Molecular and Clinical Characterization of a Novel Prognostic and Immunologic Biomarker GPSM3 in Low-Grade Gliomas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Processing
2.2. Differential Expression Analysis and Survival Analysis
2.3. Gene Set Enrichment Analysis (GSEA), Gene Set Variation Analysis (GSVA), and Functional Enrichment Analysis
2.4. Immune Cell Infiltration Analysis
2.5. Clinical Samples and RT-qPCR Analysis
2.6. Statistical Analysis and Plot Generation
3. Results
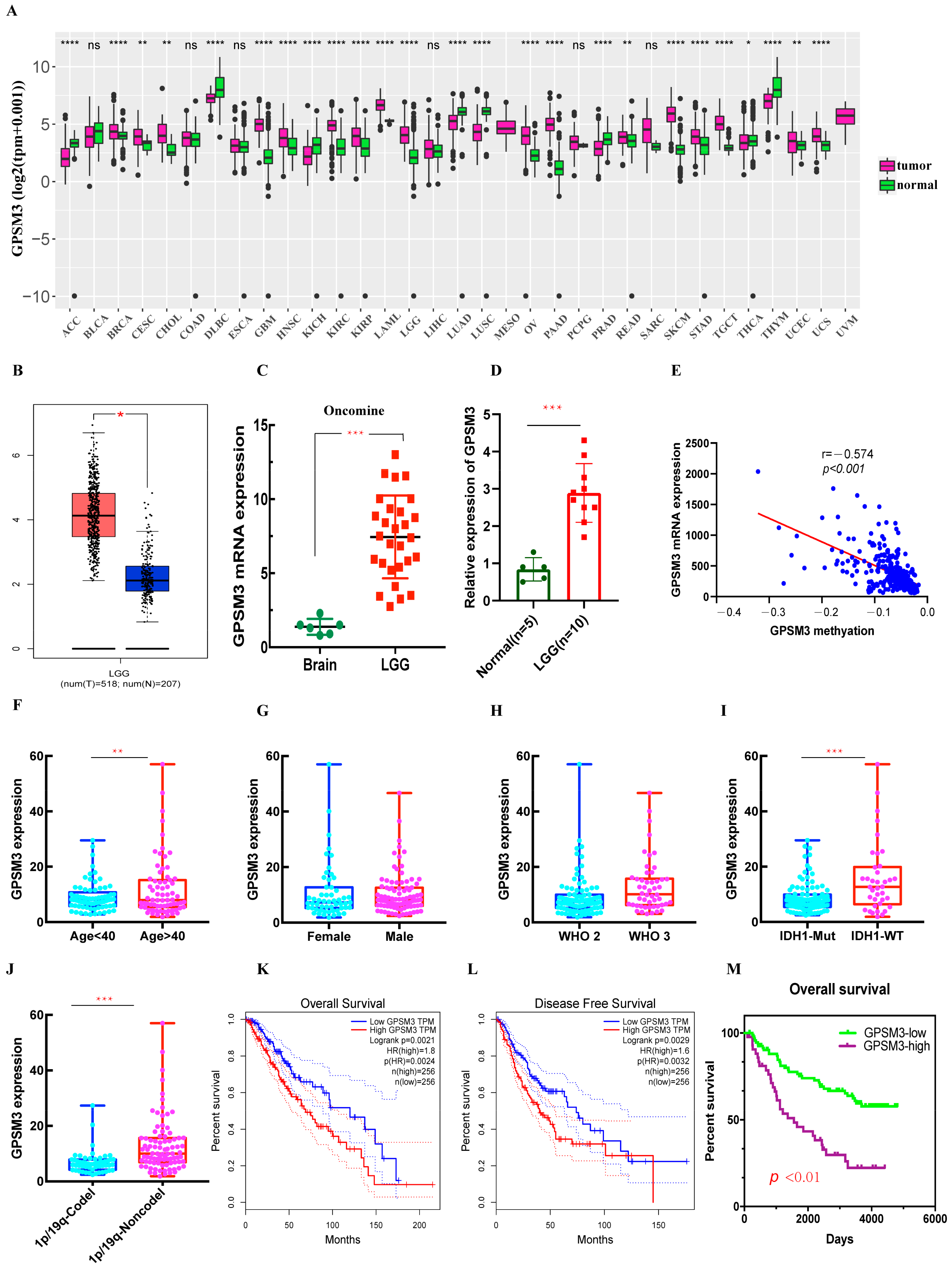
3.1. The mRNA Expression of GPSM3 in LGGs
3.2. High Level of Expression of GPSM3 Predicted an Unfavorable Prognosis in the Patients with LGG
3.3. The Potential Functions of GPSM3
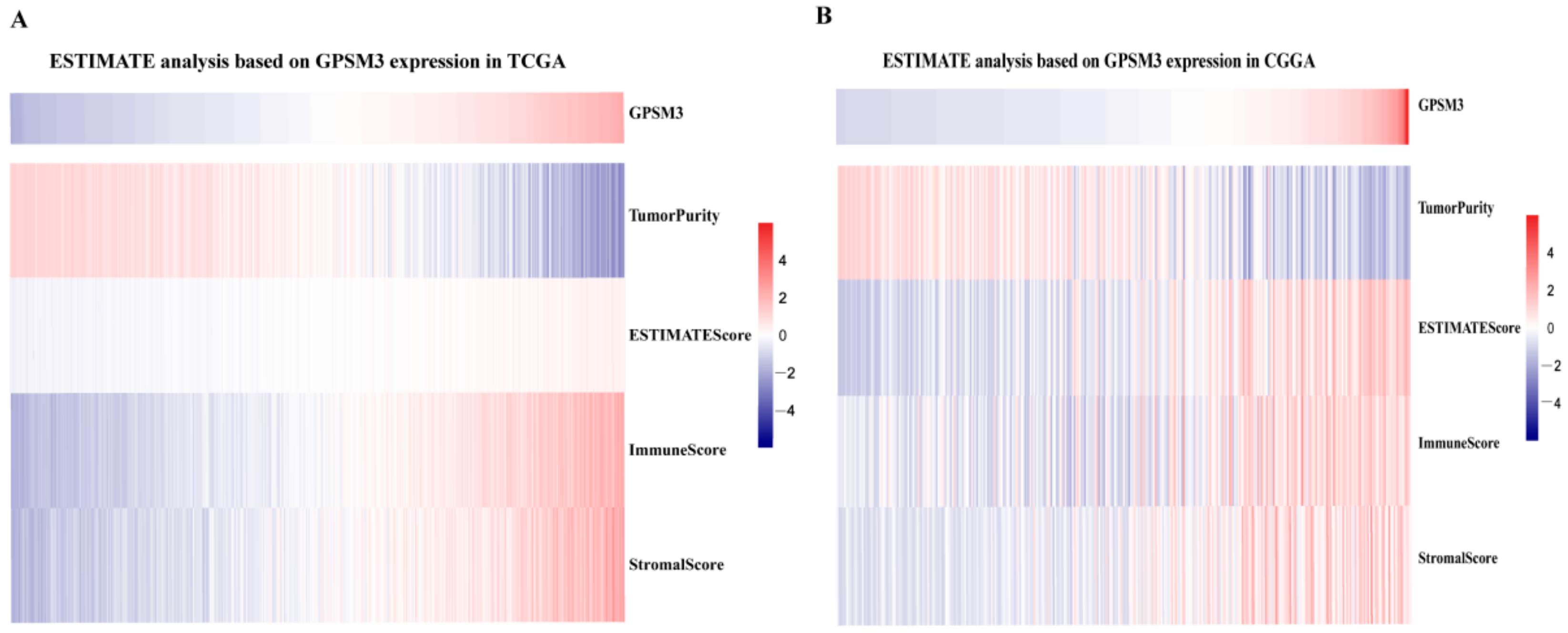
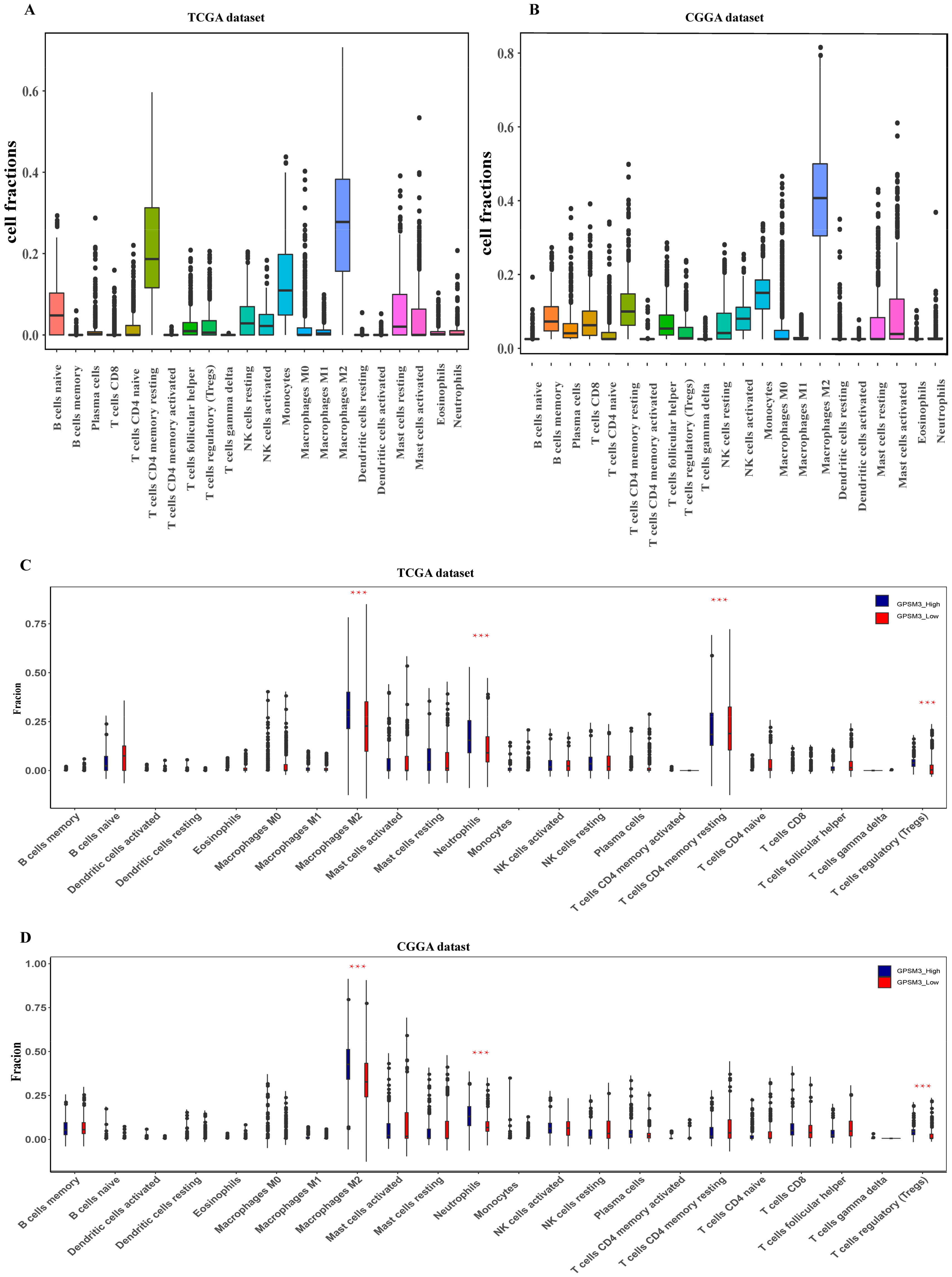
3.4. GPSM3 Regulated the Infiltration of Immune Cells in the LGGs
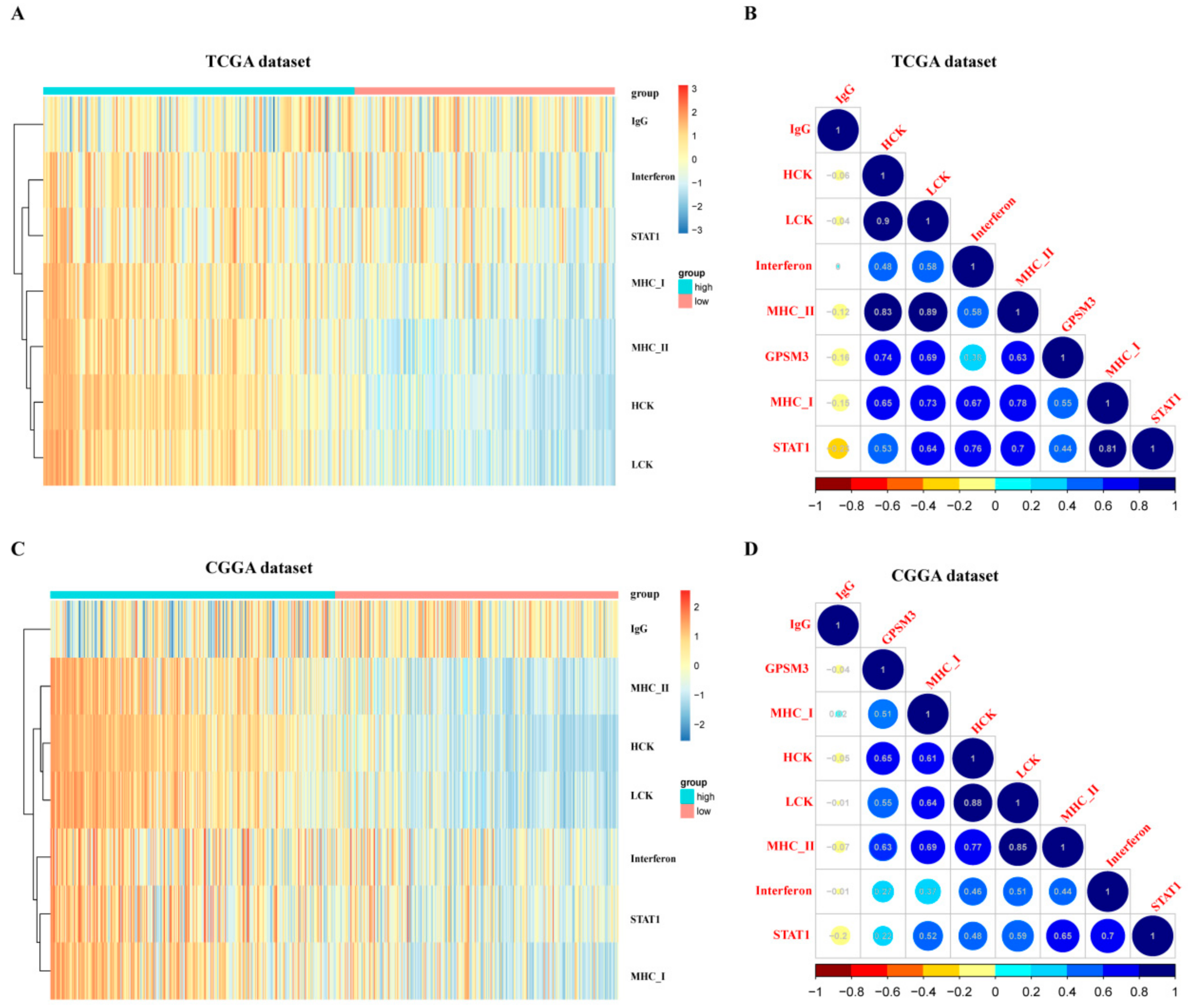
3.5. GPSM3-Related Inflammatory Responses
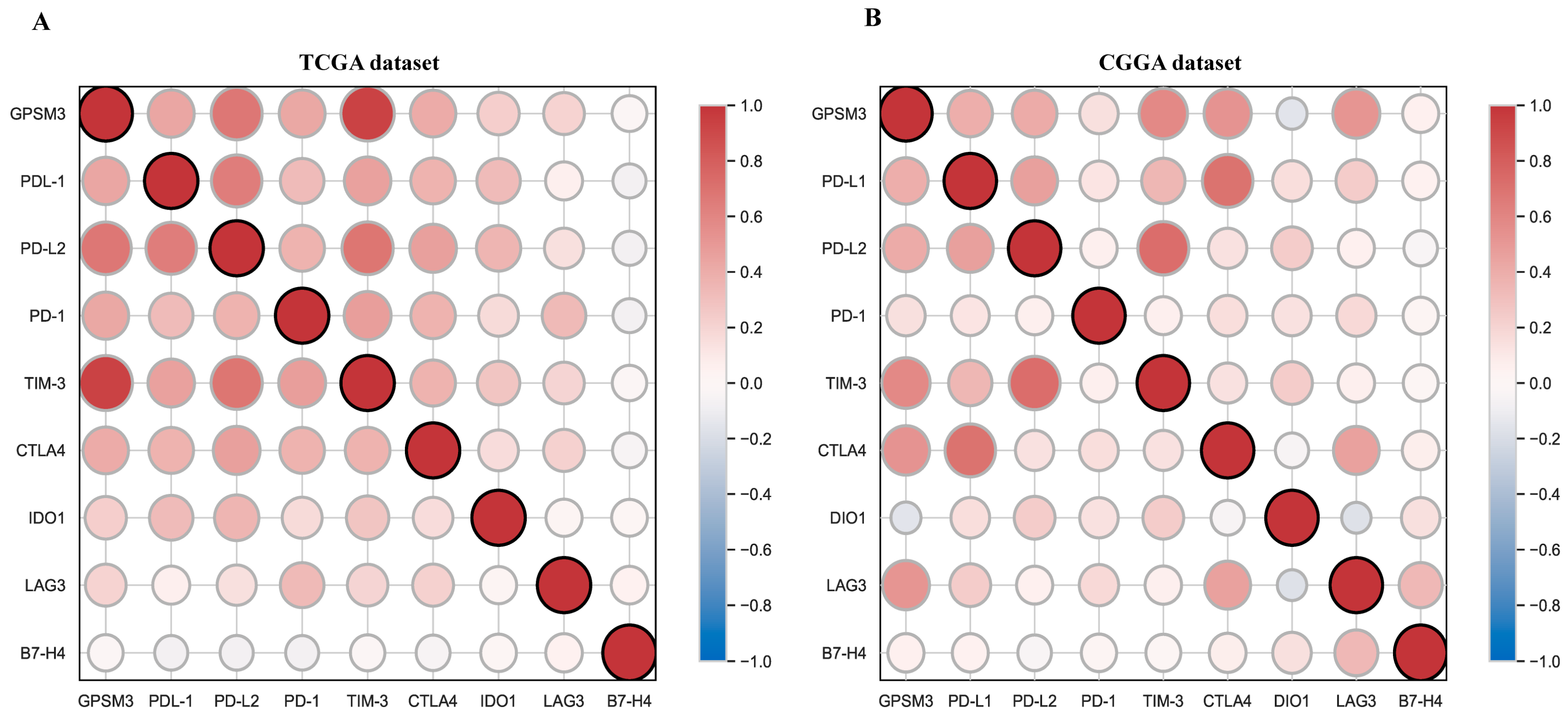
3.6. Correlation Analysis between GPSM3 and Immune Checkpoints
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Stetson, L.; Virk, S.; Barnholtz-Sloan, J.S. Epidemiology of Intracranial Gliomas. Prog. Neurol. Surg. 2018, 30, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Zhang, S.; Gong, Z.; Li, X.; Cao, K.; Deng, H.; He, Y.; et al. The role of microenvironment in tumor angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [Green Version]
