Glyoxalase 2: Towards a Broader View of the Second Player of the Glyoxalase System
Abstract
:1. Introduction
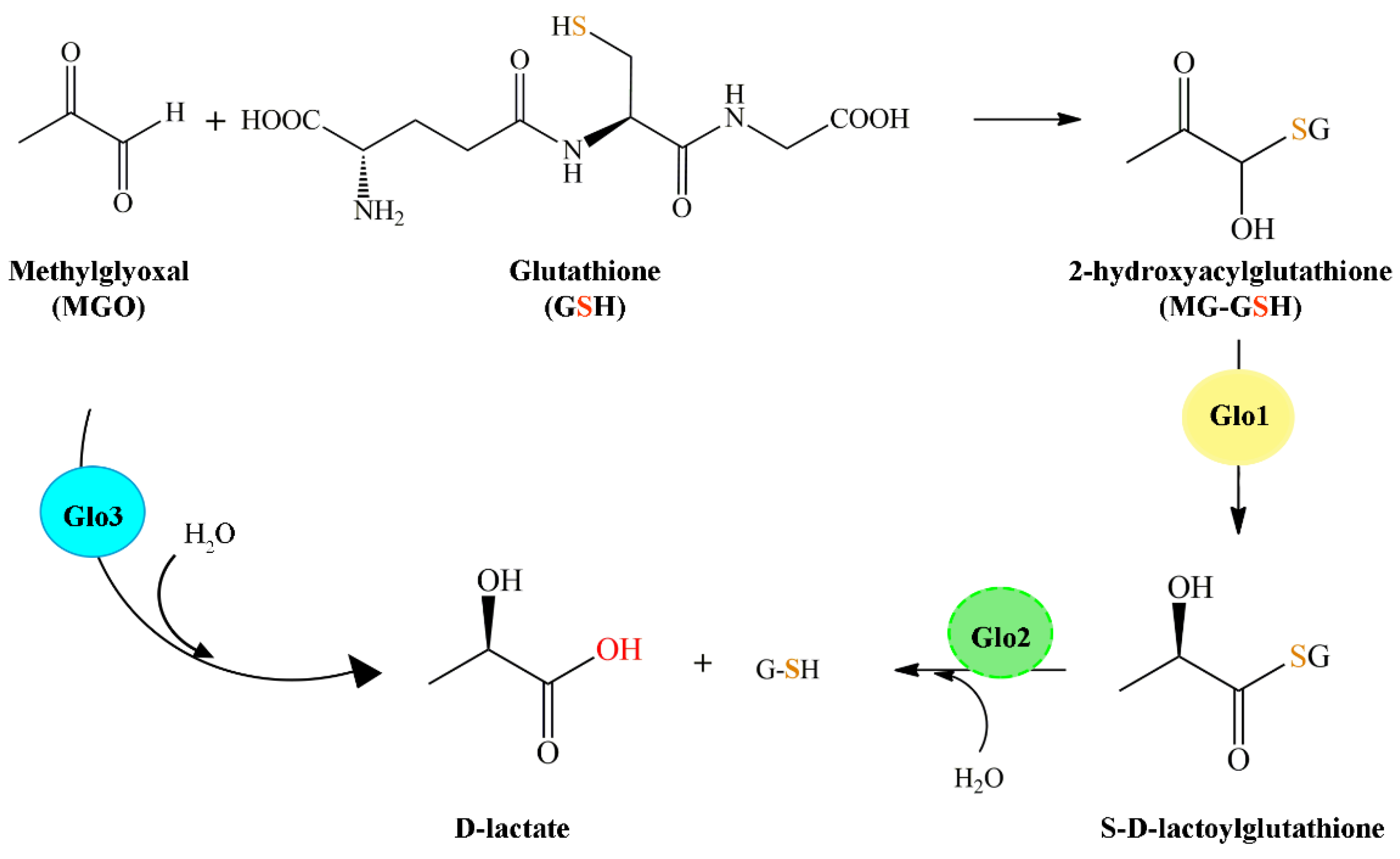
2. The Glyoxalase System
3. Genetics and Molecular Properties of Glyoxalases 2
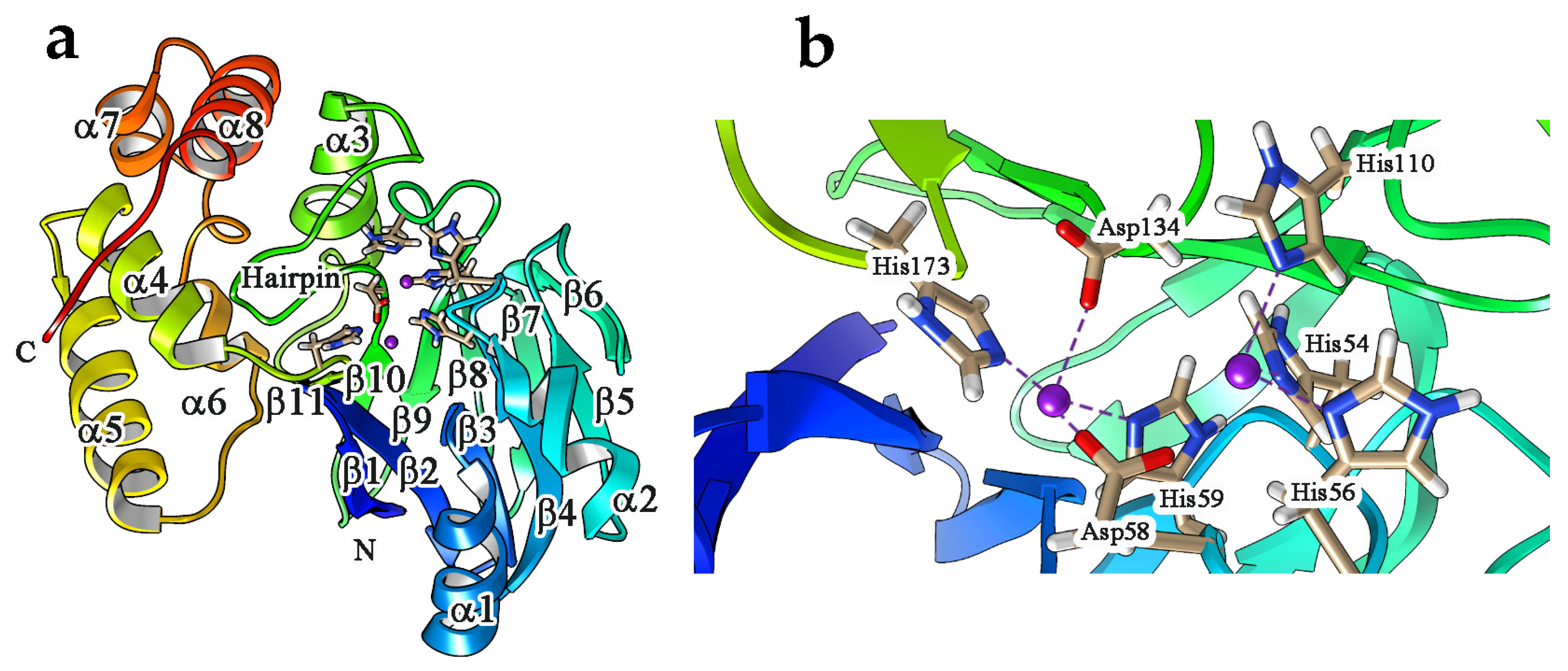
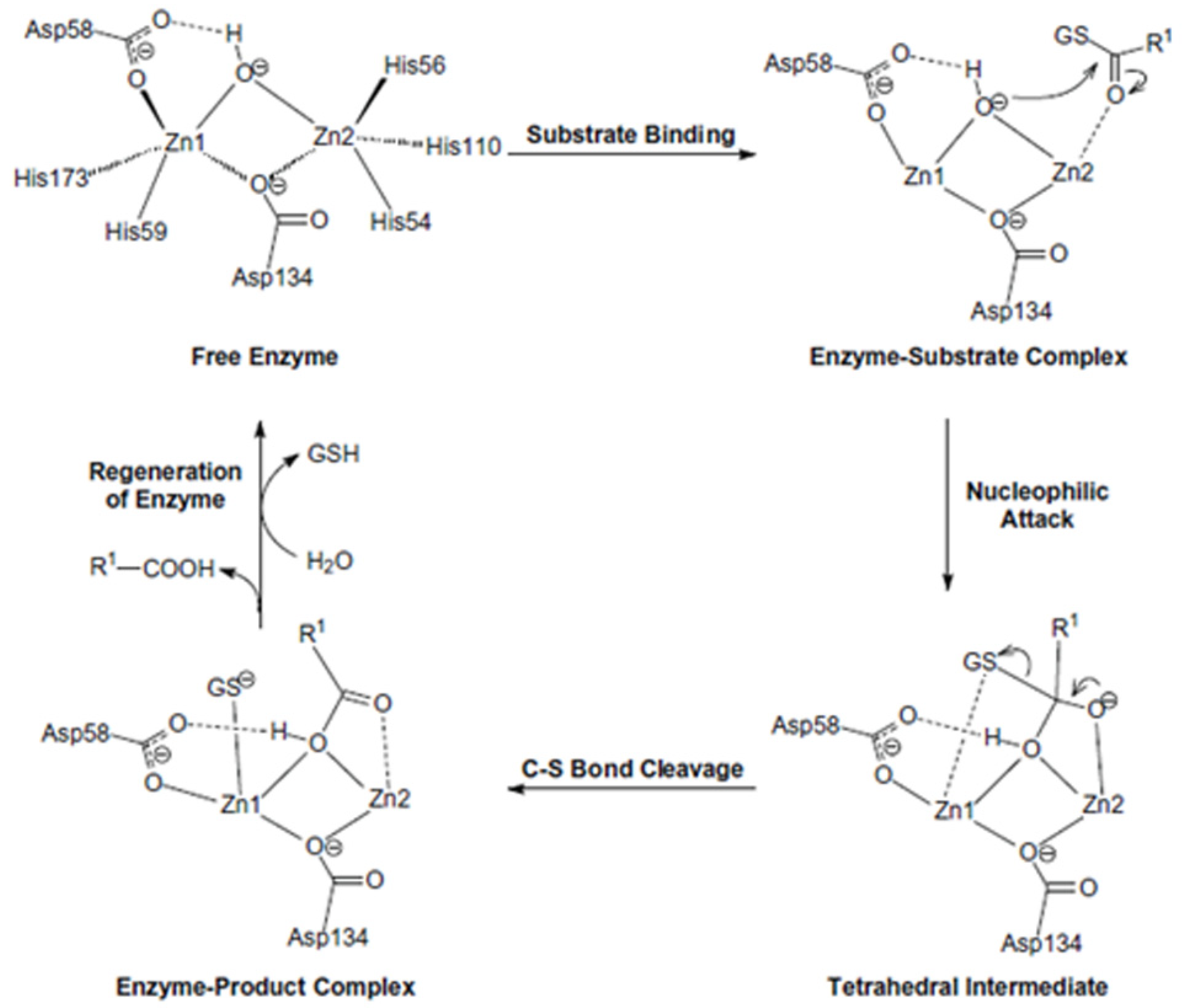
4. Enzyme Activity, Structural Features and Catalytic Mechanism
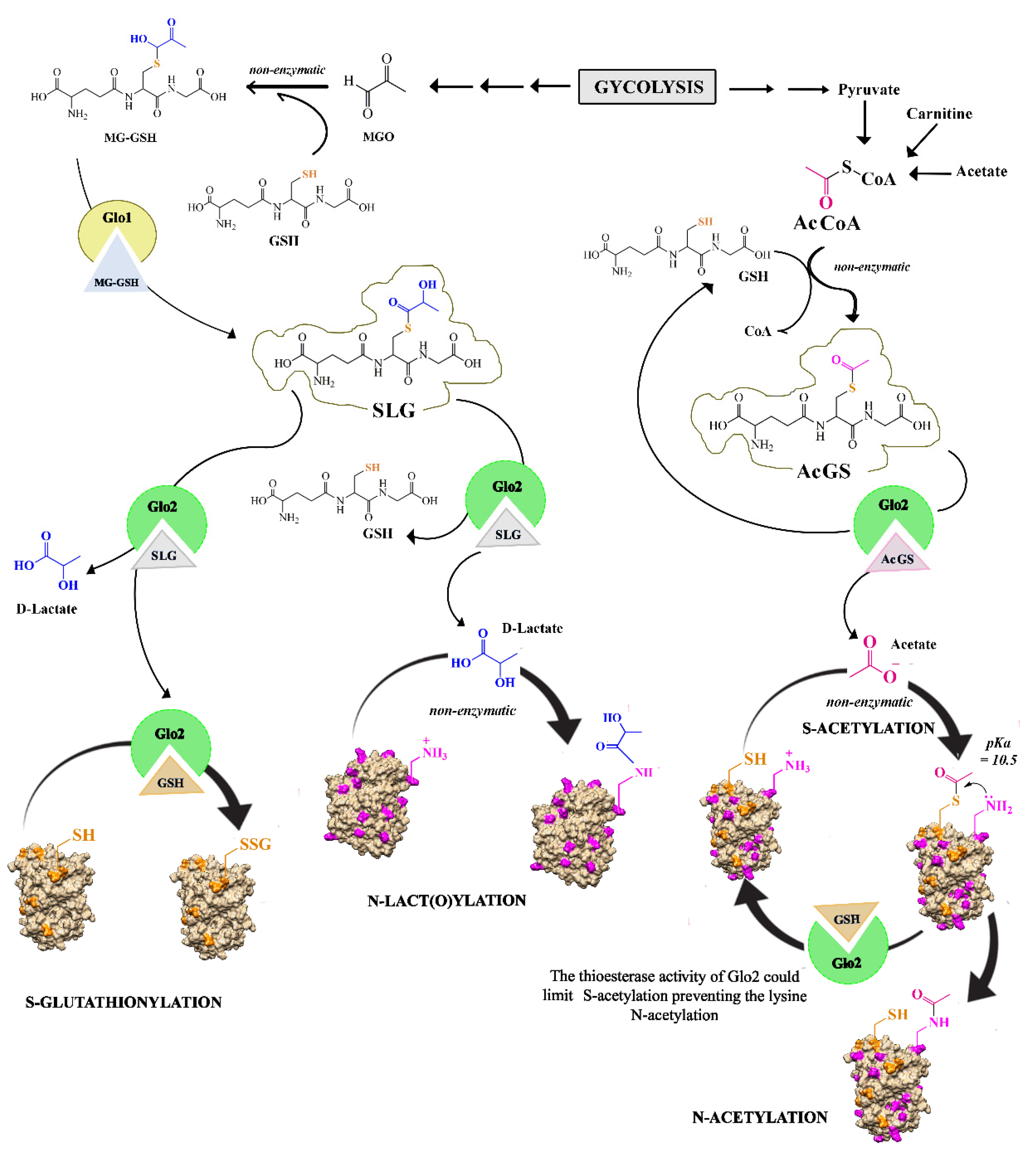
5. Glo2 Role in Post-Translational Modifications
6. Glo2 Metabolic Interactions
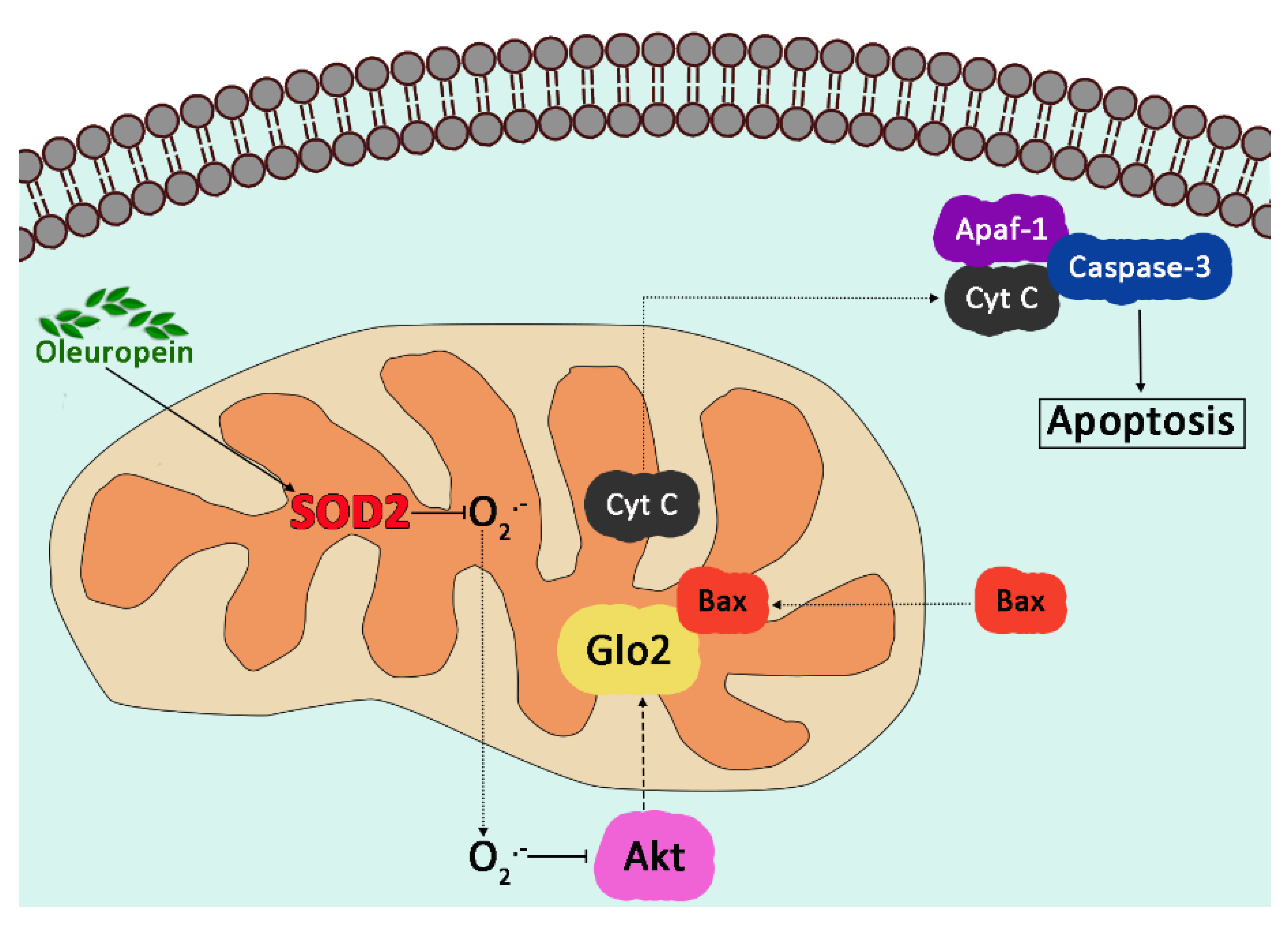
6.1. Glyoxalase 2 and Signaling Pathways
6.2. Glyoxalase 2 and Microtubules Interaction
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dakin, H.D.; Dudley, H.W. An enzyme concerned with the formation of hydroxy acids from ketonic aldehydes. J. Biol. Chem. 1913, 14, 155–157. [Google Scholar] [CrossRef]
- Neuberg, C. The destruction of lactic aldehyde and methylglyoxal by animal organs. Biochem. Z. 1913, 49, 502–506. [Google Scholar]
- Racker, E. The mechanism of action of glyoxalase. J. Biol. Chem. 1951, 190, 685–696. [Google Scholar] [CrossRef]
- Racker, E. Glutathione. In Glutathione as A Coenzyme in Intermediary Metabolism; Colowick, S., Lazarow, A., Racker, E., Schwarz, D.R., Stadtman, E., Waelsch, H., Eds.; Academic Press: Cambridge, MA, USA, 1954; pp. 165–183. [Google Scholar]
- Ekwall, K.; Mannervik, B. The stereochemical configuration of the lactoyl group of S-lactoylglutathionine formed by the action of glyoxalase I from porcine erythrocytes and yeast. Biochim. Biophys. Acta 1973, 297, 297–299. [Google Scholar] [CrossRef]
- Honek, J.F. Glyoxalase biochemistry. Biomol. Concepts 2015, 6, 401–414. [Google Scholar] [CrossRef]
- Xue, M.; Rabbani, N.; Thornalley, P.J. Glyoxalase in ageing. Semin. Cell Dev. Biol. 2011, 22, 293–301. [Google Scholar] [CrossRef]
- Bacchetti, T.; Masciangelo, S.; Armeni, T.; Bicchiega, V.; Ferretti, G. Glycation of human high density lipoprotein by methylglyoxal: Effect on HDL-paraoxonase activity. Metabolism 2014, 63, 307–311. [Google Scholar] [CrossRef]
- Morresi, C.; Cianfruglia, L.; Sartini, D.; Cecati, M.; Fumarola, S.; Emanuelli, M.; Armeni, T.; Ferretti, G.; Bacchetti, T. Effect of High Glucose-Induced Oxidative Stress on Paraoxonase 2 Expression and Activity in Caco-2 Cells. Cells 2019, 8, 1616. [Google Scholar] [CrossRef] [Green Version]
- Rabbani, N.; Xue, M.; Thornalley, P.J. Dicarbonyl stress, protein glycation and the unfolded protein response. Glycoconj. J. 2021, 38, 331–340. [Google Scholar] [CrossRef]
- Maessen, D.E.; Stehouwer, C.D.; Schalkwijk, C.G. The role of methylglyoxal and the glyoxalase system in diabetes and other age-related diseases. Clin. Sci. 2015, 128, 839–861. [Google Scholar] [CrossRef]
- Thornalley, P.J. Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems—Role in ageing and disease. Drug Metab. Drug Interact 2008, 23, 125–150. [Google Scholar] [CrossRef]
- de Bari, L.; Scire, A.; Minnelli, C.; Cianfruglia, L.; Kalapos, M.P.; Armeni, T. Interplay among Oxidative Stress, Methylglyoxal Pathway and S-Glutathionylation. Antioxidants 2020, 10, 19. [Google Scholar] [CrossRef]
- Lee, J.H.; Parveen, A.; Do, M.H.; Kang, M.C.; Yumnam, S.; Kim, S.Y. Molecular mechanisms of methylglyoxal-induced aortic endothelial dysfunction in human vascular endothelial cells. Cell Death Dis. 2020, 11, 403. [Google Scholar] [CrossRef]
- Bito, A.; Haider, M.; Briza, P.; Strasser, P.; Breitenbach, M. Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase II enzymes, Glo2p and Glo4p, from Saccharomyces cerevisiae. Protein Expr. Purif. 1999, 17, 456–464. [Google Scholar] [CrossRef]
- Deponte, M. Glyoxalase diversity in parasitic protists. Biochem. Soc. Trans. 2014, 42, 473–478. [Google Scholar] [CrossRef]
- Talesa, V.; Rosi, G.; Contenti, S.; Mangiabene, C.; Lupattelli, M.; Norton, S.J.; Giovannini, E.; Principato, G.B. Presence of glyoxalase II in mitochondria from spinach leaves: Comparison with the enzyme from cytosol. Biochem. Int. 1990, 22, 1115–1120. [Google Scholar]
- Hasanuzzaman, M.; Nahar, K.; Hossain, M.S.; Mahmud, J.A.; Rahman, A.; Inafuku, M.; Oku, H.; Fujita, M. Coordinated Actions of Glyoxalase and Antioxidant Defense Systems in Conferring Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2017, 18, 200. [Google Scholar] [CrossRef] [Green Version]
- D’Silva, C.; Daunes, S. Structure-activity study on the in vitro antiprotozoal activity of glutathione derivatives. J. Med. Chem. 2000, 43, 2072–2078. [Google Scholar] [CrossRef]
- Thornalley, P.J.; Strath, M.; Wilson, R.J. Antimalarial activity in vitro of the glyoxalase I inhibitor diester, S-p-bromobenzylglutathione diethyl ester. Biochem. Pharm. 1994, 47, 418–420. [Google Scholar] [CrossRef]
- Cordell, P.A.; Futers, T.S.; Grant, P.J.; Pease, R.J. The Human hydroxyacylglutathione hydrolase (HAGH) gene encodes both cytosolic and mitochondrial forms of glyoxalase II. J. Biol. Chem. 2004, 279, 28653–28661. [Google Scholar] [CrossRef] [Green Version]
