Ethanol Induction of Innate Immune Signals Across BV2 Microglia and SH-SY5Y Neuroblastoma Involves Induction of IL-4 and IL-13
Abstract
:1. Introduction:
2. Methods
2.1. Cell Lines and Treatment
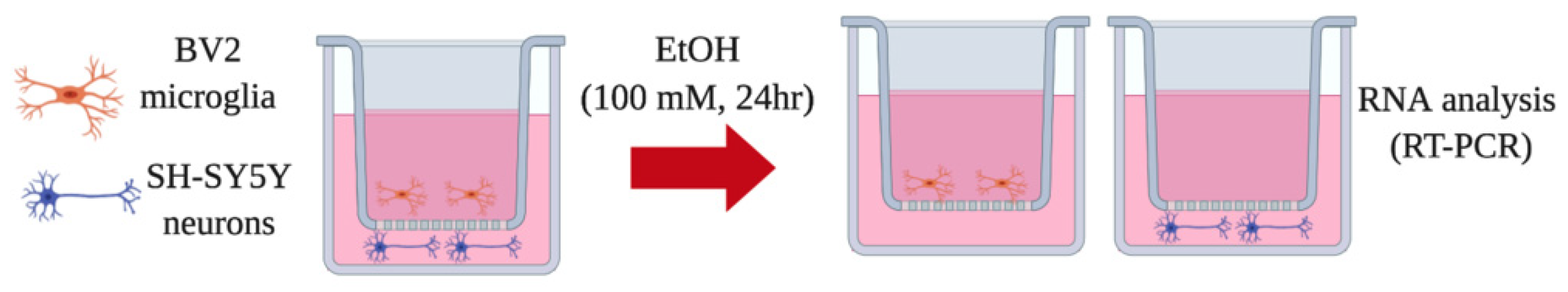
2.2. Transwell Co-Culture Model
2.3. Hippocampal Entorhinal (HEC) Slice Culture
2.4. Real-Time (RT)-PCR
2.5. Enzyme-Linked Immunosorbent Assay (Elisa)
2.6. Statistical Analysis
3. Results
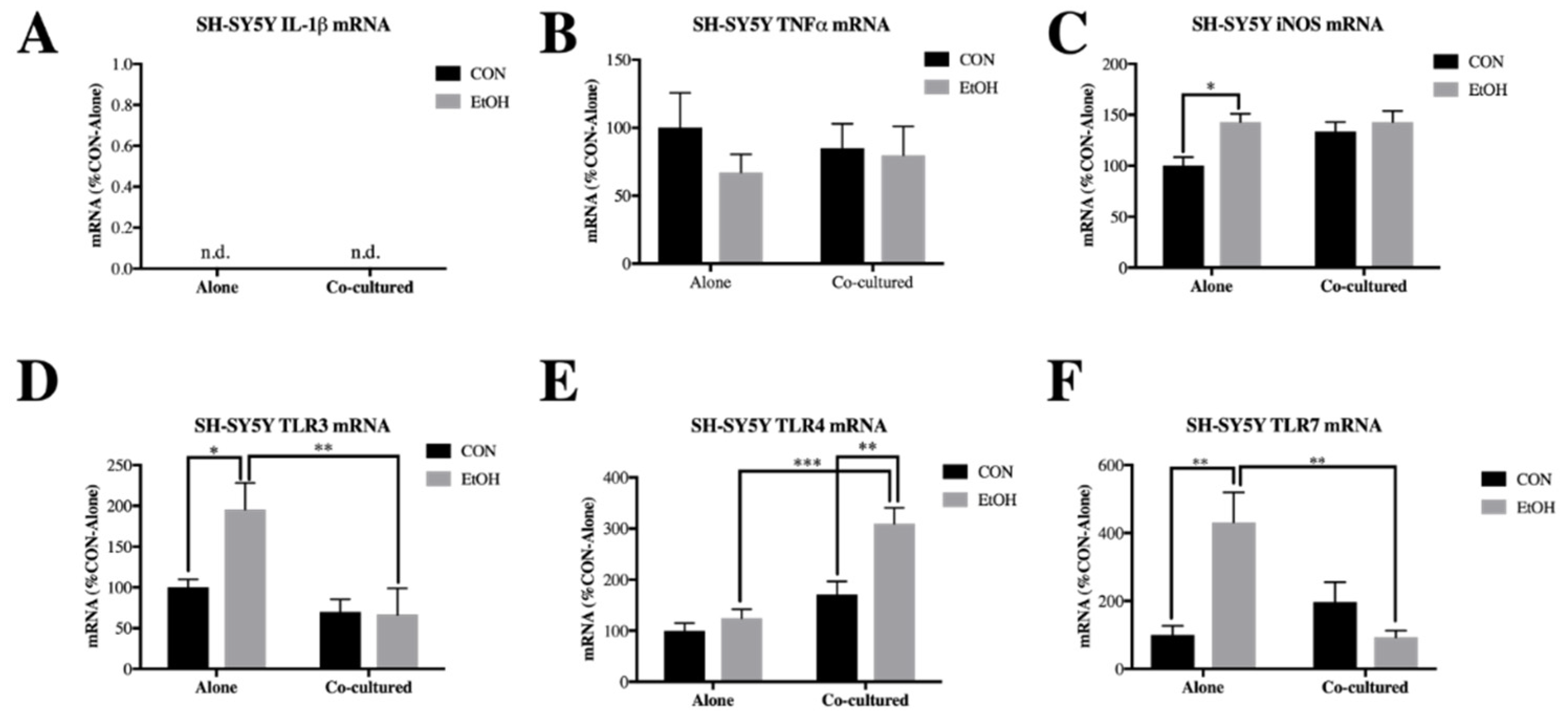
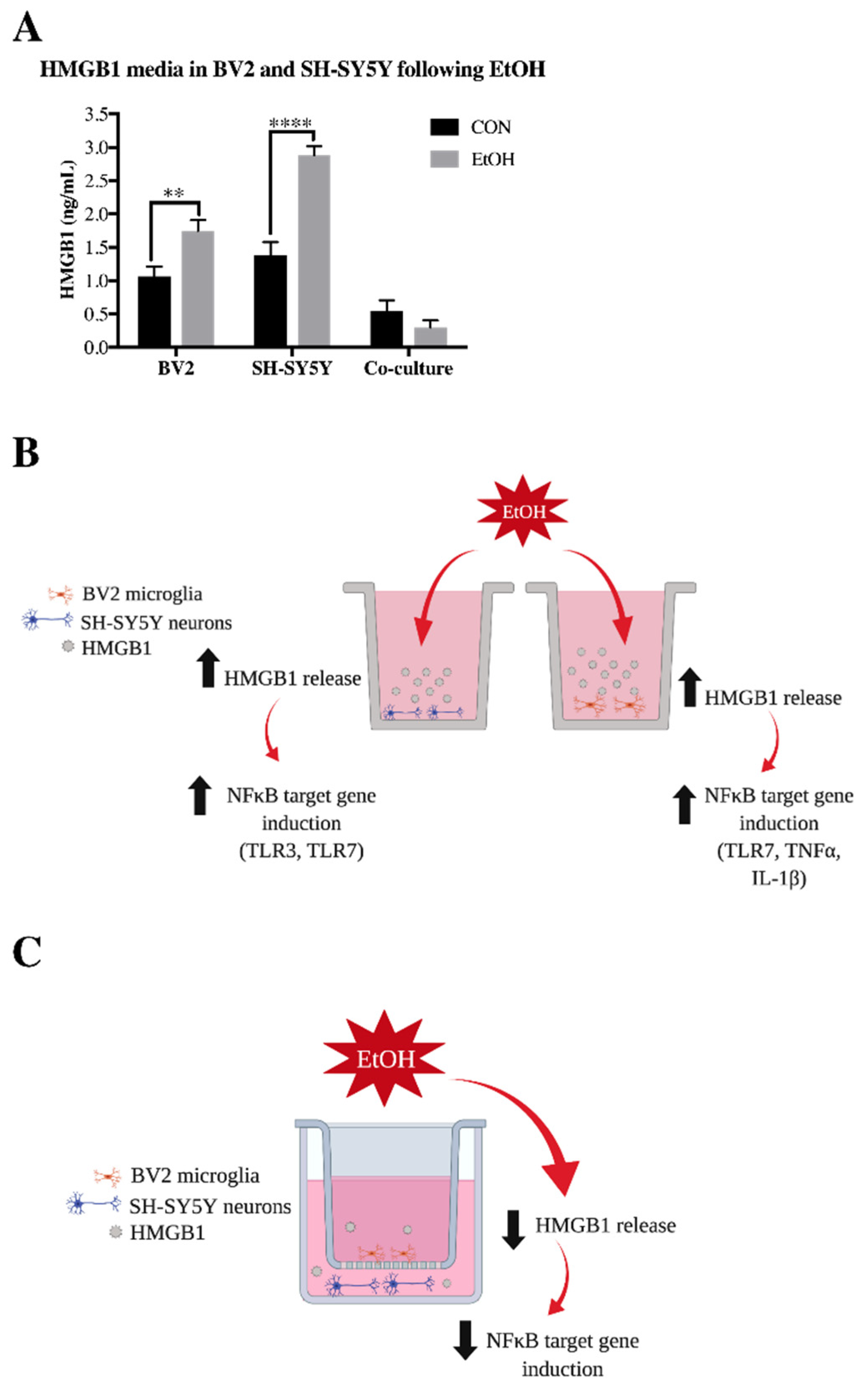
3.1. Co-Culture Modifies bv2 Microglia and SH-sy5y Gene Expression and Ethanol-Induced Cytokines and Tlrs
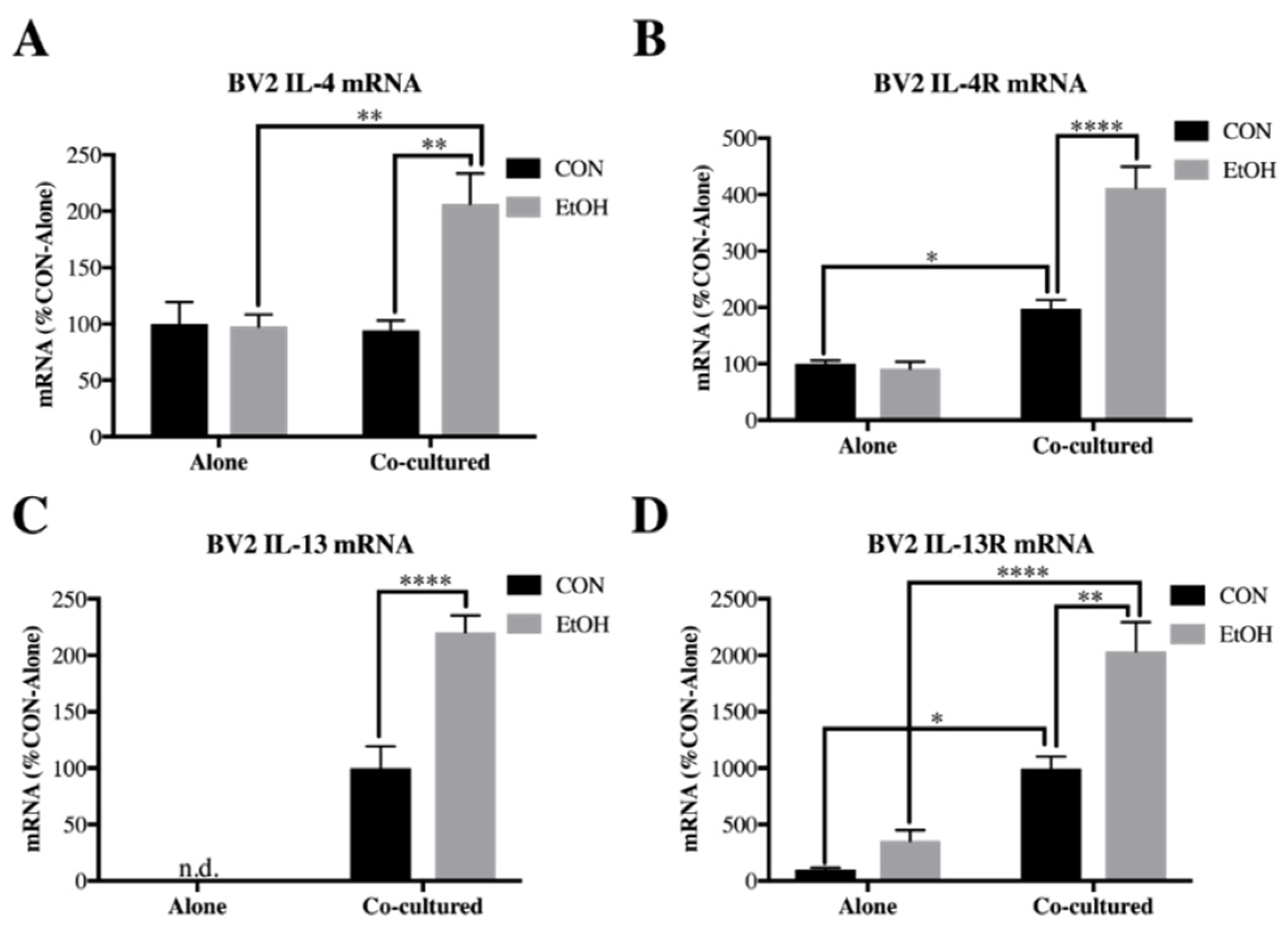
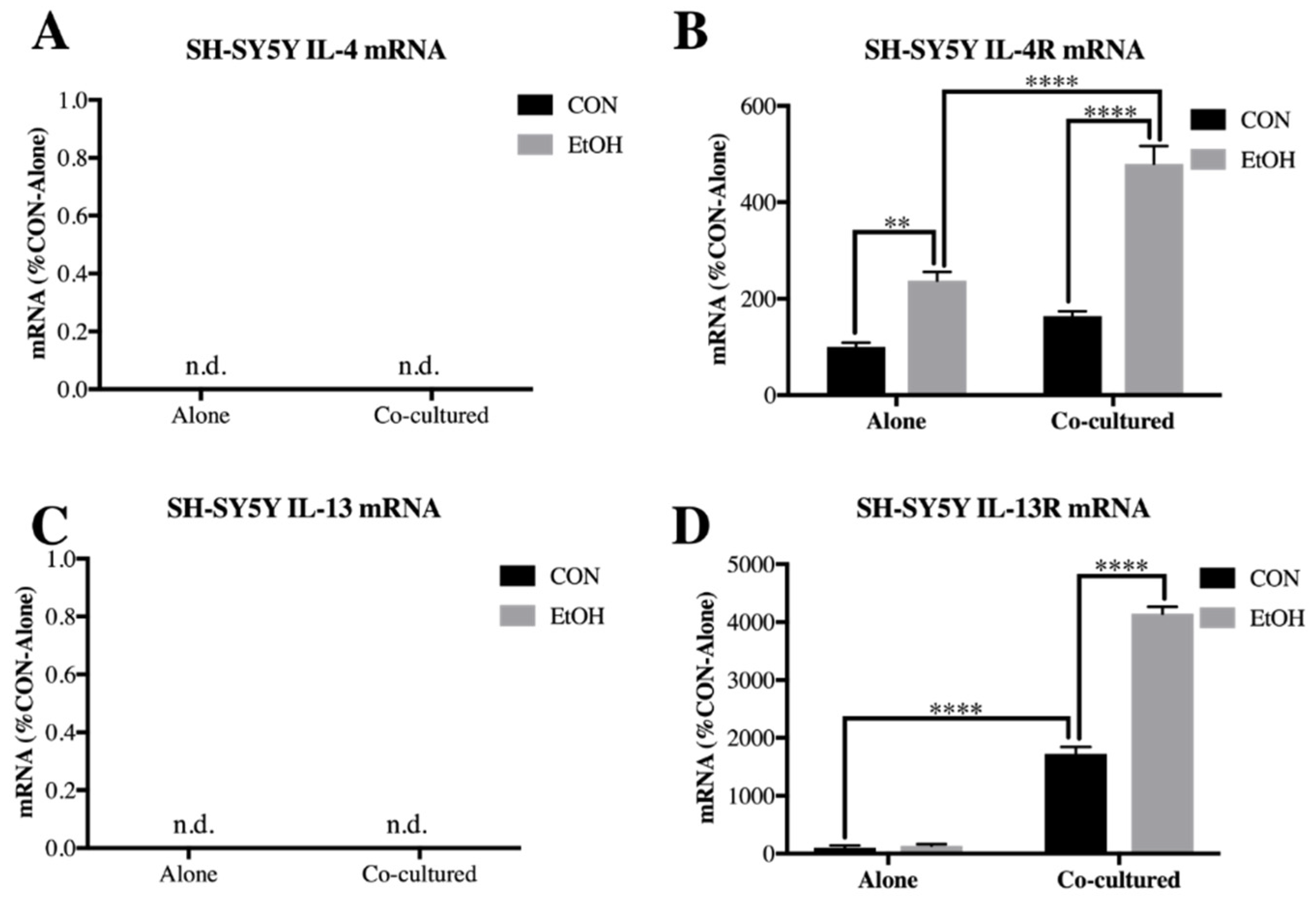
3.2. Il-4 and IL-13 Expression Is Increased by Ethanol in Co-Cultured BV2 Microglia and SH-SY5Y
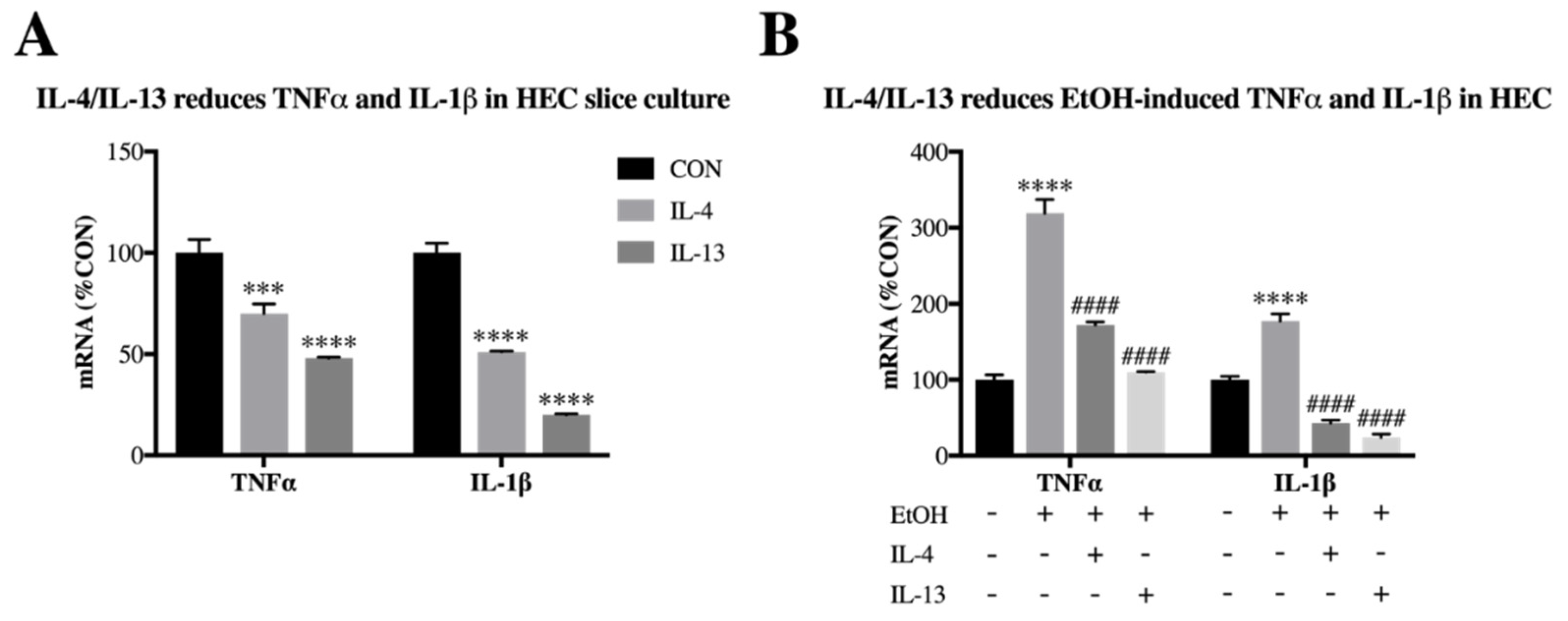
3.3. IL-4 and IL-13 Reduce Ethanol-Induced Tnfα and IL-1β in Hippocampal-Entorhinal Slice Culture
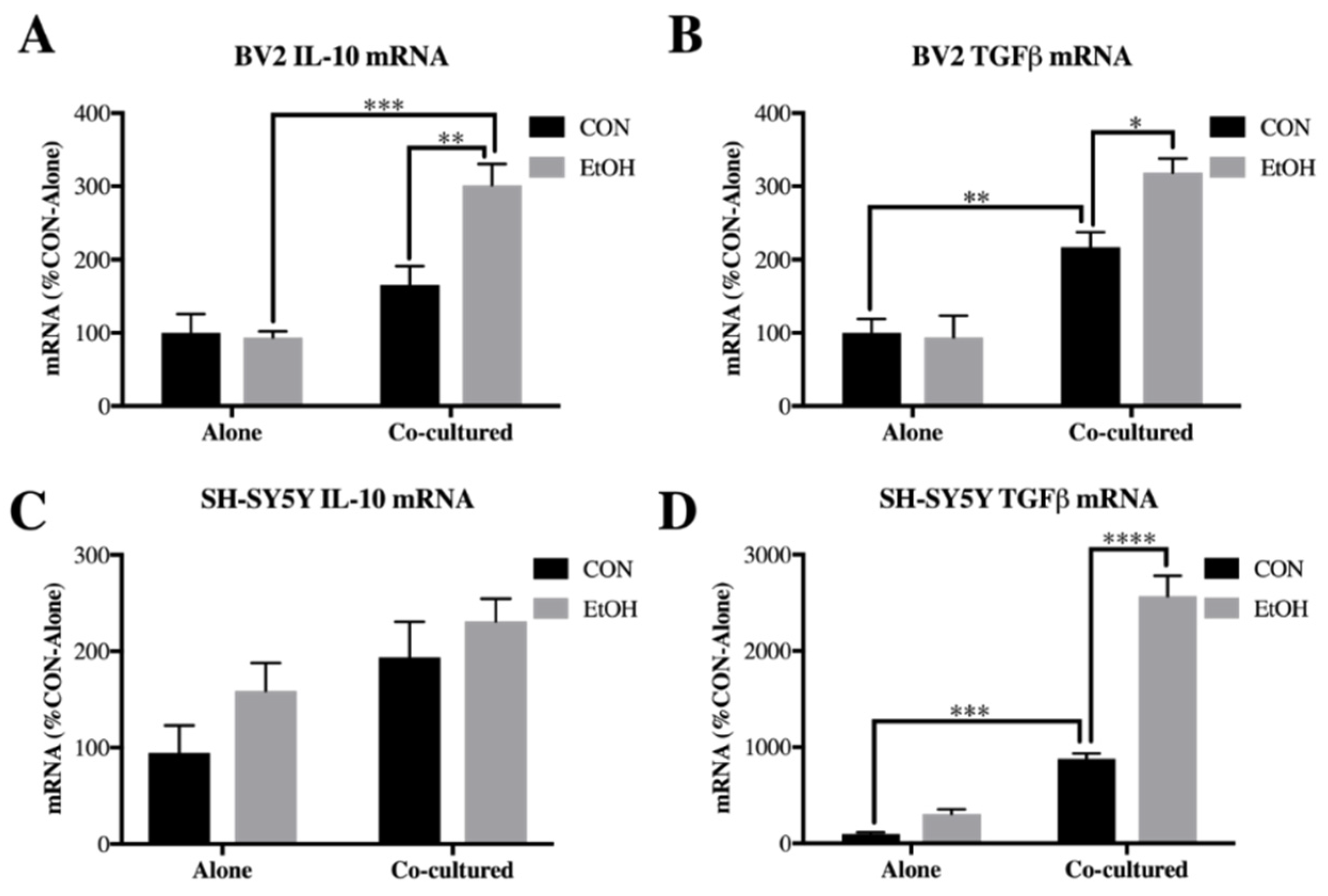
3.4. IL-10 and Tgfβ Are Increased by Ethanol in Co-Cultured BV2 Microglia and SH-SY5Y
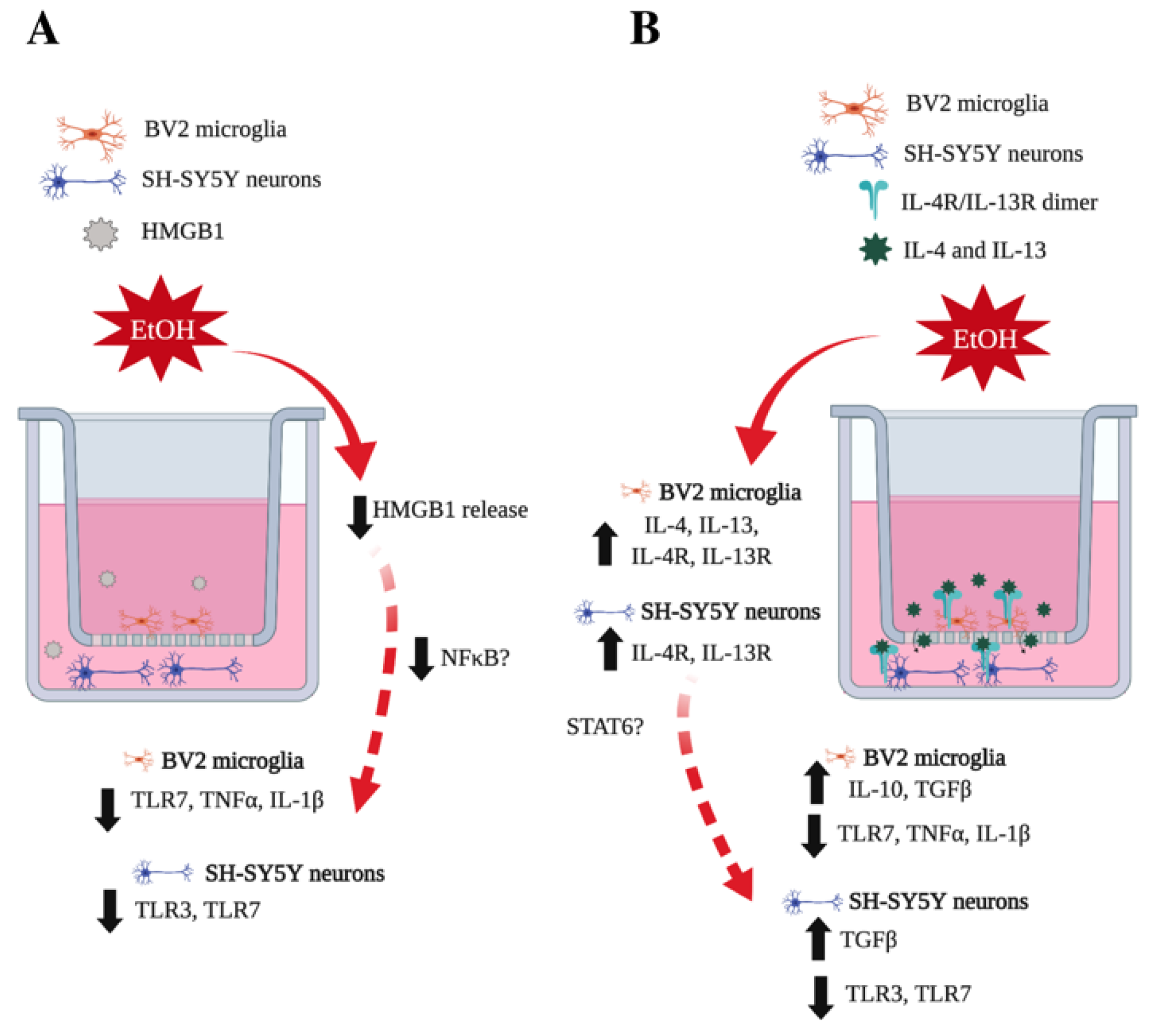
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crews, F.T.; Collins, M.A.; Dlugos, C.; Littleton, J.; Wilkins, L.; Neafsey, E.J.; Pentney, R.; Snell, L.D.; Tabakoff, B.; Zou, J.; et al. Alcohol-Induced Neurodegeneration: When, Where and Why? Alcohol. Clin. Exp. Res. 2004, 28, 350–364. [Google Scholar] [CrossRef]
- Crews, F.T.; Lawrimore, C.J.; Walter, T.J.; Coleman, L.G. The Role of Neuroimmune Signaling in Alcoholism. Neuropharmacology 2017, 122, 56–73. [Google Scholar] [CrossRef]
- Mayfield, J.; Ferguson, L.; Harris, R.A. Neuroimmune Signaling: A Key Component of Alcohol Abuse. Curr. Opin. Neurobiol. 2013, 23, 513–520. [Google Scholar] [CrossRef]
- Coleman, L.G., Jr.; Crews, F.T. Innate Immune Signaling and Alcohol Use Disorders. Hand. Experi. Pharmaco. 2018. [Google Scholar] [CrossRef]
- Park, J.S.; Svetkauskaite, D.; He, Q.; Kim, J.Y.; Strassheim, D.; Ishizaka, A.; Abraham, E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 2004, 279, 7370–7377. [Google Scholar] [CrossRef]
- Yang, H.; Antoine, D.J.; Andersson, U.; Tracey, K.J. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukoc. Boil. 2013, 93, 865–873. [Google Scholar] [CrossRef] [Green Version]
- Yanai, H.; Ban, T.; Wang, Z.; Choi, M.K.; Kawamura, T.; Negishi, H.; Nakasato, M.; Lu, Y.; Hangai, S.; Koshiba, R.; et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nat. 2009, 462, 99–103. [Google Scholar] [CrossRef]
- Narayanan, K.B.; Park, H.H. Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis 2015, 20, 196–209. [Google Scholar] [CrossRef]
- Blanco, A.M.; Vallés, S.L.; Pascual, M.; Guerri, C. Involvement of TLR4/type I IL-1 receptor signaling in the induction of inflammatory mediators and cell death induced by ethanol in cultured astrocytes. J. Immunol. 2005, 175, 6893–6899. [Google Scholar] [CrossRef]
- Fernandez-Lizarbe, S.; Pascual, M.; Guerri, C. Critical Role of TLR4 Response in the Activation of Microglia Induced by Ethanol. J. Immunol. 2009, 183, 4733–4744. [Google Scholar] [CrossRef] [Green Version]
