawa, S.; Maspoch, D.; Ameloot, R.; Evans, J.D.; Doonan, C.J. Application of metal and metal oxide nanoparticles@MOFs. Coord. Chem. Rev. 2016, 307, 237–254. [Google Scholar] [CrossRef]
- Vance, M.E.; Kuiken, T.; Vejerano, E.P.; McGinnis, S.P.; Hochella, M.F., Jr.; Rejeski, D.; Hull, M.S. Nanotechnology in the real world: Redeveloping the nanomaterial consumer products inventory. Beilstein J. Nanotechnol. 2015, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Nel, A.; Xia, T.; Mädler, L.; Li, N. Toxic Potential of Materials at the Nanolevel. Science 2006, 311, 622–627. [Google Scholar] [CrossRef] [Green Version]
- Horie, M.; Kato, H.; Fujita, K.; Endoh, S.; Iwahashi, H. In Vitro Evaluation of Cellular Response Induced by Manufactured Nanoparticles. Chem. Res. Toxicol. 2012, 25, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Kovochich, M.; Liong, M.; Mädler, L.; Gilbert, B.; Shi, H.; Yeh, J.I.; Zink, J.I.; Nel, A.E. Comparison of the Mechanism of Toxicity of Zinc Oxide and Cerium Oxide Nanoparticles Based on Dissolution and Oxidative Stress Properties. ACS Nano 2008, 2, 2121–2134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manke, A.; Wang, L.; Rojanasakul, Y. Mechanisms of Nanoparticle-Induced Oxidative Stress and Toxicity. BioMed Res. Int. 2013, 2013, 942916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahru, A.; Dubourguier, H.-C. From ecotoxicology to nanoecotoxicology. Toxicology 2010, 269, 105–119. [Google Scholar] [CrossRef]
- Passagne, I.; Morille, M.; Rousset, M.; Pujalté, I.; L’Azou, B. Implication of oxidative stress in size-dependent toxicity of silica nanoparticles in kidney cells. Toxicology 2012, 299, 112–124. [Google Scholar] [CrossRef]
- Avalos, A.; Haza, A.I.; Mateo, D.; Morales, P. Cytotoxicity and ROS production of manufactured silver nanoparticles of different sizes in hepatoma and leukemia cells. J. Appl. Toxicol. 2014, 34, 413–423. [Google Scholar] [CrossRef]
- Misawa, M.; Takahashi, J. Generation of reactive oxygen species induced by gold nanoparticles under x-ray and UV Irradiations. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 604–614. [Google Scholar] [CrossRef]
- Horie, M.; Fujita, K.; Kato, H.; Endoh, S.; Nishio, K.; Komaba, L.K.; Nakamura, A.; Miyauchi, A.; Kinugasa, S.; Hagihara, Y.; et al. Association of the physical and chemical properties and the cytotoxicity of metal oxide nanoparticles: Metal ion release, adsorption ability and specific surface area. Metallomics 2012, 4, 350–360. [Google Scholar] [CrossRef]
- Chupani, L.; Niksirat, H.; Velíšek, J.; Stará, A.; Hradilová, Š.; Kolařík, J.; Panáček, A.; Zusková, E. Chronic dietary toxicity of zinc oxide nanoparticles in common carp (Cyprinus carpio L.): Tissue accumulation and physiological responses. Ecotoxicol. Environ. Saf. 2018, 147, 110–116. [Google Scholar] [CrossRef]
- Mylona, Z.; Panteris, E.; Moustakas, M.; Kevrekidis, T.; Malea, P. Physiological, structural and ultrastructural impacts of silver nanoparticles on the seagrass Cymodocea nodosa. Chemosphere 2020, 248, 126066. [Google Scholar] [CrossRef]
- Qian, H.; Zhu, K.; Lu, H.; Lavoie, M.; Chen, S.; Zhou, Z.; Deng, Z.; Chen, J.; Fu, Z. Contrasting silver nanoparticle toxicity and detoxification strategies in Microcystis aeruginosa and Chlorella vulgaris: New insights from proteomic and physiological analyses. Sci. Total Environ. 2016, 572, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen: London, UK, 1959. [Google Scholar]
- Legislation for the Protection of Animals Used for Scientific Purposes. Available online: https://ec.europa.eu/environment/chemicals/lab_animals/legislation_en.htm (accessed on 13 August 2020).
- Erkekoglu, P.; Giray, B.K.; Basaran, N. 3R principle and alternative toxicity testing methods. Fabad J. Pharm. Sci. 2011, 36, 101–117. [Google Scholar]
- Kroeger, M. How omics technologies can contribute to the ‘3R’ principles by introducing new strategies in animal testing. Trends Biotechnol. 2006, 24, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Törnqvist, E.; Annas, A.; Granath, B.; Jalkesten, E.; Cotgreave, I.; Öberg, M. Strategic Focus on 3R Principles Reveals Major Reductions in the Use of Animals in Pharmaceutical Toxicity Testing. PLoS ONE 2014, 9, e101638. [Google Scholar] [CrossRef] [Green Version]
- OECD. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models. Available online: https://www.oecd.org/env/guidance-document-on-the-validation-of-quantitative-structure-activity-relationship-q-sar-models-9789264085442-en.htm (accessed on 13 August 2020).