- Ribas, A. Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov. 2015, 5, 915–919. [Google Scholar] [CrossRef] [Green Version]
- Abbott, M.; Ustoyev, Y. Cancer and the Immune System: The History and Background of Immunotherapy. Semin. Oncol. Nurs. 2019, 35, 150923. [Google Scholar] [CrossRef]
- Latchman, Y.E.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001, 2, 261–268. [Google Scholar] [CrossRef]
- Parry, R.V.; Chemnitz, J.M.; Frauwirth, K.A.; Lanfranco, A.R.; Braunstein, I.; Kobayashi, S.V.; Linsley, P.S.; Thompson, C.B.; Riley, J.L. CTLA-4 and PD-1 Receptors Inhibit T-Cell Activation by Distinct Mechanisms. Mol. Cell. Biol. 2005, 25, 9543–9553. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Chan, E.C.; Druey, K.M. R4 Regulator of G Protein Signaling (RGS) Proteins in Inflammation and Immunity. AAPS J. 2016, 18, 294–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arang, N.; Gutkind, J.S. G Protein-Coupled receptors and heterotrimeric G proteins as cancer drivers. FEBS Lett. 2020, 594, 4201–4232. [Google Scholar] [CrossRef] [PubMed]
- Squires, K.E.; Montanez-Miranda, C.; Pandya, R.R.; Torres, M.; Hepler, J.R. Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease. Pharmacol. Rev. 2018, 70, 446–474. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Ren, H.; Yang, X.; Zhu, H.; Yao, L.; Hang, Q.; Mao, H.; Huang, Y.; Zhang, J.; Wang, Y. Overexpression of activator of G-protein signaling 3 decreases the proliferation of esophageal squamous cell carcinoma. Pathol.-Res. Pract. 2015, 211, 449–455. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, Z.; Liu, B.; Hou, K.; Che, X.; Qu, X.; Liu, Y.; Hu, X.; Zhang, Y.; Lv, Q. Localization of GPSM2 in the Nucleus of Invasive Breast Cancer Cells Indicates a Poor Prognosis. Front. Oncol. 2020, 10, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.-Q.; Zhang, Y.-F.; Yu, J.-J.; Gan, Y.-Y.; Han, N.-N.; Zhang, M.-X.; Ge, W.; Deng, J.-J.; Zheng, Y.-F.; Xu, X.-M. High expression of G-protein signaling modulator 2 in hepatocellular carcinoma facilitates tumor growth and metastasis by activating the PI3K/AKT signaling pathway. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Dang, S.-C.; Qian, X.-B.; Jin, W.; Cui, L.; Chen, J.-X.; Gu, M. G-protein-signaling modulator 2 expression and role in a CD133+ pancreatic cancer stem cell subset. OncoTargets Ther. 2019, 12, 785–794. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Xiong, Y.; Zhao, L.; Gu, K.; Li, Y.; Zhao, F.; Li, J.; Wang, M.; Wang, H.; Tao, Z.; et al. UCSCXenaShiny: An R/CRAN Package for Interactive Analysis of UCSC Xena Data. Bioinformatics 2021, 37, btab561. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pander, A.; Chinnaiyan, A.M. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rody, A.; Holtrich, U.; Pusztai, L.; Liedtke, C.; Gaetje, R.; Ruckhaeberle, E.; Solbach, C.; Hanker, L.; Ahr, A.; Metzler, D.; et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009, 11, R15. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Delgado-López, P.D.; Corrales-García, E.M.; Martino, J.; Lastra-Aras, E.; Dueñas-Polo, M.T. Diffuse low-grade glioma: A review on the new molecular classification, natural history and current management strategies. Clin. Transl. Oncol. 2017, 19, 931–944. [Google Scholar] [CrossRef]
- Ladomersky, E.; Zhai, L.; Lauing, K.L.; Bell, A.; Xu, J.; Kocherginsky, M.; Zhang, B.; Wu, J.D.; Podojil, J.R.; Platanias, L.C.; et al. Advanced Age Increases Immunosuppression in the Brain and Decreases Immunotherapeutic Efficacy in Subjects with Glioblastoma. Clin. Cancer Res. 2020, 26, 5232–5245. [Google Scholar] [CrossRef] [PubMed]
- Court, F.; Le Boiteux, E.; Fogli, A.; Müller-Barthélémy, M.; Vaurs-Barrière, C.; Chautard, E.; Pereira, B.; Biau, J.; Kemeny, J.-L.; Khalil, T.; et al. Transcriptional alterations in glioma result primarily from DNA methylation–independent mechanisms. Genome Res. 2019, 29, 1605–1621. [Google Scholar] [CrossRef] [Green Version]