- Alfonso-Loeches, S.; Pascual-Lucas, M.; Blanco, A.M.; Sanchez-Vera, I.; Guerri, C. Pivotal Role of TLR4 Receptors in Alcohol-Induced Neuroinflammation and Brain Damage. J. Neurosci. 2010, 30, 8285–8295. [Google Scholar] [CrossRef]
- Fernández-Lizarbe, S.; Montesinos, J.; Guerri, C.; Fernandez-Lizarbe, S. Ethanol induces TLR4/TLR2 association, triggering an inflammatory response in microglial cells. J. Neurochem. 2013, 126, 261–273. [Google Scholar] [CrossRef]
- Crews, F.T.; Qin, L.; Sheedy, D.; Vetreno, R.P.; Zou, J. High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biol. psychiatry 2013, 73, 602–612. [Google Scholar] [CrossRef]
- Coleman, L.G.; Zou, J.; Crews, F.T. Microglial-derived miRNA let-7 and HMGB1 contribute to ethanol-induced neurotoxicity via TLR7. J. Neuroinflammation 2017, 14, 22. [Google Scholar] [CrossRef]
- Vetreno, R.P.; Lawrimore, C.J.; Rowsey, P.J.; Crews, F.T. Persistent Adult Neuroimmune Activation and Loss of Hippocampal Neurogenesis Following Adolescent Ethanol Exposure: Blockade by Exercise and the Anti-inflammatory Drug Indomethacin. Front. Mol. Neurosci. 2018, 12, 200. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Crews, F.T. Inflammasome-IL-1β Signaling Mediates Ethanol Inhibition of Hippocampal Neurogenesis. Front. Mol. Neurosci. 2012, 6. [Google Scholar] [CrossRef]
- Coleman, L.G., Jr.; Zou, J.; Qin, L.; Crews, F.T. HMGB1/IL-1beta complexes regulate neuroimmune responses in alcoholism. Brain Behav. Immun. 2018, 72, 61–77. [Google Scholar] [CrossRef]
- McCarthy, G.M.; Warden, A.S.; Bridges, C.R.; Blednov, Y.A.; Harris, R.A. Chronic ethanol consumption: role of TLR3/TRIF-dependent signaling. Addict. Boil. 2017, 23, 889–903. [Google Scholar] [CrossRef] [Green Version]
- Qin, L.; He, J.; Hanes, R.N.; Pluzarev, O.; Hong, J.-S.; Crews, F.T. Increased systemic and brain cytokine production and neuroinflammation by endotoxin following ethanol treatment. J. Neuroinflammation 2008, 5, 10. [Google Scholar] [CrossRef]
- Qin, L.; Crews, F.T. Chronic ethanol increases systemic TLR3 agonist-induced neuroinflammation and neurodegeneration. J. Neuroinflammation 2012, 9, 130. [Google Scholar] [CrossRef]
- Blednov, Y.A.; Bergeson, S.E.; Walker, D.; Ferreira, V.M.; Kuziel, W.A.; Harris, R.A. Perturbation of chemokine networks by gene deletion alters the reinforcing actions of ethanol. Behav. Brain Res. 2005, 165, 110–125. [Google Scholar] [CrossRef] [Green Version]
- Sheridan, G.K.; Murphy, K.J. Neuron–glia crosstalk in health and disease: fractalkine and CX3CR1 take centre stage. Open Boil. 2013, 3, 130181. [Google Scholar] [CrossRef]
- Stephan, A.H.; Barres, B.A.; Stevens, B. The Complement System: An Unexpected Role in Synaptic Pruning During Development and Disease. Annu. Rev. Neurosci. 2012, 35, 369–389. [Google Scholar] [CrossRef] [Green Version]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nat. 2017, 541, 481–487. [Google Scholar] [CrossRef]
- Haas, H.; Falcone, F.; Holland, M.; Schramm, G.; Haisch, K.; Gibbs, B.; Bufe, A.; Schlaak, M. Early Interleukin–4: Its Role in the Switch towards a Th2 Response and IgE–Mediated Allergy. Int. Arch. Allergy Immunol. 1999, 119, 86–94. [Google Scholar] [CrossRef]
