Recurrent Cholera Outbreaks in Sub-Saharan Africa: Moving beyond Epidemiology to Understand the Environmental Reservoirs and Drivers
Abstract
:1. Introduction
2. Beyond Epidemiology: Integrating Environmental Sciences into Cholera Research
2.1. Rationale for Understanding Environmental Drivers and Reservoirs
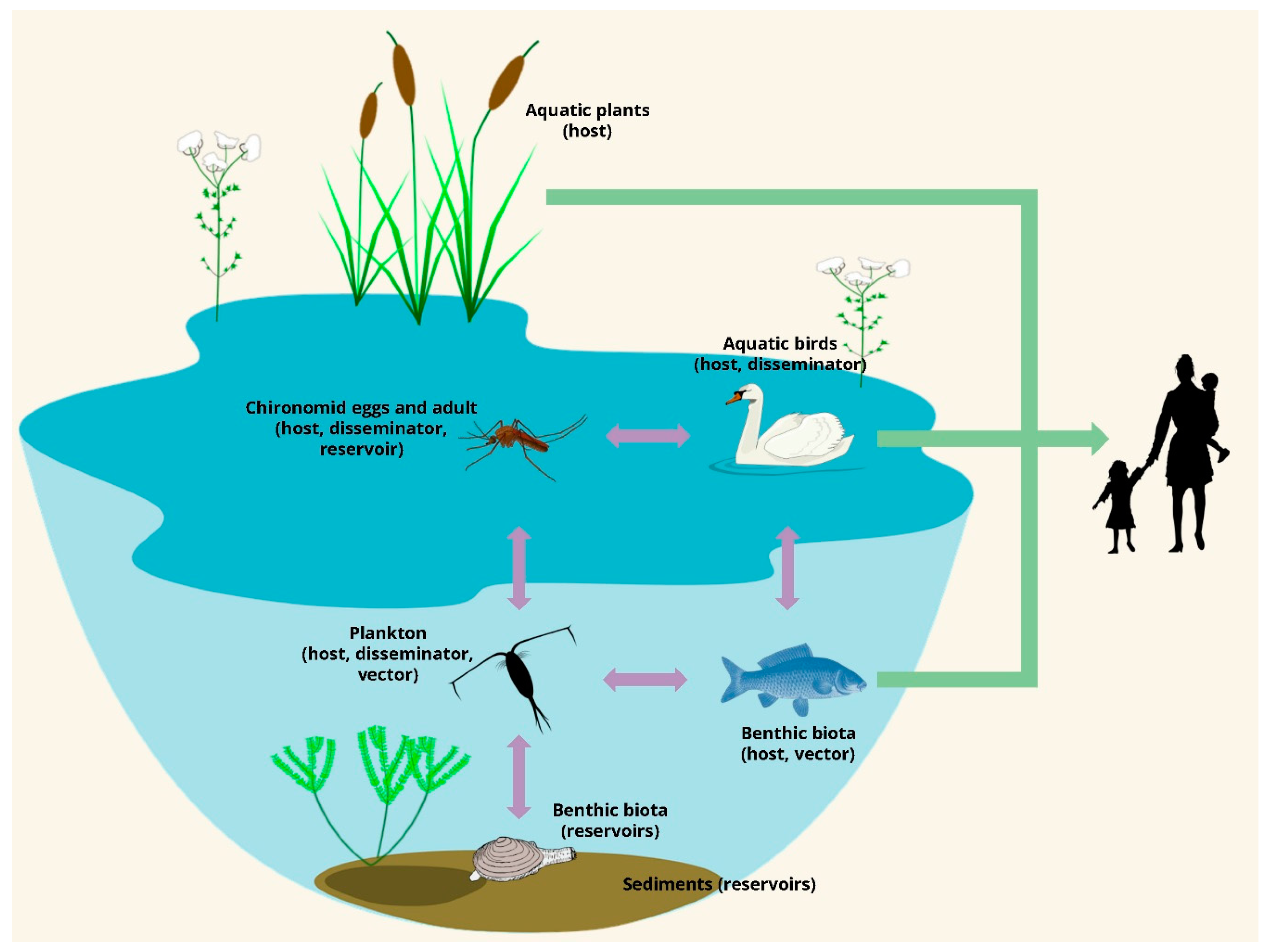
2.2. Environmental Reservoirs: Where Cholera Lurks during Inter-Epidemic Periods
2.3. What Drives Cholera Outbreaks in SSA?
3. Harnessing Emerging Research Tools and Charting Future Directions
3.1. Emerging Research Tools to Better Understand Cholera Dynamics
3.2. Knowledge Gaps and Future Research Directions
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mengel, M.A.; Delrieu, I.; Heyerdahl, L.; Gessner, B.D. Cholera Outbreaks in Africa. In Cholera Outbreaks; Nair, G.B., Takeda, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 5, pp. 117–144 . ISBN 0070-217X (Print)r0070-217X (Linking). [Google Scholar]
- Global Task Force on Cholera Control. Ending Cholera: A Global Roadmap to 2030; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Cuneo, C.N.; Sollom, R.; Beyrer, C. The Cholera Epidemic in Zimbabwe, 2008–2009: A Review and Critique of the Evidence. Health Hum. Rights 2017, 19, 249–264. [Google Scholar] [PubMed]
- Mukandavire, Z.; Liao, S.; Wang, J.; Gaff, H.; Smith, D.L.; Morris, J.G. Estimating the reproductive numbers for the 2008–2009 cholera outbreaks in Zimbabwe. Proc. Natl. Acad. Sci. USA 2011, 108, 8767–8772. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Emergencies Preparedness, Response: Cholera–Zimbabwe. Available online: http://www.who.int/csr/don/11-december-2017-cholera-kenya/en/ (accessed on 30 November 2018).
- Gaffga, N.H.; Tauxe, R.V.; Mintz, E.D. Cholera: A new homeland in Africa? Am. J. Trop. Med. Hyg. 2007, 77, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Mintz, E.D.; Tauxe, R.V. Cholera in Africa: A closer look and a time for action. J. Infect. Dis. 2013, 208, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Valia, R.; Taviani, E.; Spagnoletti, M.; Ceccarelli, D.; Cappuccinelli, P.; Colombo, M.M. Vibrio cholerae O1 epidemic variants in Angola: A retrospective study between 1992 and 2006. Front. Microbiol. 2013, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kacou-N’douba, A.; Anné, J.C.B.; Okpo, L.S.; Elogne-Kouamé, C.; Koffi, S.; Koffi, V.; N’Guessan, K.; Coulibaly, D.; Dagnan, S.N.; Eholié, S.P.; et al. Antimicrobial resistance of Vibrio cholerae O1 isolated during a cholera epidemic in 2011 in dry season in Cote d’ivoire. J. Infect. Dev. Ctries. 2012, 6, 595–597. [Google Scholar] [CrossRef] [PubMed]
- Dalsgaard, A. Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 2001, 48, 827–838. [Google Scholar] [CrossRef] [Green Version]