- Schilling, O.; Wenzel, N.; Naylor, M.; Vogel, A.; Crowder, M.; Makaroff, C.; Meyer-Klaucke, W. Flexible metal binding of the metallo-beta-lactamase domain: Glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry 2003, 42, 11777–11786. [Google Scholar] [CrossRef] [PubMed]
- Scire, A.; Cianfruglia, L.; Minnelli, C.; Bartolini, D.; Torquato, P.; Principato, G.; Galli, F.; Armeni, T. Glutathione compartmentalization and its role in glutathionylation and other regulatory processes of cellular pathways. Biofactors 2019, 45, 152–168. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Uys, J.D.; Tew, K.D.; Townsend, D.M. S-glutathionylation: From molecular mechanisms to health outcomes. Antioxid. Redox Signal. 2011, 15, 233–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ercolani, L.; Scire, A.; Galeazzi, R.; Massaccesi, L.; Cianfruglia, L.; Amici, A.; Piva, F.; Urbanelli, L.; Emiliani, C.; Principato, G.; et al. A possible S-glutathionylation of specific proteins by glyoxalase II: An in vitro and in silico study. Cell Biochem. Funct. 2016, 34, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Webster, B.R.; Li, J.H.; Sack, M.N. Identification of a molecular component of the mitochondrial acetyltransferase programme: A novel role for GCN5L1. Biochem. J. 2012, 443, 655–661. [Google Scholar] [CrossRef] [Green Version]
- Wagner, G.R.; Payne, R.M. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef] [Green Version]
- James, A.M.; Hoogewijs, K.; Logan, A.; Hall, A.R.; Ding, S.; Fearnley, I.M.; Murphy, M.P. Non-enzymatic N-acetylation of Lysine Residues by AcetylCoA Often Occurs via a Proximal S-acetylated Thiol Intermediate Sensitive to Glyoxalase II. Cell Rep. 2017, 18, 2105–2112. [Google Scholar] [CrossRef] [Green Version]
- Gaffney, D.O.; Jennings, E.Q.; Anderson, C.C.; Marentette, J.O.; Shi, T.; Schou Oxvig, A.M.; Streeter, M.D.; Johannsen, M.; Spiegel, D.A.; Chapman, E.; et al. Non-enzymatic Lysine Lactoylation of Glycolytic Enzymes. Cell Chem. Biol. 2020, 27, 206–213.e206. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, X. Glyoxalase II, a detoxifying enzyme of glycolysis byproduct methylglyoxal and a target of p63 and p73, is a pro-survival factor of the p53 family. J. Biol. Chem. 2006, 281, 26702–26713. [Google Scholar] [CrossRef] [Green Version]
- Talesa, V.N.; Ferri, I.; Bellezza, G.; Love, H.D.; Sidoni, A.; Antognelli, C. Glyoxalase 2 Is Involved in Human Prostate Cancer Progression as Part of a Mechanism Driven By PTEN/PI3K/AKT/mTOR Signaling With Involvement of PKM2 and ERalpha. Prostate 2017, 77, 196–210. [Google Scholar] [CrossRef]
- Carrington, S.J.; Douglas, K.T. The glyoxalase enigma—The biological consequences of a ubiquitous enzyme. IRCS Med. Sci. 1986, 14, 763–768. [Google Scholar]
- Thornalley, P.J. The glyoxalase system: New developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 1990, 269, 1–11. [Google Scholar] [CrossRef]
- Rabbani, N.; Xue, M.; Thornalley, P.J. Activity, regulation, copy number and function in the glyoxalase system. Biochem. Soc. Trans. 2014, 42, 419–424. [Google Scholar] [CrossRef]
- Rabbani, N.; Thornalley, P.J. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem. Biophys. Res. Commun. 2015, 458, 221–226. [Google Scholar] [CrossRef] [Green Version]
- Lyles, G.A.; Chalmers, J. The metabolism of aminoacetone to methylglyoxal by semicarbazide-sensitive amine oxidase in human umbilical artery. Biochem. Pharmacol. 1992, 43, 1409–1414. [Google Scholar] [CrossRef]
- Richard, J.P. Mechanism for the formation of methylglyoxal from triosephosphates. Biochem. Soc. Trans. 1993, 21, 549–553. [Google Scholar] [CrossRef]
- Cianfruglia, L.; Morresi, C.; Bacchetti, T.; Armeni, T.; Ferretti, G. Protection of Polyphenols against Glyco-Oxidative Stress: Involvement of Glyoxalase Pathway. Antioxidants 2020, 9, 1006. [Google Scholar] [CrossRef]
- de Bari, L.; Atlante, A.; Armeni, T.; Kalapos, M.P. Synthesis and metabolism of methylglyoxal, S-D-lactoylglutathione and D-lactate in cancer and Alzheimer’s disease. Exploring the crossroad of eternal youth and premature aging. Age. Res. Rev. 2019, 53, 100915. [Google Scholar] [CrossRef]
- Ghosh, A.; Kushwaha, H.R.; Hasan, M.R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 2016, 6, 18358. [Google Scholar] [CrossRef] [Green Version]
- Kwon, K.; Choi, D.; Hyun, J.K.; Jung, H.S.; Baek, K.; Park, C. Novel glyoxalases from Arabidopsis thaliana. FEBS J. 2013, 280, 3328–3339. [Google Scholar] [CrossRef]
- Lee, J.Y.; Song, J.; Kwon, K.; Jang, S.; Kim, C.; Baek, K.; Kim, J.; Park, C. Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 2012, 21, 3215–3225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Jagt, D.L. Glyoxalase II: Molecular characteristics, kinetics and mechanism. Biochem. Soc. Trans. 1993, 21, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.D.; Ridderstrom, M.; Olin, B.; Mannervik, B. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure 1999, 7, 1067–1078. [Google Scholar] [CrossRef]