- Zhou, X.; Spittau, B.; Krieglstein, K. TGFβ signalling plays an important role in IL4-induced alternative activation of microglia. J. Neuroinflammation 2012, 9, 210. [Google Scholar] [CrossRef]
- Shin, W.H.; Lee, D.-Y.; Park, K.W.; Kim, S.U.; Yang, M.-S.; Joe, E.-H.; Jin, B.K. Microglia expressing interleukin-13 undergo cell death and contribute to neuronal survival in vivo. Glia 2004, 46, 142–152. [Google Scholar] [CrossRef]
- Mori, S.; Sugama, S.; Nguyen, W.; Michel, T.; Sanna, M.G.; Sanchez-Alavez, M.; Cintron-Colon, R.; Moroncini, G.; Kakinuma, Y.; Maher, P.; et al. Lack of interleukin-13 receptor α1 delays the loss of dopaminergic neurons during chronic stress. J. Neuroinflammation 2017, 14, 525. [Google Scholar] [CrossRef]
- Gadani, S.P.; Cronk, J.C.; Norris, G.T.; Kipnis, J. IL-4 in the Brain: A Cytokine To Remember. J. Immun. 2012, 189, 4213–4219. [Google Scholar] [CrossRef]
- Brombacher, T.M.; Nono, J.K.; De Gouveia, K.S.; Makena, N.; Darby, M.; Womersley, J.; Tamgue, O.; Brombacher, F. IL-13–Mediated Regulation of Learning and Memory. J. Immunol. 2017, 198, 2681–2688. [Google Scholar] [CrossRef]
- Lobo-Silva, D.; Carriche, G.M.; Gil Castro, A.; Roque, S.; Saraiva, M. Balancing the immune response in the brain: IL-10 and its regulation. J. Neuroinflammation 2016, 13, 2081. [Google Scholar] [CrossRef]
- Donzis, E.J.; Tronson, N.C. Modulation of learning and memory by cytokines: Signaling mechanisms and long term consequences. Neurobiol. Learn. Mem. 2014, 115, 68–77. [Google Scholar] [CrossRef] [Green Version]
- Vivien, D.; Ali, C. Transforming growth factor-β signalling in brain disorders. Cytokine Growth Factor Rev. 2006, 17, 121–128. [Google Scholar] [CrossRef]
- Caraci, F.; Gulisano, W.; Guida, C.A.; Impellizzeri, A.A.R.; Drago, F.; Puzzo, D.; Palmeri, A. A key role for TGF-β1 in hippocampal synaptic plasticity and memory. Sci. Rep. 2015, 5, 11252. [Google Scholar] [CrossRef]
- Vetreno, R.P.; Crews, F.T. ADOLESCENT BINGE DRINKING INCREASES EXPRESSION OF THE DANGER SIGNAL RECEPTOR AGONIST HMGB1 AND TOLL-LIKE RECEPTORS IN THE ADULT PREFRONTAL CORTEX. Neurosci. 2012, 226, 475–488. [Google Scholar] [CrossRef]
- Lawrimore, C.J.; Crews, F.T. Ethanol, TLR3, and TLR4 Agonists Have Unique Innate Immune Responses in Neuron-Like SH-SY5Y and Microglia-Like BV2. Alcohol. Clin. Exp. Res. 2017, 41, 939–954. [Google Scholar] [CrossRef] [Green Version]
- Szepesi, Z.; Manouchehrian, O.; Bachiller, S.; Deierborg, T. Bidirectional Microglia–Neuron Communication in Health and Disease. Front. Cell. Neurosci. 2018, 12, 323. [Google Scholar] [CrossRef]
- Medina-Vega, L.; Martín-González, C.; González-Reimers, E.; Santolaria-Fernández, F.; Medina-García, J.; González-Pérez, J.; De La Vega-Prieto, M.; Durán-Castellón, M. TH-1 and TH-2 Cytokines in Stable Chronic Alcoholics. Alcohol Alcohol. 2012, 47, 390–396. [Google Scholar] [Green Version]
- Pandur, E.; Tamási, K.; Pap, R.; Varga, E.; Miseta, A.; Sipos, K. Fractalkine Induces Hepcidin Expression of BV-2 Microglia and Causes Iron Accumulation in SH-SY5Y Cells. Cell. Mol. Neurobiol. 2019, 39, 985–1001. [Google Scholar] [CrossRef] [Green Version]