- Marin, M.A.; Thompson, C.C.; Freitas, F.S.; Fonseca, E.L.; Aboderin, A.O.; Zailani, S.B.; Quartey, N.K.E.; Okeke, I.N.; Vicente, A.C.P. Cholera Outbreaks in Nigeria Are Associated with Multidrug Resistant Atypical El Tor and Non-O1/Non-O139 Vibrio cholerae. PLoS Negl. Trop. Dis. 2013, 7. [Google Scholar] [CrossRef]
- Lessler, J.; Moore, S.M.; Luquero, F.J.; McKay, H.S.; Grais, R.; Henkens, M.; Mengel, M.; Dunoyer, J.; M’bangombe, M.; Lee, E.C.; et al. Mapping the burden of cholera in sub-Saharan Africa and implications for control: an analysis of data across geographical scales. Lancet 2018, 6736, 1–8. [Google Scholar] [CrossRef]
- Osei, F.B.; Duker, A.A. Spatial dependency of V. cholera prevalence on open space refuse dumps in Kumasi, Ghana: A spatial statistical modelling. Int. J. Health Geogr. 2008, 7, 1–17. [Google Scholar] [CrossRef]
- Akoachere, J.F.T.K.; Masalla, T.N.; Njom, H.A. Multi-drug resistant toxigenic Vibrio cholerae O1 is persistent in water sources in New Bell-Douala, Cameroon. BMC Infect. Dis. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Rebaudet, S.; Sudre, B.; Faucher, B.; Piarroux, R. Environmental Determinants of Cholera Outbreaks in Inland Africa: A Systematic Review of Main Transmission Foci and Propagation Routes. J. Infect. Dis. 2013, 208, S46–S54. [Google Scholar] [CrossRef] [PubMed]
- Constantin de Magny, G.; Thiaw, W.; Kumar, V.; Manga, N.M.; Diop, B.M.; Gueye, L.; Kamara, M.; Roche, B.; Murtugudde, R.; Colwell, R.R. Cholera Outbreak in Senegal in 2005: Was Climate a Factor? PLoS ONE 2012, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rebaudet, S.; Sudre, B.; Faucher, B.; Piarroux, R. Cholera in Coastal Africa: A Systematic Review of Its Heterogeneous Environmental Determinants. J. Infect. Dis. 2013, 208, S98–S106. [Google Scholar] [CrossRef]
- Almagro-moreno, S.; Taylor, R.K. Cholera: Environmental Reservoirs and Impact on Disease Transmission. Microbiol. Spectr. 2013, 1, 1–12. [Google Scholar]
- Sozzi, E.; Fabre, K.; Fesselet, J.F.; Ebdon, J.E.; Taylor, H. Minimizing the risk of disease transmission in emergency settings: Novel in situ physico-chemical disinfection of pathogen-laden hospital Wastewaters. PLoS Negl. Trop. Dis. 2015, 9, e0003776. [Google Scholar] [CrossRef] [PubMed]
- Ntema, V.M.; Potgieter, N.; Barnard, T.G. Detection of Vibrio cholerae and Vibrio parahaemolyticus by molecular and culture based methods from source water to household container-stored water at the point-of-use in South African rural communities. Water Sci. Technol. 2010, 61, 3091–3101. [Google Scholar] [CrossRef] [PubMed]
- Cazelles, B.; Chavez, M.; De Magny, G.C.; Guégan, J.F.; Hales, S. Time-dependent spectral analysis of epidemiological time-series with wavelets. J. R. Soc. Interface 2007, 4, 625–636. [Google Scholar] [CrossRef] [Green Version]
- Cazelles, B.; Chavez, M.; McMichael, A.J.; Hales, S. Nonstationary influence of El Niño on the synchronous dengue epidemics in Thailand. PLoS Med. 2005, 2, 0313–0318. [Google Scholar] [CrossRef]
