Phasetime: Deep Learning Approach to Detect Nuclei in Time Lapse Phase Images
Abstract
:1. Introduction
- Establish a workflow combining traditional blob detection method and modern deep learning method to do nuclei instance segmentation which does not require labeled masks during the training time.
- Detect and segment the nuclei based on the phase contrast microscopy images instead of stained channel images.
- Perform the dynamic T-cell fluorescent imaging experiments, train a detection/segmentation model on the real data and quantify the validity of the results.
2. Materials and Methods
2.1. Immune Cell Data Generation
2.1.1. CAR T Cell Manufacturing
2.1.2. Fluorescent Staining of Nucleus
2.1.3. Nanowell Array Fabrication
2.1.4. Timelapse Imaging Microscopy in Nanowell Grids (TIMING)
2.1.5. Ground Truth
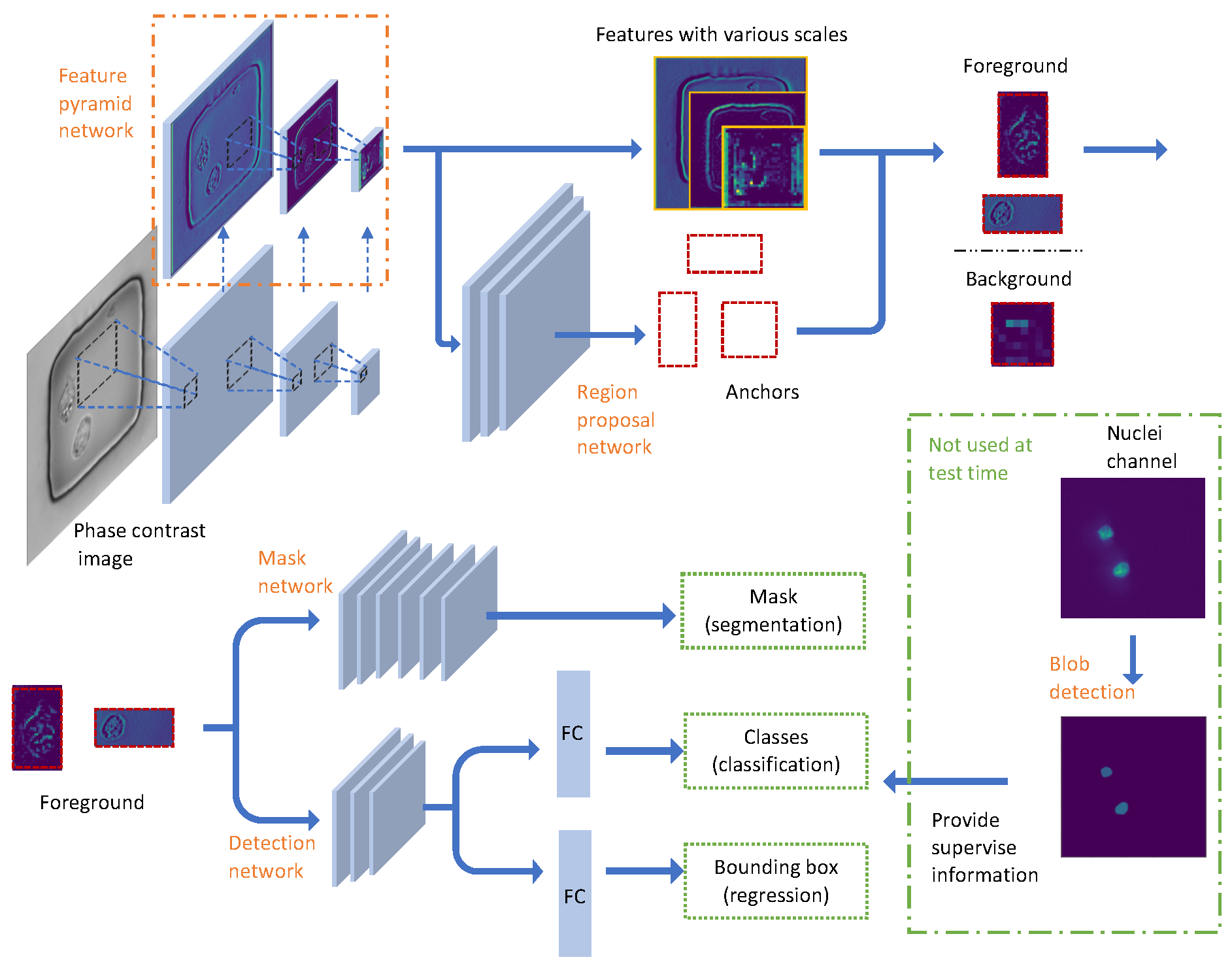
2.2. Image Data Preprocessing and Network Configuration
2.2.1. Binary Mask Generation
2.2.2. Pyramid Feature Extraction
2.2.3. Proposal Generation
2.2.4. Nuclei Detection
2.2.5. Nuclei Segmentation
2.3. Metrics for Performance Evaluation
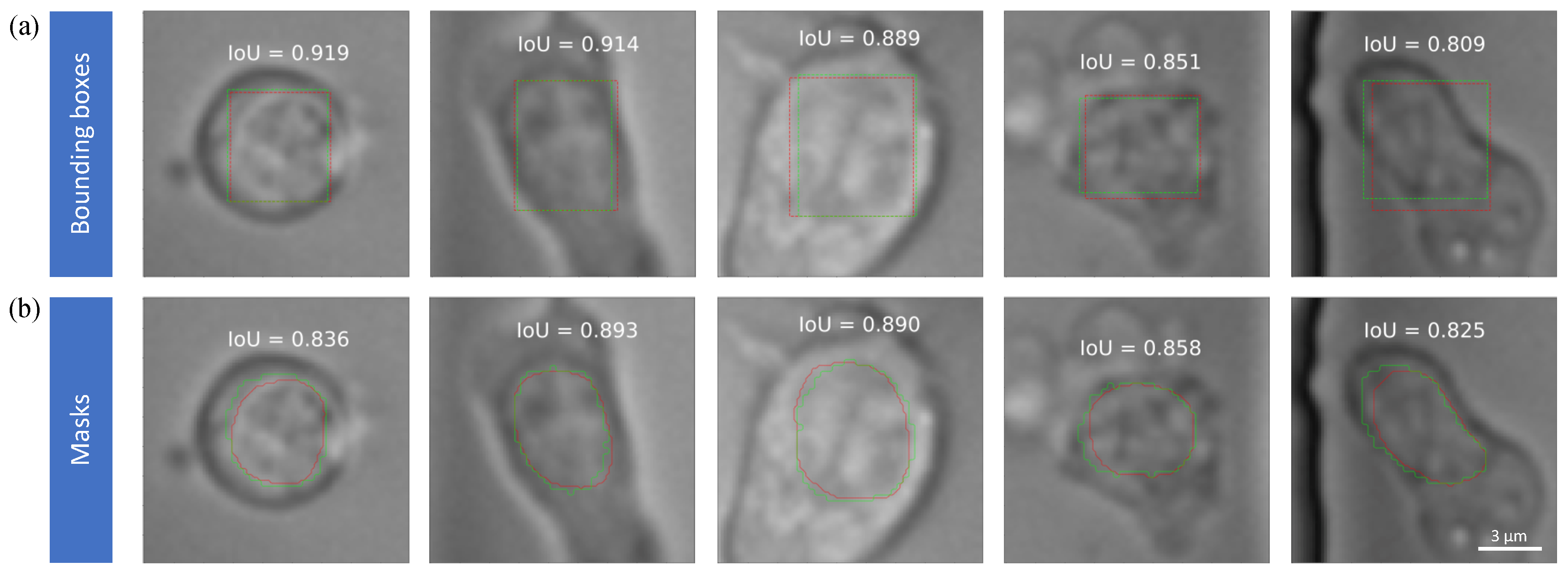
2.3.1. Intersection over Union (IoU)
2.3.2. Average Precision (AP)
- Determine whether an object exists in the image.