- Winkler, D.A. Role of Artificial Intelligence and Machine Learning in Nanosafety. Small 2020, 16, 2001883. [Google Scholar] [CrossRef] [PubMed]
- Ellison, C.M.; Piechota, P.; Madden, J.C.; Enoch, S.J.; Cronin, M.T.D. Adverse Outcome Pathway (AOP) Informed Modeling of Aquatic Toxicology: QSARs, Read-Across, and Interspecies Verification of Modes of Action. Environ. Sci. Technol. 2016, 50, 3995–4007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baskin, I.I. Machine Learning Methods in Computational Toxicology. In Computational Toxicology: Methods and Protocols; Nicolotti, O., Ed.; Springer: New York, NY, USA, 2018; pp. 119–139. [Google Scholar] [CrossRef]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Hochreiter, S. DeepTox: Toxicity Prediction using Deep Learning. Front. Environ. Sci. 2016, 3, 80. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.; Choi, J. Development of AOP relevant to microplastics based on toxicity mechanisms of chemical additives using ToxCast™ and deep learning models combined approach. Environ. Int. 2020, 137, 105557. [Google Scholar] [CrossRef]
- Xia, C.; Fu, L.; Liu, Z.; Liu, H.; Chen, L.; Liu, Y. Aquatic Toxic Analysis by Monitoring Fish Behavior Using Computer Vision: A Recent Progress. J. Toxicol. 2018, 2018, 2591924. [Google Scholar] [CrossRef]
- Karatzas, P.; Melagraki, G.; Ellis, L.-J.A.; Lynch, I.; Varsou, D.-D.; Afantitis, A.; Tsoumanis, A.; Doganis, P.; Sarimveis, H. Development of deep learning models for predicting the effects of exposure to engineered nanomaterials on Daphnia magna. Small 2020, in press. [Google Scholar] [CrossRef]
- De Morais e Silva, L.; Lorenzo, V.P.; Lopes, W.S.; Scotti, L.; Scotti, M.T. Predictive Computational Tools for Assessment of Ecotoxicological Activity of Organic Micropollutants in Various Water Sources in Brazil. Mol. Inform. 2019, 38, 1800156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satpathy, R. Computational Tools and Techniques to Predict Aquatic Toxicity of Some Halogenated Pollutants. In Handbook of Research on the Adverse Effects of Pesticide Pollution in Aquatic Ecosystems; IGI Global: Hershey, PA, USA, 2019; pp. 318–337. [Google Scholar]
- Raies, A.B.; Bajic, V.B. In silico toxicology: Computational methods for the prediction of chemical toxicity. WIREs Comput. Mol. Sci. 2016, 6, 147–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varsou, D.-D.; Tsiliki, G.; Nymark, P.; Kohonen, P.; Grafström, R.; Sarimveis, H. toxFlow: A Web-Based Application for Read-Across Toxicity Prediction Using Omics and Physicochemical Data. J. Chem. Inf. Model. 2018, 58, 543–549. [Google Scholar] [CrossRef]
- Tämm, K.; Sikk, L.; Burk, J.; Rallo, R.; Pokhrel, S.; Mädler, L.; Scott-Fordsmand, J.J.; Burk, P.; Tamm, T. Parametrization of nanoparticles: Development of full-particle nanodescriptors. Nanoscale 2016, 8, 16243–16250. [Google Scholar] [CrossRef] [PubMed]
- Puzyn, T.; Rasulev, B.; Gajewicz, A.; Hu, X.; Dasari, T.P.; Michalkova, A.; Hwang, H.M.; Toropov, A.; Leszczynska, D.; Leszczynski, J. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nanotechnol. 2011, 6, 175–178. [Google Scholar] [CrossRef]
- Toropova, A.P.; Toropov, A.A.; Rallo, R.; Leszczynska, D.; Leszczynski, J. Optimal descriptor as a translator of eclectic data into prediction of cytotoxicity for metal oxide nanoparticles under different conditions. Ecotoxicol. Environ. Saf. 2015, 112, 39–45. [Google Scholar] [CrossRef]
- Toropov, A.A.; Toropova, A.P.; Benfenati, E.; Gini, G.; Puzyn, T.; Leszczynska, D.; Leszczynski, J. Novel application of the CORAL software to model cytotoxicity of metal oxide nanoparticles to bacteria Escherichia coli. Chemosphere 2012, 89, 1098–1102. [Google Scholar] [CrossRef]
- Puzyn, T.; Leszczynska, D.; Leszczynski, J. Toward the Development of “Nano-QSARs”: Advances and Challenges. Small 2009, 5, 2494–2509. [Google Scholar] [CrossRef]
- Zhang, H.; Ji, Z.; Xia, T.; Meng, H.; Low-Kam, C.; Liu, R.; Pokhrel, S.; Lin, S.; Wang, X.; Liao, Y.-P.; et al. Use of Metal Oxide Nanoparticle Band Gap to Develop a Predictive Paradigm for Oxidative Stress and Acute Pulmonary Inflammation. ACS Nano 2012, 6, 4349–4368. [Google Scholar] [CrossRef]
- Toropov, A.A.; Achary, P.G.R.; Toropova, A.P. Quasi-SMILES and nano-QFPR: The predictive model for zeta potentials of metal oxide nanoparticles. Chem. Phys. Lett. 2016, 660, 107–110. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, S.; Toropova, A.P.; Toropov, A.A. Correlation intensity index: Mathematical modeling of cytotoxicity of metal oxide nanoparticles. Nanotoxicology 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Varsou, D.-D.; Afantitis, A.; Tsoumanis, A.; Papadiamantis, A.; Valsami-Jones, E.; Lynch, I.; Melagraki, G. Zeta-Potential Read-Across Model Utilizing Nanodescriptors Extracted via the NanoXtract Image Analysis Tool Available on the Enalos Nanoinformatics Cloud Platform. Small 2020, 16, 1906588. [Google Scholar] [CrossRef] [PubMed]
- UPCI. Nano-Image: Extracting Data from Microscopy Images. Available online: https://nanoimage.jaqpot.org/ (accessed on 20 July 2020).