- De Simone, U.; Caloni, F.; Gribaldo, L.; Coccini, T. Human Co-culture Model of Neurons and Astrocytes to Test Acute Cytotoxicity of Neurotoxic Compounds. Int. J. Toxicol. 2017, 36, 463–477. [Google Scholar] [CrossRef]
- Xicoy, H.; Wieringa, B.; Martens, G.J. The SH-SY5Y cell line in Parkinson’s disease research: a systematic review. Mol. Neurodegener. 2017, 12, 384. [Google Scholar] [CrossRef]
- Elgammal, A.; Eves, D.; Albaghli, A.; Kane, D.; Durcan, R.; Storey, D.; Gilligan, P. The Blood Alcohol Concentration Testing Emergency Room Investigation Analysis Study: A 1-Year Review of Blood Alcohol Concentration Testing in an Emergency Department. Adv. Emerg. Med. 2015, 2015, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Cartlidge, D.; Redmond, A.D. Alcohol and conscious level. Biomed. Pharmacother. 1990, 44, 205–208. [Google Scholar] [CrossRef]
- Nixon, K. Temporally Specific Burst in Cell Proliferation Increases Hippocampal Neurogenesis in Protracted Abstinence from Alcohol. J. Neurosci. 2004, 24, 9714–9722. [Google Scholar] [CrossRef]
- Walter, T.J.; Crews, F.T. Microglial depletion alters the brain neuroimmune response to acute binge ethanol withdrawal. J. Neuroinflammation 2017, 14, 386. [Google Scholar] [CrossRef]
- Renaud, J.; Martinoli, M.-G. Development of an Insert Co-culture System of Two Cellular Types in the Absence of Cell-Cell Contact. J. Vis. Exp. 2016, 113, e54356. [Google Scholar] [CrossRef]
- Yu, Z.; Yang, L.; Yang, Y.; Chen, S.; Sun, D.; Xu, H.; Fan, X. Epothilone B Benefits Nigral Dopaminergic Neurons by Attenuating Microglia Activation in the 6-Hydroxydopamine Lesion Mouse Model of Parkinson’s Disease. Front. Cell. Neurosci. 2018, 12, 324. [Google Scholar] [CrossRef]
- Gao, M.; Dong, Q.; Yao, H.; Zhang, Y.; Yang, Y.; Dang, Y.; Zhang, H.; Yang, Z.; Xu, M.; Xu, R. Induced neural stem cells modulate microglia activation states via CXCL12/CXCR4 signaling. Brain, Behav. Immun. 2017, 59, 288–299. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Uang, H.-W.; Lin, R.-J.; Chen, I.-J.; Lo, Y.-C. Neuroprotective Effects of Glyceryl Nonivamide against Microglia-Like Cells and 6-Hydroxydopamine-Induced Neurotoxicity in SH-SY5Y Human Dopaminergic Neuroblastoma Cells. J. Pharmacol. Exp. Ther. 2007, 323, 877–887. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.Y.; Crews, F.T. Release of Neuronal HMGB1 by Ethanol through Decreased HDAC Activity Activates Brain Neuroimmune Signaling. PLoS ONE 2014, 9, e87915. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D.K. What is the proper way to apply the multiple comparison test? Korean J. Anesthesiol. 2018, 71, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Altman, D.G.; Bland, J.M. Standard deviations and standard errors. BMJ 2005, 331, 903. [Google Scholar] [CrossRef]
- Bao, K.; Reinhardt, R.L. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine 2015, 75, 25–37. [Google Scholar] [CrossRef]
- Mori, S.; Maher, P.; Conti, B. Neuroimmunology of the Interleukins 13 and 4. Brain Sci. 2016, 6, 18. [Google Scholar] [CrossRef]