- Aldape, K.; Zadeh, G.; Mansouri, S.; Reifenberger, G.; von Deimling, A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 829–848. [Google Scholar] [CrossRef]
- Golebiewska, A.; Bougnaud, S.; Stieber, D.; Brons, N.H.C.; Vallar, L.; Hertel, F.; Klink, B.; Schröck, E.; Bjerkvig, R.; Niclou, S. Side population in human glioblastoma is non-tumorigenic and characterizes brain endothelial cells. Brain 2013, 136, 1462–1475. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Lei, Y.; Li, J.K.; Du, W.X.; Li, R.G.; Yang, J.; Li, J.; Li, F.; Tan, H.B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020, 470, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Fujiwara, Y.; Ohnishi, K.; Takeya, M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 2016, 99, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Liang, W.; Ferrara, N. The Complex Role of Neutrophils in Tumor Angiogenesis and Metastasis. Cancer Immunol. Res. 2016, 4, 83–91. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Piao, Y.; Holmes, L.; Fuller, G.; Henry, V.; Tiao, N.; de Groot, J.F. Neutrophils Promote the Malignant Glioma Phenotype through S100A4. Clin. Cancer Res. 2014, 20, 187–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Zhong, W.; Li, Q. Role of heterogeneous regulatory T cells in the tumor microenvironment. Pharmacol. Res. 2020, 153, 104659. [Google Scholar] [CrossRef]