- Gajewicz, A.; Rasulev, B.; Dinadayalane, T.C.; Urbaszek, P.; Puzyn, T.; Leszczynska, D.; Leszczynski, J. Advancing risk assessment of engineered nanomaterials: Application of computational approaches. Adv. Drug Deliv. Rev. 2012, 64, 1663–1693. [Google Scholar] [CrossRef] [PubMed]
- Toropov, A.A.; Toropova, A.P.; Puzyn, T.; Benfenati, E.; Gini, G.; Leszczynska, D.; Leszczynski, J. QSAR as a random event: Modeling of nanoparticles uptake in PaCa2 cancer cells. Chemosphere 2013, 92, 31–37. [Google Scholar] [CrossRef]
- Jagiello, K.; Chomicz, B.; Avramopoulos, A.; Gajewicz, A.; Mikolajczyk, A.; Bonifassi, P.; Papadopoulos, M.G.; Leszczynski, J.; Puzyn, T. Size-dependent electronic properties of nanomaterials: How this novel class of nanodescriptors supposed to be calculated? Struct. Chem. 2017, 28, 635–643. [Google Scholar] [CrossRef] [Green Version]
- Burk, J.; Sikk, L.; Burk, P.; Manshian, B.B.; Soenen, S.J.; Scott-Fordsmand, J.J.; Tamm, T.; Tämm, K. Fe-Doped ZnO nanoparticle toxicity: Assessment by a new generation of nanodescriptors. Nanoscale 2018, 10, 21985–21993. [Google Scholar] [CrossRef] [Green Version]
- Manshian, B.B.; Pokhrel, S.; Himmelreich, U.; Tämm, K.; Sikk, L.; Fernández, A.; Rallo, R.; Tamm, T.; Mädler, L.; Soenen, S.J. In Silico Design of Optimal Dissolution Kinetics of Fe-Doped ZnO Nanoparticles Results in Cancer-Specific Toxicity in a Preclinical Rodent Model. Adv. Healthc. Mater. 2017, 6, 1601379. [Google Scholar] [CrossRef]
- Xia, X.R.; Monteiro-Riviere, N.A.; Mathur, S.; Song, X.; Xiao, L.; Oldenberg, S.J.; Fadeel, B.; Riviere, J.E. Mapping the Surface Adsorption Forces of Nanomaterials in Biological Systems. ACS Nano 2011, 5, 9074–9081. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Sedykh, A.; Wang, W.; Zhao, X.; Yan, B.; Zhu, H. In silico profiling nanoparticles: Predictive nanomodeling using universal nanodescriptors and various machine learning approaches. Nanoscale 2019, 11, 8352–8362. [Google Scholar] [CrossRef]
- Xia, X.-R.; Monteiro-Riviere, N.A.; Riviere, J.E. An index for characterization of nanomaterials in biological systems. Nat. Nanotechnol. 2010, 5, 671–675. [Google Scholar] [CrossRef]
- Afantitis, A.; Melagraki, G.; Isigonis, P.; Tsoumanis, A.; Varsou, D.D.; Valsami-Jones, E.; Papadiamantis, A.; Ellis, L.-J.A.; Sarimveis, H.; Doganis, P.; et al. NanoSolveIT Project: Driving nanoinformatics research to develop innovative and integrated tools for in silico nanosafety assessment. Comput. Struct. Biotechnol. J. 2020, 18, 583–602. [Google Scholar] [CrossRef] [PubMed]
- H2020 NanoCommons e-Infrastructure Project. Available online: http://www.nanocommons.eu (accessed on 20 July 2020).