- O’Young, J.; Sukdeo, N.; Honek, J.F. Escherichia coli glyoxalase II is a binuclear zinc-dependent metalloenzyme. Arch. Biochem. Biophys. 2007, 459, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.S.; Barata, L.; Ferreira, A.E.; Romao, S.; Tomas, A.M.; Freire, A.P.; Cordeiro, C. Catalysis and structural properties of Leishmania infantum glyoxalase II: Trypanothione specificity and phylogeny. Biochemistry 2008, 47, 195–204. [Google Scholar] [CrossRef]
- Ghosh, A.; Islam, T. Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 2016, 16, 87. [Google Scholar] [CrossRef] [Green Version]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef]
- Crowder, M.W.; Maiti, M.K.; Banovic, L.; Makaroff, C.A. Glyoxalase II from A. thaliana requires Zn(II) for catalytic activity. FEBS Lett. 1997, 418, 351–354. [Google Scholar] [CrossRef] [Green Version]
- Marasinghe, G.P.; Sander, I.M.; Bennett, B.; Periyannan, G.; Yang, K.W.; Makaroff, C.A.; Crowder, M.W. Structural studies on a mitochondrial glyoxalase II. J. Biol. Chem. 2005, 280, 40668–40675. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.K.; Singla-Pareek, S.L.; Kumar, M.; Pareek, A.; Saxena, M.; Sarin, N.B.; Sopory, S.K. Characterization and functional validation of glyoxalase II from rice. Protein Expr. Purif. 2007, 51, 126–132. [Google Scholar] [CrossRef]
- Devanathan, S.; Erban, A.; Perez-Torres, R., Jr.; Kopka, J.; Makaroff, C.A. Arabidopsis thaliana glyoxalase 2-1 is required during abiotic stress but is not essential under normal plant growth. PLoS ONE 2014, 9, e95971. [Google Scholar] [CrossRef] [Green Version]
- Holdorf, M.M.; Owen, H.A.; Lieber, S.R.; Yuan, L.; Adams, N.; Dabney-Smith, C.; Makaroff, C.A. Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol. 2012, 160, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Bray, E.A.; Bailey-Serres, J.; Weretilnyk, E. Responses to abiotic stresses. In Biochemistry and Molecular Biology of Plants; Gruissem, W., Buchannan, B., Jones, R., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; pp. 1158–1249. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Hoque, T.S.; Hossain, M.A.; Mostofa, M.G.; Burritt, D.J.; Fujita, M.; Tran, L.S. Methylglyoxal: An Emerging Signaling Molecule in Plant Abiotic Stress Responses and Tolerance. Front. Plant Sci. 2016, 7, 1341. [Google Scholar] [CrossRef] [Green Version]
- Hoque, T.S.; Uraji, M.; Ye, W.; Hossain, M.A.; Nakamura, Y.; Murata, Y. Methylglyoxal-induced stomatal closure accompanied by peroxidase-mediated ROS production in Arabidopsis. J. Plant Physiol. 2012, 169, 979–986. [Google Scholar] [CrossRef]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and Methylglyoxal as Biomarkers for Plant Stress Tolerance. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Hossain, M.A.; Hossain, M.Z.; Fujita, M. Stress-induced changes of methylglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. Aust. J. Crop Sci. 2009, 3, 53–64. [Google Scholar]
- Hossain, M.A.; Mostofa, M.G.; Fujita, M. Heat-shock positively modulates oxidative protection of salt and drought-stressed mustard (Brassica campestris L.) seedlings. J. Plant Sci. Mol. Breed. 2013, 2, 2. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Alam, M.M.; Rahman, A.; Hasanuzzaman, M.; Nahar, K.; Fujita, M. Exogenous proline and glycine betaine mediated upregulation of antioxidant defense and glyoxalase systems provides better protection against salt-induced oxidative stress in two rice (Oryza sativa L.) varieties. BioMed. Res. Int. 2014, 2014, 757219. [Google Scholar] [CrossRef] [Green Version]
- Hasanuzzaman, M.; Hossain, M.A.; Fujita, M. Selenium-induced up-regulation of the antioxidant defense and methylglyoxal detoxification system reduces salinity-induced damage in rapeseed seedlings. Biol. Trace Elem. Res. 2011, 143, 1704–1721. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Pan, C.; Du, Y.; Li, D.; Liu, W. Exogenous salicylic acid regulates reactive oxygen species metabolism and ascorbate-glutathione cycle in Nitraria tangutorum Bobr. under salinity stress. Physiol. Mol. Biol. Plants 2018, 24, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Hossain, M.S.; Mahmud, J.A.; Nahar, K.; Hasanuzzaman, M.; Fujita, M. Manganese-induced salt stress tolerance in rice seedlings: Regulation of ion homeostasis, antioxidant defense and glyoxalase systems. Physiol. Mol. Biol. Plants 2016, 22, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Nahar, K.; Hasanuzzaman, M.; Fujita, M. Calcium Supplementation Improves Na(+)/K(+) Ratio, Antioxidant Defense and Glyoxalase Systems in Salt-Stressed Rice Seedlings. Front. Plant Sci. 2016, 7, 609. [Google Scholar] [CrossRef]