- Deng, G.; Song, X.; Fujimoto, S.; Piccirillo, C.A.; Nagai, Y.; Greene, M.I. Foxp3 Post-translational Modifications and Treg Suppressive Activity. Front. Immunol. 2019, 10, 2486. [Google Scholar] [CrossRef] [Green Version]
- Petitprez, F.; Meylan, M.; De Reyniès, A.; Sautes-Fridman, C.; Fridman, W.H. The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front. Immunol. 2020, 11, 784. [Google Scholar] [CrossRef]



| Datasets | Characteristic | HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|---|---|
| TCGA | Univariate | Multivariate | ||||||
| Age | 2.89 | 1.98–4.23 | 0.000 | Age | 3.24 | 2.14–4.91 | 0.000 | |
| Ethnicity | 1.26 | 0.46–3.44 | 0.645 | Ethnicity | 0.92 | 0.32–2.57 | 0.873 | |
| Gender | 1.07 | 0.75–1.53 | 0.692 | Gender | 1.19 | 0.83–1.74 | 0.336 | |
| Radiation | 2.07 | 0.89–4.79 | 0.088 | Radiation | 1.05 | 0.72–1.53 | 0.786 | |
| GPSM3 | 2.41 | 1.65–3.54 | 0.000 | GPSM3 | 2.08 | 1.36–3.21 | 0.0001 | |
| CGGA | Univariate | Multivariate | ||||||
| Gender | 2.91 | 1.92–4.41 | 0.000 | Gender | 1.61 | 1.07–2.41 | 0.019 | |
| Age | 1.27 | 0.90–1.81 | 0.016 | Age | 1.44 | 0.99–2.09 | 0.031 | |
| Radio_status | 1.37 | 0.89–2.09 | 0.145 | Radio_status | 1.27 | 0.78–2.06 | 0.328 | |