- Ha, M.K.; Trinh, T.X.; Choi, J.S.; Maulina, D.; Byun, H.G.; Yoon, T.H. Toxicity Classification of Oxide Nanomaterials: Effects of Data Gap Filling and PChem Score-based Screening Approaches. Sci. Rep. 2018, 8, 3141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uboldi, C.; Sanles Sobrido, M.; Bernard, E.; Tassistro, V.; Herlin-Boime, N.; Vrel, D.; Garcia-Argote, S.; Roche, S.; Magdinier, F.; Dinescu, G. In Vitro Analysis of the Effects of ITER-Like Tungsten Nanoparticles: Cytotoxicity and Epigenotoxicity in BEAS-2B Cells. Nanomaterials 2019, 9, 1233. [Google Scholar] [CrossRef] [Green Version]
- Batista Caixeta, M.; Sampaio Araújo, P.; Bastos Gonçalves, B.; Damacena Silva, L.; Ixchel Grano-Maldonado, M.; Lopes Rocha, T. Toxicity of engineered nanomaterials to aquatic and land snails: A scientometric and systematic review. Chemosphere 2020, 260, 127654. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Oh, E.; Liu, R.; Breger, J.C.; Medintz, I.L.; Cohen, Y. Bayesian Network Resource for Meta-Analysis: Cellular Toxicity of Quantum Dots. Small 2019, 15, 1900510. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Liu, R.; Nel, A.; Gemill, K.B.; Bilal, M.; Cohen, Y.; Medintz, I.L. Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 2016, 11, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Labouta, H.I.; Asgarian, N.; Rinker, K.; Cramb, D.T. Meta-Analysis of Nanoparticle Cytotoxicity via Data-Mining the Literature. ACS Nano 2019, 13, 1583–1594. [Google Scholar] [CrossRef]
- Polak, E.; Ribiere, G. Note sur la convergence de méthodes de directions conjuguées. ESAIM Math. Model. Numer. Anal. Modélisation Mathématique Anal. Numérique 1969, 3, 35–43. [Google Scholar]
- Buckingham, R.A.; Lennard-Jones, J.E. The classical equation of state of gaseous helium, neon and argon. Proc. R. Soc. Lond. Ser. A Math. Phys. Sci. 1938, 168, 264–283. [Google Scholar] [CrossRef]
- Wolf, D.; Keblinski, P.; Phillpot, S.R.; Eggebrecht, J. Exact method for the simulation of Coulombic systems by spherically truncated, pairwise r−1 summation. J. Chem. Phys. 1999, 110, 8254–8282. [Google Scholar] [CrossRef]
- Ewald, P.P. Die Berechnung optischer und elektrostatischer Gitterpotentiale. Ann. Phys. 1921, 369, 253–287. [Google Scholar] [CrossRef] [Green Version]
- Plimpton, S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. J. Comput. Phys. 1995, 117, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Antreas, A.; Andreas, T.; Melagraki, G. Enalos Suite of Tools: Enhance Cheminformatics and Nanoinformatics through KNIME. Curr. Med. Chem. 2020, 27, 1–13. [Google Scholar] [CrossRef]
- Melagraki, G.; Afantitis, A. Enalos InSilicoNano platform: An online decision support tool for the design and virtual screening of nanoparticles. RSC Adv. 2014, 4, 50713–50725. [Google Scholar] [CrossRef]
- Enalos+ KNIME Nodes. Available online: http://enalosplus.novamechanics.com/ (accessed on 15 August 2020).
- Varsou, D.-D.; Nikolakopoulos, S.; Tsoumanis, A.; Melagraki, G.; Afantitis, A. Enalos+ KNIME Nodes: New Cheminformatics Tools for Drug Discovery. In Rational Drug Design: Methods and Protocols; Mavromoustakos, T., Kellici, T.F., Eds.; Springer: New York, NY, USA, 2018; pp. 113–138. [Google Scholar] [CrossRef]
- Ojha, P.K.; Roy, K. Comparative QSARs for antimalarial endochins: Importance of descriptor-thinning and noise reduction prior to feature selection. Chemom. Intell. Lab. Syst. 2011, 109, 146–161. [Google Scholar] [CrossRef]
- Leach, A.R.; Gillet, V.J. An Introduction to Chemoinformatics; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- Hall, M.A. Correlation-Based Feature Selection for Machine Learning. Ph.D. Thesis, The University of Waikato, Hamilton, New Zealand, April 1999. [Google Scholar]
- Larose, D.T.; Larose, C.D. Discovering Knowledge in Data: An Introduction to Data Mining; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- ECHA. Read-Across Assessment Framework (RAAF). 2017. Available online: https://echa.europa.eu/documents/10162/13628/raaf_en.pdf (accessed on 8 November 2019).
- OECD. Validation of (Q)SAR Models. Available online: https://www.oecd.org/chemicalsafety/risk-assessment/validationofqsarmodels.htm (accessed on 8 November 2019).