- Find the corresponding location if the object exists.
3. Results
3.1. Mask Label Generation Accuracy
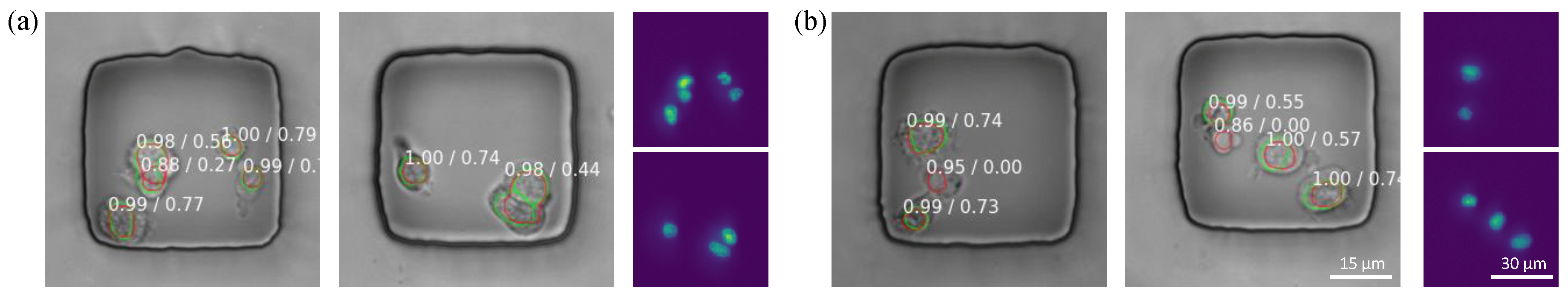
3.2. Nuclei Detection Accuracy
3.3. Nuclei Segmentation Accuracy
3.4. Error Analysis
- There is no clear boundary from the phase contrast image, illustrated by the arrows.
- Predicted masks and ground truth masks are located at the same place, but their size is different. For (b). i, the nucleus is smaller than predicted but for (b). ii, the nucleus is larger and almost overlaps with the cell boundary.
- Both boundaries from Mask RCNN and experts lie on the area where intensity changes, but they are not matched with each other. The predicted boundary may come from other organelles, thus the center may not be aligned such as (c). ii
4. Discussion
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Mask RCNN | Mask Reginal Convolutional Neural Network |
| IoU | Intersection Over Union |
| AP | Average Precision |
| FPN | Feature Pyramid Network |
| RPN | Region Proposal Network |
| FG | Foreground |
| BG | Background |
| FC | Fully-connected |
| TP | True Positive |
| FN | False Negative |
| TIMING | Timelapse Imaging Microscopy in Nanowell Grids |
References
- Rytelewski, M.; Haryutyunan, K.; Nwajei, F.; Shanmugasundaram, M.; Wspanialy, P.; Zal, M.A.; Chen, C.H.; El Khatib, M.; Plunkett, S.; Vinogradov, S.A.; et al. Merger of dynamic two-photon and phosphorescence lifetime microscopy reveals dependence of lymphocyte motility on oxygen in solid and hematological tumors. J. Immunother. Cancer 2019, 7, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liadi, I.; Singh, H.; Romain, G.; Rey-Villamizar, N.; Merouane, A.; Adolacion, J.R.T.; Kebriaei, P.; Huls, H.; Qiu, P.; Roysam, B.; et al. Individual motile CD4+ T cells can participate in efficient multikilling through conjugation to multiple tumor cells. Cancer Immunol. Res. 2015, 3, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Romain, G.; Senyukov, V.; Rey-Villamizar, N.; Merouane, A.; Kelton, W.; Liadi, I.; Mahendra, A.; Charab, W.; Georgiou, G.; Roysam, B.; et al. Antibody Fc engineering improves frequency and promotes kinetic boosting of serial killing mediated by NK cells. Blood 2014, 124, 3241–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koho, S.; Fazeli, E.; Eriksson, J.E.; Hänninen, P.E. Image quality ranking method for microscopy. Sci. Rep. 2016, 6, 28962. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, E.M.; Yang, S.J.; Ando, D.M.; Javaherian, A.; Skibinski, G.; Lipnick, S.; Mount, E.; O’Neil, A.; Shah, K.; Lee, A.K.; et al. In silico labeling: Predicting fluorescent labels in unlabeled images. Cell 2018, 173, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Liadi, I.; Roszik, J.; Romain, G.; Cooper, L.J.; Varadarajan, N. Quantitative high-throughput single-cell cytotoxicity assay for T cells. J. Vis. Exp. 2013, 72, e50058. [Google Scholar] [CrossRef] [PubMed]