- Hart, P.H.; Vitti, G.F.; Burgess, D.R.; Whitty, G.A.; Piccoli, D.S.; Hamilton, J.A. Potential antiinflammatory effects of interleukin 4: suppression of human monocyte tumor necrosis factor alpha, interleukin 1 and prostaglandin E2. Proc. Natl. Acad. Sci. USA. 1989, 86, 3803–3807. [Google Scholar] [CrossRef]
- Wong, H.L.; Costa, G.L.; Lotze, M.T.; Wahl, S.M. Interleukin (IL) 4 differentially regulates monocyte IL-1 family gene expression and synthesis in vitro and in vivo. J. Exp. Med. 1993, 177, 775–781. [Google Scholar] [CrossRef]
- Mitchell, R.E.; Hassan, M.; Burton, B.R.; Britton, G.; Hill, E.V.; Verhagen, J.; Wraith, D.C. IL-4 enhances IL-10 production in Th1 cells: implications for Th1 and Th2 regulation. Sci. Rep. 2017, 7, 11315. [Google Scholar] [CrossRef]
- Wen, F.-Q.; Kohyama, T.; Liu, X.; Zhu, Y.K.; Wang, H.; Kim, H.J.; Kobayashi, T.; Abe, S.; Spurzem, J.R.; Rennard, S.I. Interleukin-4– and Interleukin-13–Enhanced Transforming Growth Factor- β 2 Production in Cultured Human Bronchial Epithelial Cells Is Attenuated by Interferon- γ. Am. J. Respir. Cell Mol. Boil. 2002, 26, 484–490. [Google Scholar] [CrossRef]
- Zujovic, V.; Benavides, J.; Vigé, X.; Carter, C.; Taupin, V. Fractalkine modulates TNF-alpha secretion and neurotoxicity induced by microglial activation. Glia 2000, 29, 305–315. [Google Scholar] [CrossRef]
- Lyons, A.; Downer, E.J.; Crotty, S.; Nolan, Y.M.; Mills, K.H.G.; Lynch, M.A. CD200 Ligand–Receptor Interaction Modulates Microglial Activation In Vivo and In Vitro: A Role for IL-4. J. Neurosci. 2007, 27, 8309–8313. [Google Scholar] [CrossRef]
- Klegeris, A.; McGeer, P.L. Inflammatory cytokine levels are influenced by interactions between THP-1 monocytic, U-373 MG astrocytic, and SH-SY5Y neuronal cell lines of human origin. Neurosci. Lett. 2001, 313, 41–44. [Google Scholar] [CrossRef]
- Morganti, J.M.; Nash, K.R.; Grimmig, B.A.; Ranjit, S.; Small, B.; Bickford, P.C.; Gemma, C. The soluble isoform of CX3CL1 is necessary for neuroprotection in a mouse model of Parkinson’s disease. J. Neurosci. 2012, 32, 14592–14601. [Google Scholar] [CrossRef]
- Taj, S.H.; Le Blon, D.; Hoornaert, C.; Daans, J.; Quarta, A.; Praet, J.; Van Der Linden, A.; Ponsaerts, P.; Hoehn, M. Targeted intracerebral delivery of the anti-inflammatory cytokine IL13 promotes alternative activation of both microglia and macrophages after stroke. J. Neuroinflammation 2018, 15, 174. [Google Scholar]
- Liu, X.; Liu, J.; Zhao, S.; Zhang, H.; Cai, W.; Cai, M.; Ji, X.; Leak, R.K.; Gao, Y.; Chen, J.; et al. Interleukin-4 is Essential for Microglia/Macrophage M2 Polarization and Long-term Recovery after Cerebral Ischemia. Stroke 2016, 47, 498–504. [Google Scholar] [CrossRef]
- Tancredi, V.; D’Arcangelo, G.; Grassi, F.; Tarroni, P.; Palmieri, G.; Santoni, A.; Eusebi, F. Tumor necrosis factor alters synaptic transmission in rat hippocampal slices. Neurosci. Lett. 1992, 146, 176–178. [Google Scholar] [CrossRef]
- Bellinger, F.P.; Madamba, S.; Siggins, G.R. Interleukin 1 beta inhibits synaptic strength and long-term potentiation in the rat CA1 hippocampus. Brain Res. 1993, 628, 227–234. [Google Scholar] [CrossRef]
- Smith, J.A.; Das, A.; Ray, S.K.; Banik, N.L. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res. Bull. 2012, 87, 10–20. [Google Scholar] [CrossRef]