- Zhang, S.; Golbraikh, A.; Oloff, S.; Kohn, H.; Tropsha, A. A Novel Automated Lazy Learning QSAR (ALL-QSAR) Approach: Method Development, Applications, and Virtual Screening of Chemical Databases Using Validated ALL-QSAR Models. J. Chem. Inf. Model. 2006, 46, 1984–1995. [Google Scholar] [CrossRef] [Green Version]
- Puzyn, T.; Jeliazkova, N.; Sarimveis, H.; Marchese Robinson, R.L.; Lobaskin, V.; Rallo, R.; Richarz, A.-N.; Gajewicz, A.; Papadopulos, M.G.; Hastings, J.; et al. Perspectives from the NanoSafety Modelling Cluster on the validation criteria for (Q)SAR models used in nanotechnology. Food Chem. Toxicol. 2018, 112, 478–494. [Google Scholar] [CrossRef]
- Tropsha, A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef]
- Tropsha, A.; Gramatica, P.; Gombar, V.K. The Importance of Being Earnest: Validation is the Absolute Essential for Successful Application and Interpretation of QSPR Models. QSAR Comb. Sci. 2003, 22, 69–77. [Google Scholar] [CrossRef]
- Papadiamantis, A.G.; Klaessig, F.C.; Exner, T.E.; Hofer, S.; Hofstaetter, N.; Himly, M.; Williams, M.A.; Doganis, P.; Hoover, M.D.; Afantitis, A.; et al. Metadata stewardship in nanosafety research: Community-driven organisation of metadata schemas to support FAIR nanoscience data. Naomaterials 2020. submitted for publication. [Google Scholar]
- Bonaretti, S.; Willighagen, E. Two real use cases of FAIR maturity indicators in the life sciences. BioRxiv 2019, 739334. [Google Scholar] [CrossRef] [Green Version]