- Saxena, M.; Bisht, R.; Roy, S.D.; Sopory, S.K.; Bhalla-Sarin, N. Cloning and characterization of a mitochondrial glyoxalase II from Brassica juncea that is upregulated by NaCl, Zn, and ABA. Biochem. Biophys. Res. Commun. 2005, 336, 813–819. [Google Scholar] [CrossRef]
- Ekman, D.R.; Lorenz, W.W.; Przybyla, A.E.; Wolfe, N.L.; Dean, J.F. SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol. 2003, 133, 1397–1406. [Google Scholar] [CrossRef] [Green Version]
- Bito, A.; Haider, M.; Hadler, I.; Breitenbach, M. Identification and phenotypic analysis of two glyoxalase II encoding genes from Saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J. Biol. Chem. 1997, 272, 21509–21519. [Google Scholar] [CrossRef] [Green Version]
- Urscher, M.; Przyborski, J.M.; Imoto, M.; Deponte, M. Distinct subcellular localization in the cytosol and apicoplast, unexpected dimerization and inhibition of Plasmodium falciparum glyoxalases. Mol. Microbiol. 2010, 76, 92–103. [Google Scholar] [CrossRef]
- Akoachere, M.; Iozef, R.; Rahlfs, S.; Deponte, M.; Mannervik, B.; Creighton, D.J.; Schirmer, H.; Becker, K. Characterization of the glyoxalases of the malarial parasite Plasmodium falciparum and comparison with their human counterparts. Biol. Chem. 2005, 386, 41–52. [Google Scholar] [CrossRef]
- Vander Jagt, D.L.; Hunsaker, L.A.; Campos, N.M.; Baack, B.R. D-lactate production in erythrocytes infected with Plasmodium falciparum. Mol. Biochem. Parasitol. 1990, 42, 277–284. [Google Scholar] [CrossRef]
- Wendler, A.; Irsch, T.; Rabbani, N.; Thornalley, P.J.; Krauth-Siegel, R.L. Glyoxalase II does not support methylglyoxal detoxification but serves as a general trypanothione thioesterase in African trypanosomes. Mol. Biochem. Parasitol. 2009, 163, 19–27. [Google Scholar] [CrossRef]
- Florens, L.; Washburn, M.P.; Raine, J.D.; Anthony, R.M.; Grainger, M.; Haynes, J.D.; Moch, J.K.; Muster, N.; Sacci, J.B.; Tabb, D.L.; et al. A proteomic view of the Plasmodium falciparum life cycle. Nature 2002, 419, 520–526. [Google Scholar] [CrossRef]
- Le Roch, K.G.; Johnson, J.R.; Florens, L.; Zhou, Y.; Santrosyan, A.; Grainger, M.; Yan, S.F.; Williamson, K.C.; Holder, A.A.; Carucci, D.J.; et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004, 14, 2308–2318. [Google Scholar] [CrossRef] [Green Version]
- Sherman, I.W. Biochemistry of Plasmodium (Malarial parasites). Microbiol. Rev. 1979, 43, 453–495. [Google Scholar] [CrossRef]
- Kappe, S.H.; Vaughan, A.M.; Boddey, J.A.; Cowman, A.F. That was then but this is now: Malaria research in the time of an eradication agenda. Science 2010, 328, 862–866. [Google Scholar] [CrossRef]
- Ariza, A.; Vickers, T.J.; Greig, N.; Armour, K.A.; Dixon, M.J.; Eggleston, I.M.; Fairlamb, A.H.; Bond, C.S. Specificity of the trypanothione-dependent Leishmania major glyoxalase I: Structure and biochemical comparison with the human enzyme. Mol. Microbiol. 2006, 59, 1239–1248. [Google Scholar] [CrossRef]
- Irsch, T.; Krauth-Siegel, R.L. Glyoxalase II of African trypanosomes is trypanothione-dependent. J. Biol. Chem. 2004, 279, 22209–22217. [Google Scholar] [CrossRef] [Green Version]
- Sousa Silva, M.; Ferreira, A.E.; Tomas, A.M.; Cordeiro, C.; Ponces Freire, A. Quantitative assessment of the glyoxalase pathway in Leishmania infantum as a therapeutic target by modelling and computer simulation. FEBS J. 2005, 272, 2388–2398. [Google Scholar] [CrossRef]
- Fairlamb, A.H.; Cerami, A. Metabolism and functions of trypanothione in the Kinetoplastida. Annu. Rev. Microbiol. 1992, 46, 695–729. [Google Scholar] [CrossRef]
- Ariyanayagam, M.R.; Fairlamb, A.H. Ovothiol and trypanothione as antioxidants in trypanosomatids. Mol. Biochem. Parasitol. 2001, 115, 189–198. [Google Scholar] [CrossRef]
- Krauth-Siegel, R.L.; Comini, M.A. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim. Biophys. Acta 2008, 1780, 1236–1248. [Google Scholar] [CrossRef] [PubMed]
- Mandal, G.; Wyllie, S.; Singh, N.; Sundar, S.; Fairlamb, A.H.; Chatterjee, M. Increased levels of thiols protect antimony unresponsive Leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology 2007, 134, 1679–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilari, A.; Fiorillo, A.; Genovese, I.; Colotti, G. Polyamine-trypanothione pathway: An update. Future Med. Chem. 2017, 9, 61–77. [Google Scholar] [CrossRef]
- Barata, L.; Sousa Silva, M.; Schuldt, L.; Ferreira, A.E.; Gomes, R.A.; Tomas, A.M.; Weiss, M.S.; Ponces Freire, A.; Cordeiro, C. Enlightening the molecular basis of trypanothione specificity in trypanosomatids: Mutagenesis of Leishmania infantum glyoxalase II. Exp. Parasitol. 2011, 129, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chandrangsu, P.; Loi, V.V.; Antelmann, H.; Helmann, J.D. The Role of Bacillithiol in Gram-Positive Firmicutes. Antioxid. Redox Signal. 2018, 28, 445–462. [Google Scholar] [CrossRef] [Green Version]