- Zell, R.; Krumbholz, A.; Wutzler, P. Impact of global warming on viral diseases: what is the evidence? Curr. Opin. Biotechnol. 2008, 19, 652–660. [Google Scholar] [CrossRef]
- Zell, R. Global climate change and the emergence/re-emergence of infectious diseases. Int. J. Med. Microbiol. Suppl. 2004, 293, 16–26. [Google Scholar] [CrossRef]
- Liu, Z.; Alexander, M. Atmospheric bridge, oceanic tunnel, and global climatic teleconnections. Rev. Geophys. 2007, 45, RG2005. [Google Scholar] [CrossRef]
- Anyamba, A.; Linthicum, K.J.; Small, J.L.; Collins, K.M.; Tucker, C.J.; Pak, E.W.; Britch, S.C.; Eastman, J.R.; Pinzon, J.E.; Russell, K.L. Climate teleconnections and recent patterns of human and animal disease outbreaks. PLoS Negl. Trop. Dis. 2012, 6. [Google Scholar] [CrossRef] [PubMed]
- Sanganyado, E.; Lu, Z.; Fu, Q.; Schlenk, D.; Gan, J. Chiral pharmaceuticals: A review on their environmental occurrence and fate processes. Water Res. 2017, 124, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Vezzulli, L.; Pruzzo, C.; Huq, A.; Colwell, R.R. Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ. Microbiol. Rep. 2010, 2, 27–33. [Google Scholar] [CrossRef]
- Martinelli-Filho, J.E.; Lopes, R.M.; Rivera, I.N.G.; Colwell, R.R. Are natural reservoirs important for cholera surveillance? The case of an outbreak in a Brazilian estuary. Lett. Appl. Microbiol. 2016, 63, 183–188. [Google Scholar] [CrossRef]
- Halpern, M.; Broza, Y.B.; Mittler, S.; Arakawa, E.; Broza, M. Chironomid Egg Masses as a Natural Reservoir of Vibrio cholerae Non-O1 and Non-O139 in Freshwater Habitats. Microb. Ecol. 2004, 47, 341–349. [Google Scholar] [CrossRef]
- Bwire, G.; Ali, M.; Sack, D.A.; Nakinsige, A.; Naigaga, M.; Debes, A.K.; Ngwa, M.C.; Brooks, W.A.; Garimoi Orach, C. Identifying cholera “hotspots” in Uganda: An analysis of cholera surveillance data from 2011 to 2016. PLoS Negl. Trop. Dis. 2017, 11, e0006118. [Google Scholar] [CrossRef]
- Nyenje, P.M.; Foppen, J.W.; Uhlenbrook, S.; Kulabako, R.; Muwanga, A. Eutrophication and nutrient release in urban areas of sub-Saharan Africa—A review. Sci. Total Environ. 2010, 408, 447–455. [Google Scholar] [CrossRef]
- Gwenzi, W.; Chaukura, N. Organic contaminants in African aquatic systems: Current knowledge, health risks, and future research directions. Sci. Total Environ. 2018, 619–620, 1493–1514. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Obi, L.C.; Tom, M.; Okoh, A.I. Detection of potential risk of wastewater effluents for transmission of antibiotic resistance from Vibrio species as a reservoir in a peri-urban community in South Africa. Int. J. Environ. Health Res. 2011, 21, 402–414. [Google Scholar] [CrossRef]
- Gwenzi, W.; Musiyiwa, K.; Mangori, L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: A hotspot reservoir. J. Environ. Chem. Eng. 2018. [Google Scholar] [CrossRef]