- Låg, M.; Skuland, T.; Godymchuk, A.; Nguyen, T.H.T.; Pham, H.L.T.; Refsnes, M. Silica Nanoparticle-induced Cytokine Responses in BEAS-2B and HBEC3-KT Cells: Significance of Particle Size and Signalling Pathways in Different Lung Cell Cultures. Basic Clin. Pharmacol. Toxicol. 2018, 122, 620–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Hu, H.; Jiang, L.; Zou, Y.; Duan, J.; Sun, Z. Cytotoxicity and autophagy dysfunction induced by different sizes of silica particles in human bronchial epithelial BEAS-2B cells. Toxicol. Res. 2016, 5, 1216–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makama, S.; Kloet, S.K.; Piella, J.; van den Berg, H.; de Ruijter, N.C.A.; Puntes, V.F.; Rietjens, I.M.C.M.; van den Brink, N.W. Effects of Systematic Variation in Size and Surface Coating of Silver Nanoparticles on Their In Vitro Toxicity to Macrophage RAW 264.7 Cells. Toxicol. Sci. 2017, 162, 79–88. [Google Scholar] [CrossRef]
- Ali, S.A.; Rizk, M.Z.; Hamed, M.A.; Aboul-Ela, E.I.; El-Rigal, N.S.; Aly, H.F.; Abdel-Hamid, A.-H.Z. Assessment of titanium dioxide nanoparticles toxicity via oral exposure in mice: Effect of dose and particle size. Biomarkers 2019, 24, 492–498. [Google Scholar] [CrossRef]
- Loan, T.T.; Do, L.T.; Yoo, H. Platinum Nanoparticles Induce Apoptosis on Raw 264.7 Macrophage Cells. J. Nanosci. Nanotechnol. 2018, 18, 861–864. [Google Scholar] [CrossRef]
- Nishanth, R.P.; Jyotsna, R.G.; Schlager, J.J.; Hussain, S.M.; Reddanna, P. Inflammatory responses of RAW 264.7 macrophages upon exposure to nanoparticles: Role of ROS-NFκB signaling pathway. Nanotoxicology 2011, 5, 502–516. [Google Scholar] [CrossRef]
- Huk, A.; Izak-Nau, E.; Reidy, B.; Boyles, M.; Duschl, A.; Lynch, I.; Dušinska, M. Is the toxic potential of nanosilver dependent on its size? Part. Fibre Toxicol. 2014, 11, 65. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.K.; Bello, D.; Cohen, J.; Demokritou, P. Implications of in vitro dosimetry on toxicological ranking of low aspect ratio engineered nanomaterials. Nanotoxicology 2015, 9, 871–885. [Google Scholar] [CrossRef] [Green Version]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Roohi, F.; Lohrke, J.; Ide, A.; Schütz, G.; Dassler, K. Studying the effect of particle size and coating type on the blood kinetics of superparamagnetic iron oxide nanoparticles. Int. J. Nanomed. 2012, 7, 4447–4458. [Google Scholar] [CrossRef] [Green Version]
- Abakumov, M.A.; Semkina, A.S.; Skorikov, A.S.; Vishnevskiy, D.A.; Ivanova, A.V.; Mironova, E.; Davydova, G.A.; Majouga, A.G.; Chekhonin, V.P. Toxicity of iron oxide nanoparticles: Size and coating effects. J. Biochem. Mol. Toxicol. 2018, 32, e22225. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Oberdörster, G.; Biswas, P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J. Nanopart. Res. 2009, 11, 77–89. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Simchi, A.; Milani, A.S.; Stroeve, P. Cell toxicity of superparamagnetic iron oxide nanoparticles. J. Colloid Interface Sci. 2009, 336, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Atkins, P.W.; de Paula, J. Physical Chemistry: Thermodynamics, Structure, and Change; W.H. Freeman: New York, NY, USA, 2014. [Google Scholar]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.O.; Di Maio, A.; Guggenheim, E.J.; Chetwynd, A.J.; Pencross, D.; Tang, S.; Belinga-Desaunay, M.-F.A.; Thomas, S.G.; Rappoport, J.Z.; Lynch, I. Surface Chemistry-Dependent Evolution of the Nanomaterial Corona on TiO2 Nanomaterials Following Uptake and Sub-Cellular Localization. Nanomaterials 2020, 10, 401. [Google Scholar] [CrossRef] [Green Version]