- Oh, C.K.; Geba, G.P.; Molfino, N. Investigational therapeutics targeting the IL-4/IL-13/STAT-6 pathway for the treatment of asthma. Eur. Respir. Rev. 2010, 19, 46–54. [Google Scholar] [CrossRef]
- Li, L.; Wu, Y.; Wang, Y.; Wu, J.; Song, L.; Xian, W.; Yuan, S.; Pei, L.; Shang, Y. Resolvin D1 promotes the interleukin-4-induced alternative activation in BV-2 microglial cells. J. Neuroinflammation 2014, 11, 72. [Google Scholar] [CrossRef]
- Korecka, J.A.; Van Kesteren, R.E.; Blaas, E.; Spitzer, S.O.; Kamstra, J.H.; Smit, A.B.; Swaab, D.F.; Verhaagen, J.; Bossers, K. Phenotypic Characterization of Retinoic Acid Differentiated SH-SY5Y Cells by Transcriptional Profiling. PLoS ONE 2013, 8, e63862. [Google Scholar] [CrossRef]
- Henn, A. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX 2009, 26, 83–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene | Species | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| ADAM10 | Human | ATGGGAGGTCAGTATGGGAATC | ACTGCTCTTTTGGCACGCT |
| β-Actin | Human | GAT GCA GAA GGA GAT CAC TGC | ATA CTC CTG CTT GCT GAT CCA |
| Caspase 3 | Human | CTCTGGTTTTCGGTGGGTGT | GCTGCATCGACATCTGTACC |
| CCR2 | Human | CCACATCTCGTTCTCGGTTTATC | CAGGGAGCACCGTAATCATAATC |
| CD200 | Human | ACGTCTGTTACCAGCATCCTC | CTTAAAGTCGGTCACAGTCCC |
| ChAT | Human | GGA GGT GGA GGG TTT GTG AC | ATT TCC TTG GCA CCC TGA GG |
| CX3CL1 | Human | ACCACGGTGTGACGAAATG | TGTTGATAGTGGATGAGCAAAGC |
| CXCL10 | Human | GACTCTGAGTGGAACTCAAGGAAT | GTGGCAATGATCTCAACACG |
| DAP12 | Human | ACTGAGACCGAGTCGCCTTAT | ATACGGCCTCTGTGTGTTGAG |
| DCX | Human | TTCAAGGGGATTGTGTACGCT | GTCAGACAGAGATCGCGTCAG |
| DR3 | Human | AGAGATACTGACTGTGGGACC | CCCAGAACACACCTACTCTGC |
| FADD | Human | GAAAACGCGCTCTTGTCGAT | GCCCGAGGCATAGGAACTTG |
| Fas | Human | GTCTCCTGCGATGTTTGGC | TTCAAGGAAAGCTGATACCTATTTC |
| HDAC1 | Human | CCGCATGACTCATAATTTGCTG | ATTGGCTTTGTGAGGGCGATA |
| HDAC2 | Human | ATGGCGTACAGTCAAGGAGG | TGCGGATTCTATGAGGCTTCA |
| HMGB1 | Human | GGA GAT CCT AAG AAG CCG AGA | CAT GGT CTT CCA CCT CTC TGA |
| HMOX1 | Human | AAGACTGCGTTCCTGCTCAAC | AAAGCCCTACAGCAACTGTCG |
| IKKβ | Human | CTGGCCTTTGAGTGCATCAC | CGCTAACAACAATGTCCACCT |
| IL-10 | Human | GACTTTAAGGGTTACCTGGGTTG | TCACATGCGCCTTGATGTCTG |
| IL-13 | Human | GAGGATGCTGAGCGGATTCTG | CACCTCGATTTTGGTGTCTCG |
| IL-13R | Human | ACAACCTGAGCTACATGAAGTG | GGCTTCTGTGCCAATAGTAGAG |
| IL-1β | Human | ATG ATG GCT TAT TAC AGT GGC AA | GTCGGAGATTCGTAGCTGGA |
| IL-4 | Human | CCAACTGCTTCCCCCTCTG | TCTGTTACGGTCAACTCGGTG |
| IL-4R | Human | ACACCAATGTCTCCGACACTC | TGTTGACTGCATAGGTGAGATGA |
| iNOS | Human | TTCAGTATCACAACCTCAGCAAG | TGGACCTGCAAGTTAAAATCCC |
| MCP1 | Human | CTCTCGCCTCCAGCATGAAA | AGGGTGTCTGGGGAAAGCTA |
| MD-2 | Human | GAA GCT CAG AAG CAG TAT TGG GTC | GGT TGG TGT AGG ATG ACA AAC TCC |
| NGFR | Human | CCTACGGCTACTACCAGGATG | CACACGGTGTTCTGCTTGT |
| RAGE | Human | CTA CCG AGT CCG TGT CTA CCA | CAT CCA AGT GCC AGC TAA GAG |
| TBK1 | Human | TGGGTGGAATGAATCATCTACGA | GCTGCACCAAAATCTGTGAGT |
| TGFβ | Human | CAATTCCTGGCGATACCTCAG | GCACAACTCCGGTGACATCAA |
| TH | Human | GGAAGGCCGTGCTAAACCT | GGATTTTGGCTTCAAACGTCTC |
| TL1A | Human | TGCAGGACTCACCACATA | CTTGGCTTATCTCCGTCT |