- Akoachere, J.F.T.K.; Omam, L.A.; Massalla, T.N. Assessment of the relationship between bacteriological quality of dug-wells, hygiene behaviour and well characteristics in two cholera endemic localities in Douala, Cameroon. BMC Public Health 2013, 13. [Google Scholar] [CrossRef]
- Bwire, G.; Mwesawina, M.; Baluku, Y.; Kanyanda, S.S.E.; Orach, C.G. Cross-border cholera outbreaks in Sub-Saharan Africa, the mystery behind the silent illness: What needs to be done? PLoS ONE 2016, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.; Bouma, M.J.; Dobson, A.P. Cholera and climate: revisiting the quantitative evidence. Microbes Infect. 2002, 4, 237–245. [Google Scholar] [CrossRef]
- Akanda, A.S.; Jutla, A.S.; Islam, S. Dual peak cholera transmission in Bengal Delta: A hydroclimatological explanation. Geophys. Res. Lett. 2009, 36, 1–6. [Google Scholar] [CrossRef]
- Schwartz, B.S.; Harris, J.B.; Khan, A.I.; LaRocque, R.C.; Sack, D.A.; Malek, M.A.; Faruque, A.S.G.; Qadri, F.; Calderwood, S.B.; Luby, S.P.; et al. Diarrheal epidemics in Dhaka, Bangladesh, during three consecutive floods: 1988, 1998, and 2004. Am. J. Trop. Med. Hyg. 2006, 74, 1067–1073. [Google Scholar] [CrossRef]
- Lobitz, B.; Beck, L.; Huq, A.; Wood, B.; Fuchs, G.; Faruque, A.S.G.; Colwell, R. Climate and infectious disease: Use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. USA 2000, 97, 1438–1443. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.J.; Drasar, B.S.; Feachem, R.G. Cholera and Estuarine Salinity in Calcutta and London. Lancet 1982, 319, 1216–1218. [Google Scholar] [CrossRef]
- Sirajul Islam, M.; Brooks, A.; Kabir, M.S.; Jahid, I.K.; Shafiqul Islam, M.; Goswami, D.; Nair, G.B.; Larson, C.; Yukiko, W.; Luby, S. Faecal contamination of drinking water sources of Dhaka city during the 2004 flood in Bangladesh and use of disinfectants for water treatment. J. Appl. Microbiol. 2007, 103, 80–87. [Google Scholar] [CrossRef] [Green Version]
- Bertuzzo, E.; Mari, L.; Righetto, L.; Gatto, M.; Casagrandi, R.; Rodriguez-Iturbe, I.; Rinaldo, A. Hydroclimatology of dual-peak annual cholera incidence: Insights from a spatially explicit model. Geophys. Res. Lett. 2012, 39, 1–6. [Google Scholar] [CrossRef]
- Hashizume, M.; Armstrong, B.; Hajat, S.; Wagatsuma, Y.; Faruque, A.S.G.; Hayashi, T.; Sack, D.A. The effect of rainfall on the incidence of cholera in Bangladesh. Epidemiology 2008, 19, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Boraschi, D.; Abebe Alemayehu, M.; Aseffa, A.; Chiodi, F.; Chisi, J.; Del Prete, G.; Doherty, T.M.; Elhassan, I.; Engers, H.; Gyan, B.; et al. Immunity against HIV/AIDS, Malaria, and Tuberculosis during Co-Infections with Neglected Infectious Diseases: Recommendations for the European Union Research Priorities. PLoS Negl. Trop. Dis. 2008, 2, e255. [Google Scholar] [CrossRef] [PubMed]