- Gliga, A.R.; Skoglund, S.; Odnevall Wallinder, I.; Fadeel, B.; Karlsson, H.L. Size-dependent cytotoxicity of silver nanoparticles in human lung cells: The role of cellular uptake, agglomeration and Ag release. Part Fibre Toxicol. 2014, 11, 11. [Google Scholar] [CrossRef] [Green Version]
- Varsou, D.-D.; Tsoumanis, A.; Afantitis, A.; Melagraki, G. Enalos Cloud Platform: Nanoinformatics and Cheminformatics Tools. In Ecotoxicological QSARs; Roy, K., Ed.; Springer: New York, NY, USA, 2020; pp. 789–800. [Google Scholar] [CrossRef]
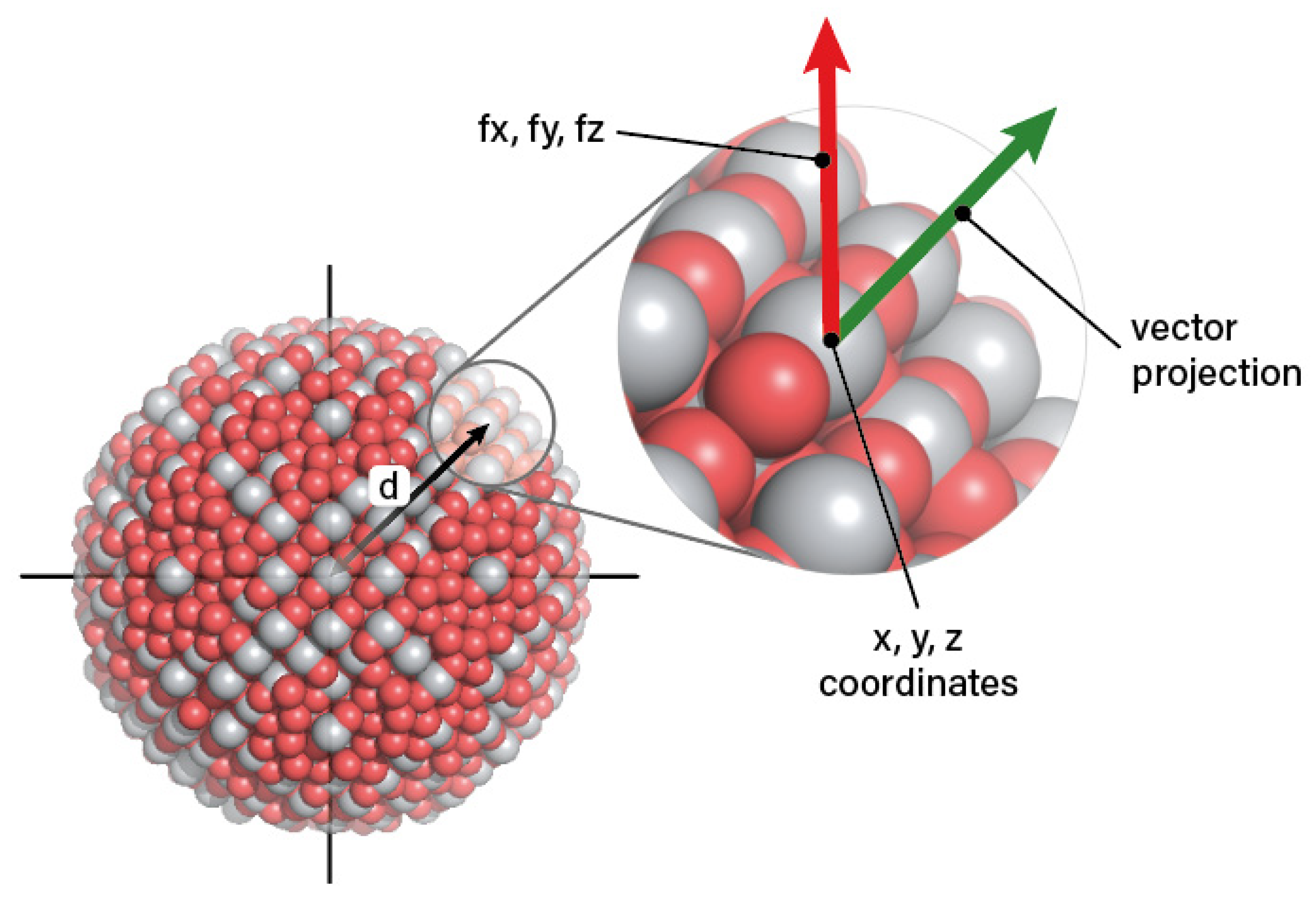
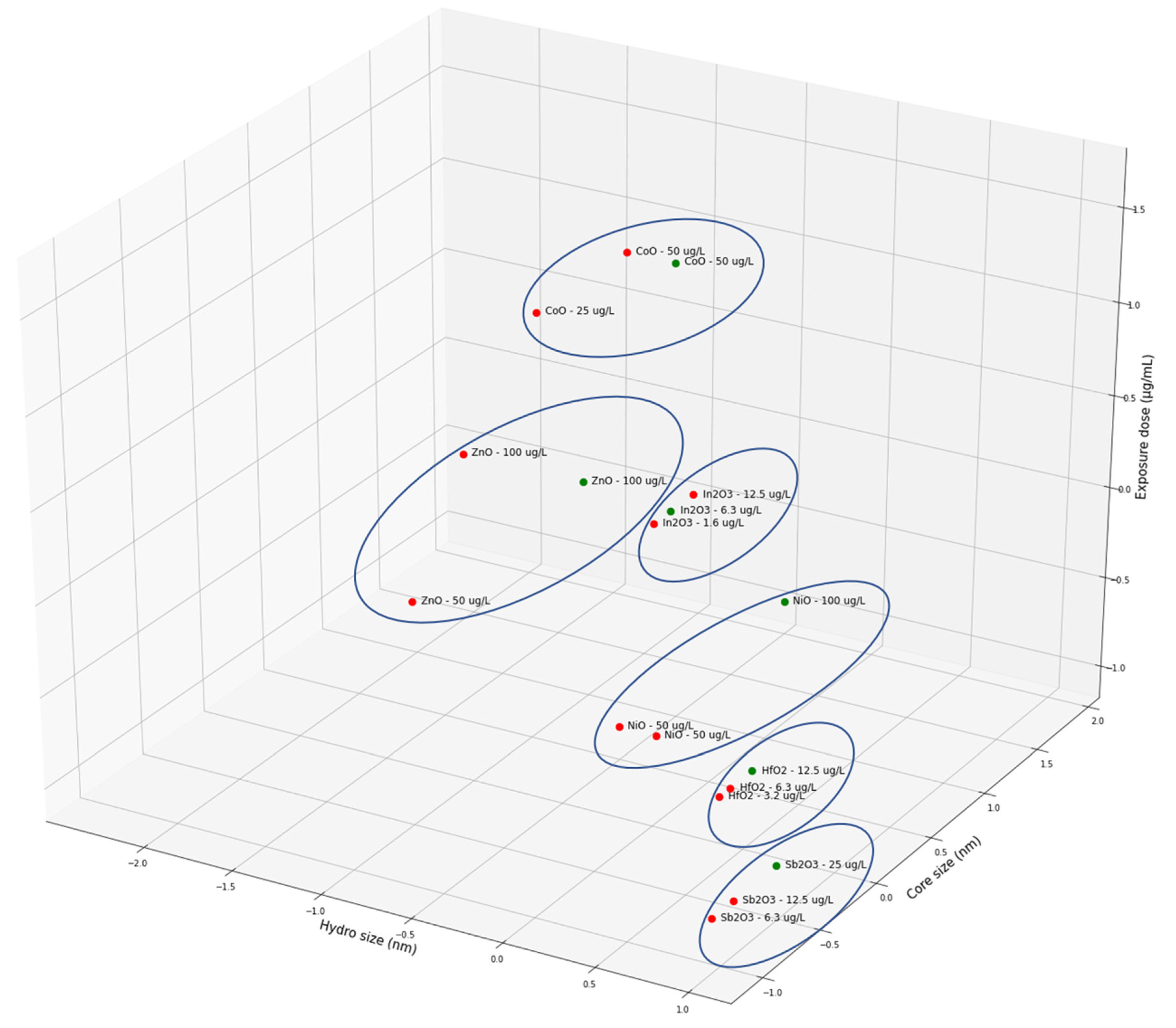
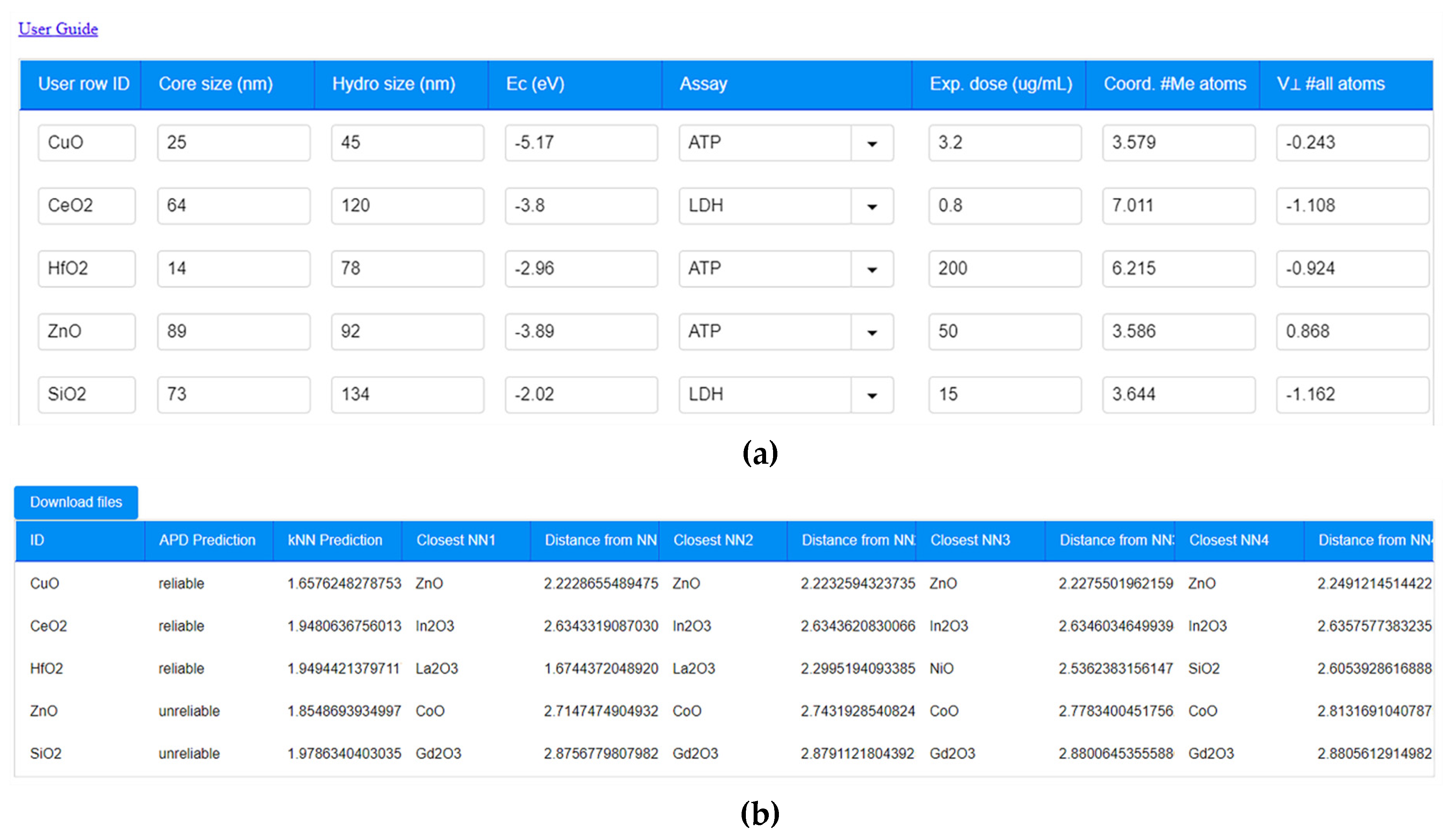
| Criterion | Result | Assessment |
|---|---|---|
| R2 > 0.6 | 0.91 | Pass |
| Rcvext > 0.5 | 0.904 | Pass |
| 0.022 | Pass | |
| 0.002 | Pass | |
| 0.018 | Pass | |
| 0.85 < k < 1.15 | 0.994 | Pass |
| 0.85 < k’ < 1.15 | 1.005 | Pass |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadiamantis, A.G.; Jänes, J.; Voyiatzis, E.; Sikk, L.; Burk, J.; Burk, P.; Tsoumanis, A.; Ha, M.K.; Yoon, T.H.; Valsami-Jones, E.; et al. Predicting Cytotoxicity of Metal Oxide Nanoparticles Using Isalos Analytics Platform. Nanomaterials 2020, 10, 2017. https://doi.org/10.3390/nano10102017
Papadiamantis AG, Jänes J, Voyiatzis E, Sikk L, Burk J, Burk P, Tsoumanis A, Ha MK, Yoon TH, Valsami-Jones E, et al. Predicting Cytotoxicity of Metal Oxide Nanoparticles Using Isalos Analytics Platform. Nanomaterials. 2020; 10(10):2017. https://doi.org/10.3390/nano10102017
Chicago/Turabian StylePapadiamantis, Anastasios G., Jaak Jänes, Evangelos Voyiatzis, Lauri Sikk, Jaanus Burk, Peeter Burk, Andreas Tsoumanis, My Kieu Ha, Tae Hyun Yoon, Eugenia Valsami-Jones, and et al. 2020. "Predicting Cytotoxicity of Metal Oxide Nanoparticles Using Isalos Analytics Platform" Nanomaterials 10, no. 10: 2017. https://doi.org/10.3390/nano10102017