| Chemo_status | 0.94 | 0.64–1.38 | 0.788 | Chemo_status | 0.64 | 0.41–1.00 | 0.448 | |
| IDH_mutation | 2.18 | 1.50–3.16 | 0.000 | IDH_mutation | 2.34 | 1.50–3.66 | 0.000 | |
| X1p19q_codeletion | 2.78 | 1.78–4.33 | 0.000 | X1p19q_codeletion | 1.08 | 0.51–2.28 | 0.831 | |
| MGMTp_methylation | 1.16 | 0.82–1.64 | 0.038 | MGMTp_methylation | 0.91 | 0.62–1.34 | 0.041 | |
| GPSM3 | 1.24 | 0.878–1.76 | 0.000 | GPSM3 | 1.09 | 0.73–1.63 | 0.005 |
| TCGA-KEGG | SIZE | ES | NES | NOM p-Value | CGGA-KEGG | SIZE | ES | NES | NOM p-Value |
|---|---|---|---|---|---|---|---|---|---|
| LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION | 116 | 0.61 | 2.19 | 0 | ANTIGEN_PROCESSING_AND_PRESENTATION | 80 | 0.68 | 2.38 | 0 |
| B_CELL_RECEPTOR_SIGNALING_PATHWAY | 75 | 0.61 | 2.05 | 0 | PRIMARY_IMMUNODEFICIENCY | 65 | 0.77 | 2.36 | 0 |
| PRIMARY_IMMUNODEFICIENCY | 65 | 0.74 | 2.01 | 0 | INTESTINAL_IMMUNE_NETWORK_FOR_IGA | 56 | 0.62 | 2.29 | 0 |
| ACUTE_MYELOID_LEUKEMIA | 57 | 0.52 | 1.77 | 0.001 | CYTOKINE_RECEPTOR_INTERACTION | 115 | 0.63 | 2.26 | 0 |
| CELL_ADHESION_MOLECULES_CAMS | 131 | 0.56 | 1.93 | 0.001 | B_CELL_RECEPTOR_SIGNALING_PATHWAY | 74 | 0.61 | 2.22 | 0 |
| NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 132 | 0.54 | 1.96 | 0.004 | TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 101 | 0.58 | 2.19 | 0.001 |