- Weill, F.X.; Domman, D.; Njamkepo, E.; Tarr, C.; Rauzier, J.; Fawal, N.; Keddy, K.H.; Salje, H.; Moore, S.; Mukhopadhyay, A.K.; et al. Genomic history of the seventh pandemic of cholera in Africa. Science 2017, 358, 785–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De, R.; Ghosh, J.B.; Sen Gupta, S.; Takeda, Y.; Nair, G.B. The Role of Vibrio cholerae Genotyping in Africa. J. Infect. Dis. 2013, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldor, M.K.; RayChaudhuri, D. Treasure trove for cholera research. Nature 2000, 406, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Dziejman, M.; Balon, E.; Boyd, D.; Fraser, C.M.; Heidelberg, J.F.; Mekalanos, J.J. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 2002, 99, 1556–1561. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.; Grim, C.J.; Hasan, N.A.; Lee, J.H.; Choi, S.Y.; Haley, B.J.; Taviani, E.; Jeon, Y.-S.; Kim, D.W.; Lee, J.-H.; et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2009, 106, 15442–15447. [Google Scholar] [CrossRef] [Green Version]
- Kaboré, S.; Cecchi, P.; Mosser, T.; Toubiana, M.; Traoré, O.; Ouattara, A.S.; Traoré, A.S.; Barro, N.; Colwell, R.R.; Monfort, P. Occurrence of Vibrio cholerae in water reservoirs of Burkina Faso. Res. Microbiol. 2017, 169. [Google Scholar] [CrossRef]
- Jutla, A.; Aldaach, H.; Billian, H.; Akanda, A.; Huq, A.; Colwell, R. Satellite based assessment of hydroclimatic conditions related to cholera in Zimbabwe. PLoS ONE 2015, 10, 1–17. [Google Scholar] [CrossRef]
- Sanganyado, E.; Teta, C.; Masiri, B. Impact of African traditional worldviews on climate change adaptation. Integr. Environ. Assess. Manag. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Patching, H.M.M.; Hudson, L.M.; Cooke, W.; Garcia, A.J.; Hay, S.I.; Roberts, M.; Moyes, C.L. A Supervised Learning Process to Validate Online Disease Reports for Use in Predictive Models. Big Data 2015, 3, 230–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Midani, F.S.; Weil, A.A.; Chowdhury, F.; Begum, Y.A.; Khan, A.I.; Debela, M.D.; Durand, H.K.; Reese, A.T.; Nimmagadda, S.N.; Silverman, J.D.; et al. Human Gut Microbiota Predicts Susceptibility to Vibrio cholerae Infection. J. Infect. Dis. 2018, 218, 645–653. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Thompson, C.C.; Vicente, A.C.P.; Marin, M.A.; Lee, C.; Silva, G.G.Z.; Schmieder, R.; Andrade, B.G.N.; Chimetto, L.; Cuevas, D.; et al. Comparative genomics of 274 Vibrio cholerae genomes reveals mobile functions structuring three niche dimensions. BMC Genomics 2014, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nasr-Azadani, F.; Khan, R.; Rahimikollu, J.; Unnikrishnan, A.; Akanda, A.; Alam, M.; Huq, A.; Jutla, A.; Colwell, R. Hydroclimatic sustainability assessment of changing climate on cholera in the Ganges-Brahmaputra basin. Adv. Water Resour. 2017, 108, 332–344. [Google Scholar] [CrossRef]
- Lee, E.C.; Asher, J.M.; Goldlust, S.; Kraemer, J.D.; Lawson, A.B.; Bansal, S. Mind the scales: Harnessing spatial big data for infectious disease surveillance and inference. J. Infect. Dis. 2016, 214, S409–S413. [Google Scholar] [CrossRef] [PubMed]