| TLR3 | Human | TTGCCTTGTATCTACTTTTGGGG | TCAACACTGTTATGTTTGTGGGT |
| TLR4 | Human | CCTGCGTGGAGGTGGTTCC | AGAGGTGGCTTAGGCTCTGATA |
| TLR7 | Human | GATAACAATGTCACAGCCGTCC | GTTCCTGGAGTTTGTTGATGTTC |
| TNFα | Human | CCC AGG CAG TCA GAT CAT CTT CT | ATG AGG TAC AGG CCC TCT GAT |
| TREM1 | Human | GAACTCCGAGCTGCAACTAAA | TCTAGCGTGTAGTCACATTTCAC |
| TREM2 | Human | GGTCAGCACGCACAACTTG | CGCAGCGTAATGGTGAGAGT |
| Arg1 | Mouse | TTAGGCCAAGGTGCTTGCTGCC | TACCATGGCCCTGAGGAGGTTC |
| β-Actin | Mouse | GTA TGA CTC CAC TCA CGG CAA A | GGT CTC GCT CCT GGA AGA TG |
| CCR2 | Mouse | ATCCACGGCATACTATCAACATC | CAAGGCTCACCATCATCGTAG |
| CD200R | Mouse | TAAGGTGGAGGCATTTCCAGT | GATTCCAATGGCCGACAAAGTA |
| CX3CR1 | Mouse | TCTTCACGTTCGGTCTGGTG | TGCACTGTCCGGTTGTTCAT |
| DAP12 | Mouse | GAGTGACACTTTCCCAAGATGC | CCTTGACCTCGGGAGACCA |
| DR3 | Mouse | CTCACCTTTCTCTTGTGTCCC | GTGCAGTCATTGCCACGTAT |
| HMGB1 | Mouse | CGC GGA GGA AAA TCA ACT AA | TCA TAA CGA GCC TTG TCA GC |
| IKKβ | Mouse | GGAGCCTGGGAAATGAAAGAA | GCCAGAGCCCTACCTGATTG |
| IL-10 | Mouse | GGC AGA GAA GCA TGG CCC AGA A | GGC TGA GGC GCT GTC ATC GAT T |
| IL-13 | Mouse | CCTGGCTCTTGCTTGCCTT | GGTCTTGTGTGATGTTGCTCA |
| IL-13R | Mouse | TCAGCCACCTGTGACGAATTT | TGAGAGTGCAATTTGGACTGG |
| IL-1β | Mouse | CTG GTG TGT GAC GTT CCC ATT A | CCG ACA GCA CGA GGC TTT |
| IL-4 | Mouse | TGGGTCTCAACCCCCAGCTAGT | TGCATGGCGTCCCTTCTCCTGT |
| IL-4R | Mouse | TCTGCATCCCGTTGTTTTGC | GCACCTGTGCATCCTGAATG |
| iNOS | Mouse | GTGGTGACAAGCACATTTGG | CATGGTGAACACGTTCTTGG |
| MCP1 | Mouse | ACT GAA GCC AGC TCT CTC TTC CTC | TTC CTT CTT GGG GTC AGC ACA GAC |
| MD-2 | Mouse | CGC TGC TTT CTC CCA TAT TGA | CCT CAG TCT TAT GCA GGG TTC A |
| RAGE | Mouse | GAA GGC TCT GTG GGT GAG TC | CCG CTT CCT CTG ACT GAT TC |
| TBK1 | Mouse | ACTGGTGATCTCTATGCTGTCA | TTCTGGAAGTCCATACGCATTG |
| TGFβ | Mouse | CTCCCGTGGCTTCTAGTGC | GCCTTAGTTTGGACAGGATCTG |
| TLR3 | Mouse | GTG AGA TAC AAC GTA GCT GAC TG | TCC TGC ATC CAA GAT AGC AAG T |
| TLR4 | Mouse | ATG GCA TGG CTT ACA CCA CC | GAG GCC AAT TTT GTC TCC ACA |
| TLR7 | Mouse | ATG TGG ACA CGG AAG AGA CAA | GGT AAG GGT AAG ATT GGT GGT G |
| TNFα | Mouse | GAC CCT CAC ACT CAG ATC ATC TTC T | CCT CCA CTT GGT GGT TTG CT |
| TREM1 | Mouse | GACTGCTGTGCGTGTTCTTTG | GCCAAGCCTTCTGGCTGTT |
| TREM2 | Mouse | CTGGAACCGTCACCATCACTC | CGAAACTCGATGACTCCTCGG |
| β-Actin | Rat | CTACAATGAGCTGCGTGT | CAGGTCCAGACGCAGGAT |
| IL-1β | Rat | TTGTGCAAGTGTCTGAAGCA | TGTCAGCCTCAAAGAACAGG |
| TNFα | Rat | ATGTGGAACTGGCAGAGGAG | ACGAGCAGGAATGAGAAGAGG |
| BV2 Microglia | ||||
|---|---|---|---|---|
| Gene | CON-Alone | EtOH-Alone | CON-CC | EtOH-CC |
| NFκB signaling genes | ||||
| CCR2 | 100 ± 11 | 185 ± 29 * | 177 ± 23 ns | 294 ± 16@@,## |
| HMGB1 | 100 ± 12 | 109 ± 13 | 107 ± 6.9 | 67 ± 8.3 |
| IKKβ | 100 ± 29 | 90 ± 21 | 367 ± 11 **** | 500 ± 45@@@@,# |
| MCP1 | 100 ± 15 | 62 ± 21 | 58 ± 5.6 | 19 ± 0.7# |
| MD-2 | 100 ± 21 | 147 ± 22 ns | 187 ± 5.9 * | 173 ± 22 |
| RAGE | 100 ± 8.5 | 117 ± 12 | 342 ± 55 *** | 340 ± 33@@@ |
| TBK1 | 100 ± 23 | 106 ± 22 | 631 ± 61 **** | 666 ± 65@@@@ |
| Microglial markers | ||||
| Arg1 | 100 ± 21 | 73 ± 15 | 160 ± 5.6 * | 137 ± 5.3@ |