- Newton, G.L.; Rawat, M.; La Clair, J.J.; Jothivasan, V.K.; Budiarto, T.; Hamilton, C.J.; Claiborne, A.; Helmann, J.D.; Fahey, R.C. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 2009, 5, 625–627. [Google Scholar] [CrossRef]
- Perera, V.R.; Newton, G.L.; Pogliano, K. Bacillithiol: A key protective thiol in Staphylococcus aureus. Expert Rev. Anti. Infect. Ther. 2015, 13, 1089–1107. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.V.; Arbach, M.; Roberts, A.A.; Macdonald, C.J.; Groom, M.; Hamilton, C.J. Biophysical features of bacillithiol, the glutathione surrogate of Bacillus subtilis and other firmicutes. Chembiochem 2013, 14, 2160–2168. [Google Scholar] [CrossRef] [Green Version]
- Gaballa, A.; Newton, G.L.; Antelmann, H.; Parsonage, D.; Upton, H.; Rawat, M.; Claiborne, A.; Fahey, R.C.; Helmann, J.D. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. USA 2010, 107, 6482–6486. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.A.; Sharma, S.V.; Strankman, A.W.; Duran, S.R.; Rawat, M.; Hamilton, C.J. Mechanistic studies of FosB: A divalent-metal-dependent bacillithiol-S-transferase that mediates fosfomycin resistance in Staphylococcus aureus. Biochem. J. 2013, 451, 69–79. [Google Scholar] [CrossRef] [Green Version]
- Chi, B.K.; Gronau, K.; Mader, U.; Hessling, B.; Becher, D.; Antelmann, H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteom. 2011, 10, M111.009506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, B.K.; Roberts, A.A.; Huyen, T.T.; Basell, K.; Becher, D.; Albrecht, D.; Hamilton, C.J.; Antelmann, H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid. Redox Signal. 2013, 18, 1273–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrangsu, P.; Dusi, R.; Hamilton, C.J.; Helmann, J.D. Methylglyoxal resistance in Bacillus subtilis: Contributions of bacillithiol-dependent and independent pathways. Mol. Microbiol. 2014, 91, 706–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talesa, V.; Uotila, L.; Koivusalo, M.; Principato, G.; Giovannini, E.; Rosi, G. Isolation of glyoxalase II from two different compartments of rat liver mitochondria. Kinetic and immunochemical characterization of the enzymes. Biochim. Biophys. Acta 1989, 993, 7–11. [Google Scholar] [CrossRef]
- Talesa, V.; Principato, G.B.; Norton, S.J.; Contenti, S.; Mangiabene, C.; Rosi, G. Isolation of glyoxalase II from bovine liver mitochondria. Biochem. Int. 1990, 20, 53–58. [Google Scholar] [PubMed]
- Scire, A.; Tanfani, F.; Saccucci, F.; Bertoli, E.; Principato, G. Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of liposomes results in the inhibition of the cytosolic enzyme only. Proteins 2000, 41, 33–39. [Google Scholar] [CrossRef]
- Armeni, T.; Cianfruglia, L.; Piva, F.; Urbanelli, L.; Luisa Caniglia, M.; Pugnaloni, A.; Principato, G. S-D-Lactoylglutathione can be an alternative supply of mitochondrial glutathione. Free. Radic. Biol. Med. 2014, 67, 451–459. [Google Scholar] [CrossRef]
- Ashour, A.; Xue, M.; Al-Motawa, M.; Thornalley, P.J.; Rabbani, N. Glycolytic overload-driven dysfunction of periodontal ligament fibroblasts in high glucose concentration, corrected by glyoxalase 1 inducer. BMJ Open Diabetes Res. Care 2020, 8, e001458. [Google Scholar] [CrossRef]
- Antognelli, C.; Ferri, I.; Bellezza, G.; Siccu, P.; Love, H.D.; Talesa, V.N.; Sidoni, A. Glyoxalase 2 drives tumorigenesis in human prostate cells in a mechanism involving androgen receptor and p53–p21 axis. Mol. Carcinog. 2017, 56, 2112–2126. [Google Scholar] [CrossRef]
- Campos-Bermudez, V.A.; Leite, N.R.; Krog, R.; Costa-Filho, A.J.; Soncini, F.C.; Oliva, G.; Vila, A.J. Biochemical and structural characterization of Salmonella typhimurium glyoxalase II: New insights into metal ion selectivity. Biochemistry 2007, 46, 11069–11079. [Google Scholar] [CrossRef]
- Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J. 2014, 80, 93–105. [Google Scholar] [CrossRef] [Green Version]
- Limphong, P.; McKinney, R.M.; Adams, N.E.; Makaroff, C.A.; Bennett, B.; Crowder, M.W. The metal ion requirements of Arabidopsis thaliana Glx2-2 for catalytic activity. J. Biol. Inorg. Chem. 2010, 15, 249–258. [Google Scholar] [CrossRef]
- Wenzel, N.F.; Carenbauer, A.L.; Pfiester, M.P.; Schilling, O.; Meyer-Klaucke, W.; Makaroff, C.A.; Crowder, M.W. The binding of iron and zinc to glyoxalase II occurs exclusively as di-metal centers and is unique within the metallo-beta-lactamase family. J. Biol. Inorg. Chem. 2004, 9, 429–438. [Google Scholar] [CrossRef]
- Zang, T.M.; Hollman, D.A.; Crawford, P.A.; Crowder, M.W.; Makaroff, C.A. Arabidopsis glyoxalase II contains a zinc/iron binuclear metal center that is essential for substrate binding and catalysis. J. Biol. Chem. 2001, 276, 4788–4795. [Google Scholar] [CrossRef]
- Ridderstrom, M.; Mannervik, B. Molecular cloning and characterization of the thiolesterase glyoxalase II from Arabidopsis thaliana. Biochem. J. 1997, 322 Pt 2, 449–454. [Google Scholar] [CrossRef] [Green Version]