- Chunara, R.; Andrews, J.R.; Brownstein, J.S. Social and news media enable estimation of epidemiological patterns early in the 2010 Haitian cholera outbreak. Am. J. Trop. Med. Hyg. 2012, 86, 39–45. [Google Scholar] [CrossRef]
- Traore, B.B.; Tangara, F. Data mining techniques on satellite images for discovery of risk areas. Expert Syst. Appl. 2017, 72, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Semenza, J.C.; Sudre, B.; Oni, T.; Suk, J.E.; Giesecke, J. Linking Environmental Drivers to Infectious Diseases: The European Environment and Epidemiology Network. PLoS Negl. Trop. Dis. 2013, 7, e2323. [Google Scholar] [CrossRef]
| Environmental Reservoirs and Drivers | Development of Cholera Prediction Tools |
|---|---|
| 1. What are the key environmental reservoirs of cholera during the inter-epidemic periods? | 1. Using predictive tools, what are the hotspot areas and hot moments in terms of cholera outbreaks? |
| 2. What anthropogenic and environmental variables drives the population dynamics of Vibrio cholerae and its predators (i.e., vibriophages)? | 2. What anthropogenic and hydroclimatic variables are best predictors of cholera outbreaks? |
| 3. For each epidemic, what were the specific transfer mechanisms of cholera from reservoirs into the human body? | 3. How can existing spatial explicit and other models based on systems analysis tools be further developed into predictive tools |
| 4. What precursor anthropogenic and hydroclimatic events triggered a cholera outbreak? | 4. What environmental variables should be monitored during cholera surveillance to support the development of predictive models? |
| 5. In addition to local anthropogenic and hydroclimatic drivers, what is the role of climatic teleconnections in cholera outbreaks? | |
| 6. How are cholera outbreaks linked to other epidemics such as malaria, tuberculosis, and HIV/AIDS in SSA? | |
| 7. What dominant factors explain the high CFR ratios of cholera in Africa, and can this be attributed to the development of antibiotic resistance? |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gwenzi, W.; Sanganyado, E. Recurrent Cholera Outbreaks in Sub-Saharan Africa: Moving beyond Epidemiology to Understand the Environmental Reservoirs and Drivers. Challenges 2019, 10, 1. https://doi.org/10.3390/challe10010001
Gwenzi W, Sanganyado E. Recurrent Cholera Outbreaks in Sub-Saharan Africa: Moving beyond Epidemiology to Understand the Environmental Reservoirs and Drivers. Challenges. 2019; 10(1):1. https://doi.org/10.3390/challe10010001
Chicago/Turabian StyleGwenzi, Willis, and Edmond Sanganyado. 2019. "Recurrent Cholera Outbreaks in Sub-Saharan Africa: Moving beyond Epidemiology to Understand the Environmental Reservoirs and Drivers" Challenges 10, no. 1: 1. https://doi.org/10.3390/challe10010001