| CD200R | 100 ± 27 | 217 ± 46 ns | 320 ± 27 * | 658 ± 71@@@@,### |
| CX3CR1 | 100 ± 23 | 122 ± 20 | 203 ± 32 * | 234 ± 20@ |
| TREM signaling genes | ||||
| DAP12 | 100 ± 6.6 | 187 ± 8.2 **** | 75 ± 2.3 ** | 124 ± 4.4@@@@,#### |
| TREM1 | 100 ± 11 | 114 ± 18 | 401 ± 34 ns | 1144 ± 199@@@,## |
| TREM2 | 100 ± 13 | 138 ± 3 | 86 ± 8.9 | 125 ± 7.1 |
| TNF superfamily genes | ||||
| DR3 | 100 ± 12 | 199 ± 36 ns | 341 ± 24 ns | 2261 ± 429@@@@,#### |
| SH-SY5Y neuroblastoma | ||||
|---|---|---|---|---|
| Gene | CON-Alone | EtOH-Alone | CON-CC | EtOH-CC |
| NFκB signaling genes | ||||
| CCR2 | 100 ± 24 | 187 ± 26 | 98 ± 14 | 142 ± 45 |
| CXCL10 | 100 ± 32 | 84 ± 12 | 210 ± 32 * | 85 ± 18# |
| HMGB1 | 100 ± 5.9 | 71 ± 8.9 | 42 ± 12 | 102 ± 22# |
| HMOX1 | 100 ± 8.5 | 580 ± 95 **** | 126 ± 7.5 | 186 ± 25 ns |
| IKKβ | 100 ± 7.5 | 258 ± 57 * | 202 ± 12 | 490 ± 47@@,### |
| MCP1 | 100 ± 29 | 42 ± 11 | 180 ± 19 * | 77 ± 7.7## |
| MD-2 | n.d. | n.d. | n.d. | n.d. |
| RAGE | 100 ± 6.2 | 233 ± 38 ns | 810 ± 63 **** | 796 ± 114@@@ |
| TBK1 | 100 ± 33 | 185 ± 47 | 243 ± 24 ns | 642 ± 107@@@,## |
| TNF superfamily and death receptor pathway genes | ||||
| Caspase 3 | 100 ± 19 | 243 ± 34 ns | 525 ± 78 ** | 636 ± 134@@ |
| DR3 | 100 ± 33 | 178 ± 33 | 561 ± 28 **** | 702 ± 70@@@@ |
| FADD | 100 ± 7.1 | 124 ± 11.5 | 171 ± 18 | 330 ± 58@@@,# |
| Fas | 100 ± 11.8 | 861 ± 352 ns | 1203 ± 357 ns | 2422 ± 238@ |
| TL1A | n.d. | n.d. | 100 ± 19 | 43 ± 7.1# |
| NGFR | 100 ± 12 | 5127 ± 471**** | 42 ± 1.7 | 331 ± 80@@@@ |
| Neuron-specific genes | ||||
| CD200 | 100 ± 26 | 183 ± 61 | 290 ± 14 ** | 604 ± 67@@@@,### |
| CX3CL1 | 100 ± 6.3 | 123 ± 7.0 | 213 ± 17 **** | 274 ± 12@@@@, ## |
| ChAT | 100 ± 16 | 144 ± 43 | 177 ± 29 | 234 ± 38 |
| DCX | 100 ± 26 | 108 ± 32 | 729 ± 40 **** | 732 ± 54@@@@ |
| TH | 100 ± 15 | 124 ± 40 | 668 ± 113 **** | 110 ± 27#### |
| TREM signaling genes | ||||
| DAP12 | 100 ± 20 | 70 ± 5.2 | 13701 ± 3140 *** | 11800 ± 1552@@@ |
| TREM1 | 100 ± 23 | 177 ± 45 | 485 ± 221 | 1506 ± 343@@@,# |
| TREM2 | 100 ± 23 | 140 ± 40 | 100 ± 14 | 172 ± 25 |
| Proteases | ||||
| ADAM10 | 100 ± 33 | 199 ± 65 | 700 ± 56 ** | 1429 ± 161@@@@,### |
| Histone deacetylases | ||||
| HDAC1 | 100 ± 11.7 | 131 ± 7.0 | 185 ± 8.8 | 336 ± 64@@ |
| HDAC2 | 100 ± 11.7 | 193 ± 23 | 354 ± 72 * | 378 ± 84 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lawrimore, C.J.; Coleman, L.G.; Zou, J.; Crews, F.T. Ethanol Induction of Innate Immune Signals Across BV2 Microglia and SH-SY5Y Neuroblastoma Involves Induction of IL-4 and IL-13. Brain Sci. 2019, 9, 228. https://doi.org/10.3390/brainsci9090228
Lawrimore CJ, Coleman LG, Zou J, Crews FT. Ethanol Induction of Innate Immune Signals Across BV2 Microglia and SH-SY5Y Neuroblastoma Involves Induction of IL-4 and IL-13. Brain Sciences. 2019; 9(9):228. https://doi.org/10.3390/brainsci9090228
Chicago/Turabian StyleLawrimore, Colleen J., Leon G. Coleman, Jian Zou, and Fulton T. Crews. 2019. "Ethanol Induction of Innate Immune Signals Across BV2 Microglia and SH-SY5Y Neuroblastoma Involves Induction of IL-4 and IL-13" Brain Sciences 9, no. 9: 228. https://doi.org/10.3390/brainsci9090228