- Ridderstrom, M.; Saccucci, F.; Hellman, U.; Bergman, T.; Principato, G.; Mannervik, B. Molecular cloning, heterologous expression, and characterization of human glyoxalase II. J. Biol. Chem. 1996, 271, 319–323. [Google Scholar] [CrossRef] [Green Version]
- Urscher, M.; Deponte, M. Plasmodium falciparum glyoxalase II: Theorell-Chance product inhibition patterns, rate-limiting substrate binding via Arg(257)/Lys(260), and unmasking of acid-base catalysis. Biol. Chem. 2009, 390, 1171–1183. [Google Scholar] [CrossRef] [Green Version]
- Uotila, L. Purification and characterization of S-2-hydroxyacylglutathione hydrolase (glyoxalase II) from human liver. Biochemistry 1973, 12, 3944–3951. [Google Scholar] [CrossRef]
- Allen, R.E.; Lo, T.W.; Thornalley, P.J. Purification and characterisation of glyoxalase II from human red blood cells. Eur. J. Biochem. 1993, 213, 1261–1267. [Google Scholar] [CrossRef]
- Ball, J.C.; Vander Jagt, D.L. S-2-hydroxyacylglutathione hydrolase (glyoxalase II): Active-site mapping of a nonserine thiolesterase. Biochemistry 1981, 20, 899–905. [Google Scholar] [CrossRef]
- Bush, P.E.; Norton, S.J. S-(nitrocarbobenzoxy)glutathiones: Potent competitive inhibitors of mammalian glyoxalase II. J. Med. Chem. 1985, 28, 828–830. [Google Scholar] [CrossRef]
- Chyan, M.K.; Elia, A.C.; Principato, G.B.; Giovannini, E.; Rosi, G.; Norton, S.J. S-fluorenylmethoxycarbonyl glutathione and diesters: Inhibition of mammalian glyoxalase II. Enzym. Protein 1994, 48, 164–173. [Google Scholar] [CrossRef]
- Hsu, Y.R.; Norton, S.J. S-carbobenzoxyglutathione: A competitive inhibitor of mammalian glyoxalase II. J. Med. Chem. 1983, 26, 1784–1785. [Google Scholar] [CrossRef] [PubMed]
- Trincao, J.; Sousa Silva, M.; Barata, L.; Bonifacio, C.; Carvalho, S.; Tomas, A.M.; Ferreira, A.E.; Cordeiro, C.; Ponces Freire, A.; Romao, M.J. Purification, crystallization and preliminary X-ray diffraction analysis of the glyoxalase II from Leishmania infantum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 805–807. [Google Scholar] [CrossRef]
- Carfi, A.; Pares, S.; Duee, E.; Galleni, M.; Duez, C.; Frere, J.M.; Dideberg, O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995, 14, 4914–4921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limphong, P.; McKinney, R.M.; Adams, N.E.; Bennett, B.; Makaroff, C.A.; Gunasekera, T.; Crowder, M.W. Human glyoxalase II contains an Fe(II)Zn(II) center but is active as a mononuclear Zn(II) enzyme. Biochemistry 2009, 48, 5426–5434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragani, B.; Cocco, R.; Ridderstrom, M.; Stenberg, G.; Mannervik, B.; Aceto, A. Unfolding and refolding of human glyoxalase II and its single-tryptophan mutants. J. Mol. Biol. 1999, 291, 481–490. [Google Scholar] [CrossRef]
- Chen, S.L.; Fang, W.H.; Himo, F. Reaction mechanism of the binuclear zinc enzyme glyoxalase II—A theoretical study. J. Inorg. Biochem. 2009, 103, 274–281. [Google Scholar] [CrossRef]
- Limphong, P.; Crowder, M.W.; Bennett, B.; Makaroff, C.A. Arabidopsis thaliana GLX2-1 contains a dinuclear metal binding site, but is not a glyoxalase 2. Biochem. J. 2009, 417, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Campos-Bermudez, V.A.; Moran-Barrio, J.; Costa-Filho, A.J.; Vila, A.J. Metal-dependent inhibition of glyoxalase II: A possible mechanism to regulate the enzyme activity. J. Inorg. Biochem. 2010, 104, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Galeazzi, R.; Laudadio, E.; Falconi, E.; Massaccesi, L.; Ercolani, L.; Mobbili, G.; Minnelli, C.; Scire, A.; Cianfruglia, L.; Armeni, T. Protein-protein interactions of human glyoxalase II: Findings of a reliable docking protocol. Org. Biomol. Chem. 2018, 16, 5167–5177. [Google Scholar] [CrossRef] [PubMed]
- Antognelli, C.; Perrelli, A.; Armeni, T.; Nicola Talesa, V.; Retta, S.F. Dicarbonyl Stress and S-Glutathionylation in Cerebrovascular Diseases: A Focus on Cerebral Cavernous Malformations. Antioxidants 2020, 9, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armeni, T.; Ercolani, L.; Urbanelli, L.; Magini, A.; Magherini, F.; Pugnaloni, A.; Piva, F.; Modesti, A.; Emiliani, C.; Principato, G. Cellular redox imbalance and changes of protein S-glutathionylation patterns are associated with senescence induced by oncogenic H-ras. PLoS ONE 2012, 7, e52151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mieyal, J.J.; Gallogly, M.M.; Qanungo, S.; Sabens, E.A.; Shelton, M.D. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 2008, 10, 1941–1988. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.; Pinto, J.T.; Callery, P.S. Reversible and irreversible protein glutathionylation: Biological and clinical aspects. Expert Opin. Drug Metab. Toxicol. 2011, 7, 891–910. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Boja, E.S.; Tan, W.; Tekle, E.; Fales, H.M.; English, S.; Mieyal, J.J.; Chock, P.B. Reversible glutathionylation regulates actin polymerization in A431 cells. J. Biol. Chem. 2001, 276, 47763–47766. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Huff, L.P.; Fujii, M.; Griendling, K.K. Redox regulation of the actin cytoskeleton and its role in the vascular system. Free Radic. Biol. Med. 2017, 109, 84–107. [Google Scholar] [CrossRef]