- Merouane, A.; Rey-Villamizar, N.; Lu, Y.; Liadi, I.; Romain, G.; Lu, J.; Singh, H.; Cooper, L.J.; Varadarajan, N.; Roysam, B. Automated profiling of individual cell–cell interactions from high-throughput time-lapse imaging microscopy in nanowell grids (TIMING). Bioinformatics 2015, 31, 3189–3197. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 740–755. [Google Scholar]
- Abdulla, W. Mask R-CNN for Object Detection and Instance Segmentation on Keras and TensorFlow. 2017. Available online: https://github.com/matterport/Mask_RCNN (accessed on 23 April 2019).
- Van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Kong, H.; Akakin, H.C.; Sarma, S.E. A generalized Laplacian of Gaussian filter for blob detection and its applications. IEEE Trans. Cybern. 2013, 43, 1719–1733. [Google Scholar] [CrossRef]
- Shafarenko, L.; Petrou, M.; Kittler, J. Automatic watershed segmentation of randomly textured color images. IEEE Trans. Image Process. 1997, 6, 1530–1544. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Neubeck, A.; Van Gool, L. Efficient non-maximum suppression. In Proceedings of the 18th International Conference on Pattern Recognition (ICPR’06), Hong Kong, China, 20–24 August 2006; Volume 3, pp. 850–855. [Google Scholar]
- Beucher, S. Use of watersheds in contour detection. In Proceedings of the International Workshop on Image Processing, Trieste, Italy, 4–8 June 1979. [Google Scholar]
- Wählby, C.; Sintorn, I.M.; Erlandsson, F.; Borgefors, G.; Bengtsson, E. Combining intensity, edge and shape information for 2D and 3D segmentation of cell nuclei in tissue sections. J. Microsc. 2004, 215, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Caicedo, J.C.; Roth, J.; Goodman, A.; Becker, T.; Karhohs, K.W.; Broisin, M.; Csaba, M.; McQuin, C.; Singh, S.; Theis, F.; et al. Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images. BioRxiv 2019, 2019, 335216. [Google Scholar] [CrossRef]
- Kemper, B.; Von Bally, G. Digital holographic microscopy for live cell applications and technical inspection. Appl. Opt. 2008, 47, A52–A61. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K. Principles and techniques of digital holographic microscopy. SPIE Rev. 2010, 1, 018005. [Google Scholar] [CrossRef]
- Lee, K.; Kim, K.; Jung, J.; Heo, J.; Cho, S.; Lee, S.; Chang, G.; Jo, Y.; Park, H.; Park, Y. Quantitative phase imaging techniques for the study of cell pathophysiology: From principles to applications. Sensors 2013, 13, 4170–4191. [Google Scholar] [CrossRef] [PubMed]
- Marquet, P.; Depeursinge, C.; Magistretti, P.J. Review of quantitative phase-digital holographic microscopy: Promising novel imaging technique to resolve neuronal network activity and identify cellular biomarkers of psychiatric disorders. Neurophotonics 2014, 1, 020901. [Google Scholar] [CrossRef]