| ANTIGEN_PROCESSING_AND_PRESENTATION | 81 | 0.66 | 2.02 | 0.004 | CELL_ADHESION_MOLECULES_CAMS | 128 | 0.55 | 2.06 | 0.004 |
| T_CELL_RECEPTOR_SIGNALING_PATHWAY | 107 | 0.52 | 1.84 | 0.005 | NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 131 | 0.55 | 2.04 | 0.004 |
| CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 263 | 0.55 | 1.92 | 0.006 | JAK_STAT_SIGNALING_PATHWAY | 151 | 0.55 | 2.04 | 0.005 |
| NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 62 | 0.56 | 1.84 | 0.009 | T_CELL_RECEPTOR_SIGNALING_PATHWAY | 105 | 0.54 | 1.99 | 0.006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Jia, J.; Cui, Y.; Peng, Y.; Jiang, Y. Molecular and Clinical Characterization of a Novel Prognostic and Immunologic Biomarker GPSM3 in Low-Grade Gliomas. Brain Sci. 2021, 11, 1529. https://doi.org/10.3390/brainsci11111529
Wang M, Jia J, Cui Y, Peng Y, Jiang Y. Molecular and Clinical Characterization of a Novel Prognostic and Immunologic Biomarker GPSM3 in Low-Grade Gliomas. Brain Sciences. 2021; 11(11):1529. https://doi.org/10.3390/brainsci11111529
Chicago/Turabian StyleWang, Ming, Jiaoying Jia, Yan Cui, Yong Peng, and Yugang Jiang. 2021. "Molecular and Clinical Characterization of a Novel Prognostic and Immunologic Biomarker GPSM3 in Low-Grade Gliomas" Brain Sciences 11, no. 11: 1529. https://doi.org/10.3390/brainsci11111529