- Oldenburg, J.; de Rooij, J. Mechanical control of the endothelial barrier. Cell Tissue Res. 2014, 355, 545–555. [Google Scholar] [CrossRef]
- Shasby, D.M.; Shasby, S.S.; Sullivan, J.M.; Peach, M.J. Role of endothelial cell cytoskeleton in control of endothelial permeability. Circ. Res. 1982, 51, 657–661. [Google Scholar] [CrossRef] [Green Version]
- Pollard, T.D.; Blanchoin, L.; Mullins, R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 545–576. [Google Scholar] [CrossRef] [Green Version]
- Dalle-Donne, I.; Giustarini, D.; Rossi, R.; Colombo, R.; Milzani, A. Reversible S-glutathionylation of Cys 374 regulates actin filament formation by inducing structural changes in the actin molecule. Free Radic. Biol. Med. 2003, 34, 23–32. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Colombo, R.; Milzani, A. Actin S-glutathionylation: Evidence against a thiol-disulphide exchange mechanism. Free Radic. Biol. Med. 2003, 35, 1185–1193. [Google Scholar] [CrossRef]
- Lassing, I.; Schmitzberger, F.; Bjornstedt, M.; Holmgren, A.; Nordlund, P.; Schutt, C.E.; Lindberg, U. Molecular and structural basis for redox regulation of beta-actin. J. Mol. Biol. 2007, 370, 331–348. [Google Scholar] [CrossRef]
- Cha, S.J.; Kim, H.; Choi, H.J.; Lee, S.; Kim, K. Protein Glutathionylation in the Pathogenesis of Neurodegenerative Diseases. Oxid. Med. Cell Longev. 2017, 2017, 2818565. [Google Scholar] [CrossRef]
- Kruyer, A.; Ball, L.E.; Townsend, D.M.; Kalivas, P.W.; Uys, J.D. Post-translational S-glutathionylation of cofilin increases actin cycling during cocaine seeking. PLoS ONE 2019, 14, e0223037. [Google Scholar] [CrossRef] [Green Version]
- Pastore, A.; Tozzi, G.; Gaeta, L.M.; Bertini, E.; Serafini, V.; Di Cesare, S.; Bonetto, V.; Casoni, F.; Carrozzo, R.; Federici, G.; et al. Actin glutathionylation increases in fibroblasts of patients with Friedreich’s ataxia: A potential role in the pathogenesis of the disease. J. Biol. Chem. 2003, 278, 42588–42595. [Google Scholar] [CrossRef] [Green Version]
- Varland, S.; Vandekerckhove, J.; Drazic, A. Actin Post-translational Modifications: The Cinderella of Cytoskeletal Control. Trends. Biochem. Sci. 2019, 44, 502–516. [Google Scholar] [CrossRef] [Green Version]
- Agro, A.F.; Mavelli, I.; Cannella, C.; Federici, G. Activation of porcine heart mitochondrial malate dehydrogenase by zero valence sulfur and rhodanese. Biochem. Biophys. Res. Commun. 1976, 68, 553–560. [Google Scholar] [CrossRef]
- Ruelland, E.; Lemaire-Chamley, M.; Le Marechal, P.; Issakidis-Bourguet, E.; Djukic, N.; Miginiac-Maslow, M. An internal cysteine is involved in the thioredoxin-dependent activation of sorghum leaf NADP-malate dehydrogenase. J. Biol. Chem. 1997, 272, 19851–19857. [Google Scholar] [CrossRef] [Green Version]
- Kojer, K.; Riemer, J. Balancing oxidative protein folding: The influences of reducing pathways on disulfide bond formation. Biochim. Biophys. Acta 2014, 1844, 1383–1390. [Google Scholar] [CrossRef]
- Iglesias, A.A.; Andreo, C.S. NADP-dependent malate dehydrogenase (decarboxylating) from sugar cane leaves. Kinetic properties of different oligomeric structures. Eur. J. Biochem. 1990, 192, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.R.; Hirschey, M.D. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 2014, 54, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinert, B.T.; Moustafa, T.; Iesmantavicius, V.; Zechner, R.; Choudhary, C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 2015, 34, 2620–2632. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.N.; Kjalarsdottir, L.; Thompson, J.W.; Dubois, L.G.; Stevens, R.D.; Ilkayeva, O.R.; Brosnan, M.J.; Rolph, T.P.; Grimsrud, P.A.; Muoio, D.M. The Acetyl Group Buffering Action of Carnitine Acetyltransferase Offsets Macronutrient-Induced Lysine Acetylation of Mitochondrial Proteins. Cell Rep. 2016, 14, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Bracher, P.J.; Snyder, P.W.; Bohall, B.R.; Whitesides, G.M. The relative rates of thiol-thioester exchange and hydrolysis for alkyl and aryl thioalkanoates in water. Orig. Life Evol. Biosph. 2011, 41, 399–412. [Google Scholar] [CrossRef]
- Edwards, L.G.; Adesida, A.; Thornalley, P.J. Inhibition of human leukaemia 60 cell growth by S-D-lactoylglutathione in vitro. Mediation by metabolism to N-D-lactoylcysteine and induction of apoptosis. Leuk. Res. 1996, 20, 17–26. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Khadka, S.; Barekatain, Y.; Muller, F. Re-Evaluating the Mechanism of Histone Lactylation. OSF Preprints. Available online: https://osf.io/kyab5 (accessed on 22 May 2020).