- Park, H.S.; Rinehart, M.T.; Walzer, K.A.; Chi, J.T.A.; Wax, A. Automated detection of P. falciparum using machine learning algorithms with quantitative phase images of unstained cells. PLoS ONE 2016, 11, e0163045. [Google Scholar] [CrossRef]
- Pavillon, N.; Hobro, A.J.; Akira, S.; Smith, N.I. Noninvasive detection of macrophage activation with single-cell resolution through machine learning. Proc. Natl. Acad. Sci. USA 2018, 115, E2676–E2685. [Google Scholar] [CrossRef]
- Mobiny, A.; Lu, H.; Nguyen, H.V.; Roysam, B.; Varadarajan, N. Automated Classification of Apoptosis in Phase Contrast Microscopy Using Capsule Network. IEEE Trans. Med Imaging 2019. [Google Scholar] [CrossRef]
- Anderson, J.K.; Mehta, A. A review of chimeric antigen receptor T-cells in lymphoma. Expert Rev. Hematol. 2019, 12, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Labanieh, L.; Majzner, R.G.; Mackall, C.L. Programming CAR-T cells to kill cancer. Nat. Biomed. Eng. 2018, 2, 377. [Google Scholar] [CrossRef] [PubMed]
- Zabel, M.; Tauber, P.A.; Pickl, W.F. The making and function of CAR cells. Immunol. Lett. 2019, 212, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Weinkove, R.; George, P.; Dasyam, N.; McLellan, A.D. Selecting costimulatory domains for chimeric antigen receptors: functional and clinical considerations. Clin. Transl. Immunol. 2019, 8, e1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahmasebi, S.; Elahi, R.; Esmaeilzadeh, A. Solid Tumors Challenges and New Insights of CAR T Cell Engineering. Stem Cell Rev. Rep. 2019, 2019, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Li, J.; Martinez-Paniagua, M.A.; Bandey, I.N.; Amritkar, A.; Singh, H.; Mayerich, D.; Varadarajan, N.; Roysam, B. TIMING 2.0: High-throughput single-cell profiling of dynamic cell–cell interactions by time-lapse imaging microscopy in nanowell grids. Bioinformatics 2018, 35, 706–708. [Google Scholar] [CrossRef] [PubMed]





| Otsu | E0 | E4 | E6 | ||
|---|---|---|---|---|---|
| Mask generation | IoU | 0.653 | 0.538 | 0.842 | 0.738 |
| Instance segmentation | No | Yes | Yes | Yes |
| Based on | AP_30 | AP_40 | AP_50 | AP_60 | AP_70 | |
|---|---|---|---|---|---|---|
| Mask RCNN | Bounding boxes | 0.930 | 0.906 | 0.821 | 0.668 | 0.365 |
| Mask RCNN | Masks | 0.922 | 0.893 | 0.793 | 0.568 | 0.247 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, P.; Rezvan, A.; Li, X.; Varadarajan, N.; Van Nguyen, H. Phasetime: Deep Learning Approach to Detect Nuclei in Time Lapse Phase Images. J. Clin. Med. 2019, 8, 1159. https://doi.org/10.3390/jcm8081159
Yuan P, Rezvan A, Li X, Varadarajan N, Van Nguyen H. Phasetime: Deep Learning Approach to Detect Nuclei in Time Lapse Phase Images. Journal of Clinical Medicine. 2019; 8(8):1159. https://doi.org/10.3390/jcm8081159
Chicago/Turabian StyleYuan, Pengyu, Ali Rezvan, Xiaoyang Li, Navin Varadarajan, and Hien Van Nguyen. 2019. "Phasetime: Deep Learning Approach to Detect Nuclei in Time Lapse Phase Images" Journal of Clinical Medicine 8, no. 8: 1159. https://doi.org/10.3390/jcm8081159