- Kulkarni, C.A.; Brookes, P. Many Routes from Glycolysis to Histone PTMs. Nature “Matters Arising” response to: Zhang et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 7779, OSF Preprints. Available online: https://osf.io/sba8j (accessed on 8 May 2020).
- Donnellan, L.; Young, C.; Simpson, B.S.; Acland, M.; Dhillon, V.S.; Costabile, M.; Fenech, M.; Hoffmann, P.; Deo, P. Proteomic Analysis of Methylglyoxal Modifications Reveals Susceptibility of Glycolytic Enzymes to Dicarbonyl Stress. Int. J. Mol. Sci. 2022, 23, 3689. [Google Scholar] [CrossRef]
- Antognelli, C.; Talesa, V.N. Glyoxalases in Urological Malignancies. Int. J. Mol. Sci. 2018, 19, 415. [Google Scholar] [CrossRef] [Green Version]
- Alexandrova, E.M.; Moll, U.M. Role of p53 family members p73 and p63 in human hematological malignancies. Leuk. Lymphoma 2012, 53, 2116–2129. [Google Scholar] [CrossRef]
- Moll, U.M.; Slade, N. p63 and p73: Roles in development and tumor formation. Mol. Cancer Res. 2004, 2, 371–386. [Google Scholar] [CrossRef]
- Rulli, A.; Antognelli, C.; Prezzi, E.; Baldracchini, F.; Piva, F.; Giovannini, E.; Talesa, V. A possible regulatory role of 17beta-estradiol and tamoxifen on glyoxalase I and glyoxalase II genes expression in MCF7 and BT20 human breast cancer cells. Breast Cancer Res. Treat. 2006, 96, 187–196. [Google Scholar] [CrossRef]
- Antognelli, C.; Del Buono, C.; Baldracchini, F.; Talesa, V.; Cottini, E.; Brancadoro, C.; Zucchi, A.; Mearini, E. Alteration of glyoxalase genes expression in response to testosterone in LNCaP and PC3 human prostate cancer cells. Cancer Biol. Ther. 2007, 6, 1880–1888. [Google Scholar] [CrossRef] [Green Version]
- Dafre, A.L.; Schmitz, A.E.; Maher, P. Methylglyoxal-induced AMPK activation leads to autophagic degradation of thioredoxin 1 and glyoxalase 2 in HT22 nerve cells. Free Radic. Biol. Med. 2017, 108, 270–279. [Google Scholar] [CrossRef]
- Antognelli, C.; Frosini, R.; Santolla, M.F.; Peirce, M.J.; Talesa, V.N. Oleuropein-Induced Apoptosis Is Mediated by Mitochondrial Glyoxalase 2 in NSCLC A549 Cells: A Mechanistic Inside and a Possible Novel Nonenzymatic Role for an Ancient Enzyme. Oxid. Med. Cell Longev. 2019, 2019, 8576961. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, E.; Lichtenstein, L.M. Histamine release from human leukocytes: Studies with deuterium oxide, colchicine, and cytochalasin B. J. Clin. Invest. 1972, 51, 2941–2947. [Google Scholar] [CrossRef]
- Norton, S.J.; Elia, A.C.; Chyan, M.K.; Gillis, G.; Frenzel, C.; Principato, G.B. Inhibitors and inhibition studies of mammalian glyoxalase II activity. Biochem. Soc. Trans. 1993, 21, 545–549. [Google Scholar] [CrossRef] [Green Version]
- Clellan, J.D.; Thornalley, P.J. The potentiation of GTP-dependent assembly of microtubules by S-D-lactoylglutathione. Biochem. Soc. Trans. 1993, 21, 160S. [Google Scholar] [CrossRef]
- Di Simplicio, P.; Vignani, R.; Talesa, V.; Principato, G. Evidence of glyoxalase II activity associated with microtubule polymerization in bovine brain. Pharmacol. Res. 1990, 22, 172. [Google Scholar] [CrossRef]
- Chen, W.; Seefeldt, T.; Young, A.; Zhang, X.; Zhao, Y.; Ruffolo, J.; Kaushik, R.S.; Guan, X. Microtubule S-glutathionylation as a potential approach for antimitotic agents. BMC Cancer 2012, 12, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carletti, B.; Passarelli, C.; Sparaco, M.; Tozzi, G.; Pastore, A.; Bertini, E.; Piemonte, F. Effect of protein glutathionylation on neuronal cytoskeleton: A potential link to neurodegeneration. Neuroscience 2011, 192, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, E. Effects of S-lactoylglutathione and inhibitors of glyoxalase I on histamine release from human leukocytes. Nature 1979, 277, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, E. Concanavalin A increases glyoxalase enzyme activities in polymorphonuclear leukocytes and lymphocytes. J. Immunol. 1978, 121, 923–925. [Google Scholar]
- Gillespie, E. Cell-free microtubule assembly: Evidence for control by glyoxalase. Fed. Proc. 1975, 34, 541. [Google Scholar]
- Li, X.; Fargue, S.; Challa, A.K.; Poore, W.; Knight, J.; Wood, K.D. Generation of a GLO-2 deficient mouse reveals its effects on liver carbonyl and glutathione levels. Biochem. Biophys. Rep. 2021, 28, 101138. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scirè, A.; Cianfruglia, L.; Minnelli, C.; Romaldi, B.; Laudadio, E.; Galeazzi, R.; Antognelli, C.; Armeni, T. Glyoxalase 2: Towards a Broader View of the Second Player of the Glyoxalase System. Antioxidants 2022, 11, 2131. https://doi.org/10.3390/antiox11112131
Scirè A, Cianfruglia L, Minnelli C, Romaldi B, Laudadio E, Galeazzi R, Antognelli C, Armeni T. Glyoxalase 2: Towards a Broader View of the Second Player of the Glyoxalase System. Antioxidants. 2022; 11(11):2131. https://doi.org/10.3390/antiox11112131
Chicago/Turabian StyleScirè, Andrea, Laura Cianfruglia, Cristina Minnelli, Brenda Romaldi, Emiliano Laudadio, Roberta Galeazzi, Cinzia Antognelli, and Tatiana Armeni. 2022. "Glyoxalase 2: Towards a Broader View of the Second Player of the Glyoxalase System" Antioxidants 11, no. 11: 2131. https://doi.org/10.3390/antiox11112131





