Regression Networks for Neurophysiological Indicator Evaluation in Practicing Motor Imagery Tasks
Abstract
:1. Introduction
2. Material & Methods
2.1. Electrophysiological Indicators in Mi Tasks
2.2. Regression Analysis between Classifier Performance and Electrophysiological Indicators
- –
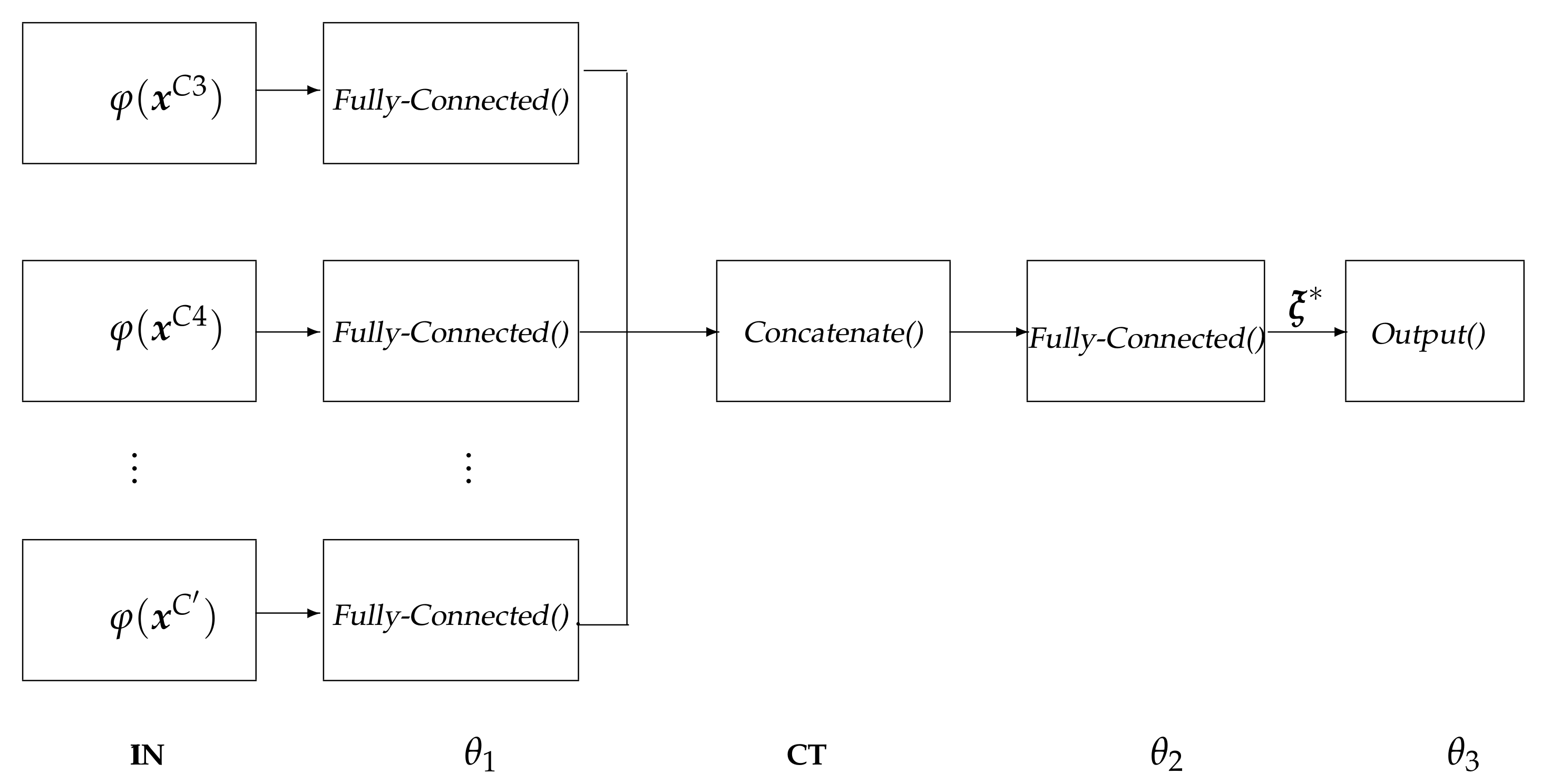
- IN: input layer that holds the extracted relevant patterns .
- –
- : fully-Connected layer that is used for extracting robust and epileptic relevant patterns that are mapped into a high-dimensional latent space [44], holding neurons, being ⌜·⌝ the ceiling operator.
- –
- CT: a concatenate layer that condenses the resulting feature sets of all electrodes into a single block, sizing .
- –
- : a fully-connected layer with size that is linked to each output-layer neuron.
- –
- : the one-neuron regression equipped with a linear activation function to predict the response.
- –
- The set of relevant patterns that holds elements extracted by the following statistical moments: mean, median, variance, minimal, and maximal values. For every subject, the moments are estimated over data using a short-time window lasting 1 s with a overlap. All time-varying moments are concatenated to form a single set per channel.
- –
- Both layers, and , employ a hyperbolic tangent (tanh) as the activation function.
- –
- During learning, Adam algorithm optimizer and loss function are used, measuring the Mean Absolute Error and fixing the learning rate to . In addition, the weight values (empirically set to ) are regularized while using the Elastic Net regularization.
- –
- The backpropagation algorithm solves the parameter set optimization of with auto differentiation under a Wide Deep Neural Network framework that includes two hidden layers under elastic-net regularization.
- –
- As the function mapping , two operators over the response vectors are tested: (a) the mean accuracy (noted as mean) that is averaged across the extraction window lengths and weighted by the subject variance performed at each window; (b) first PCA component of the accuracy vectors (noted as PCA). The set is the subject accuracy values evaluated at four lengths of feature extraction s, and performed over the whole trail MI data set, as explained before in Section 3.2.
- –
- For evaluation purposes, we also contrast the DRN-based regression analysis with the case of avoiding the data-driven indicator extraction. Which is, the estimator in Equation (5) is directly fed by the scalar-valued neurophysiological indicators devised in Equations (1a) and (3), fixing each individual vector element of to and removing the concatenation layer CT.
3. Experimental Set-Up
3.1. MI Database Description and Pre-Processing
3.2. Bi-Class Accuracy Estimation as a Response Variable
4. Results and Discussion
4.1. Computation of Pre-Training Desynchronization Indicator
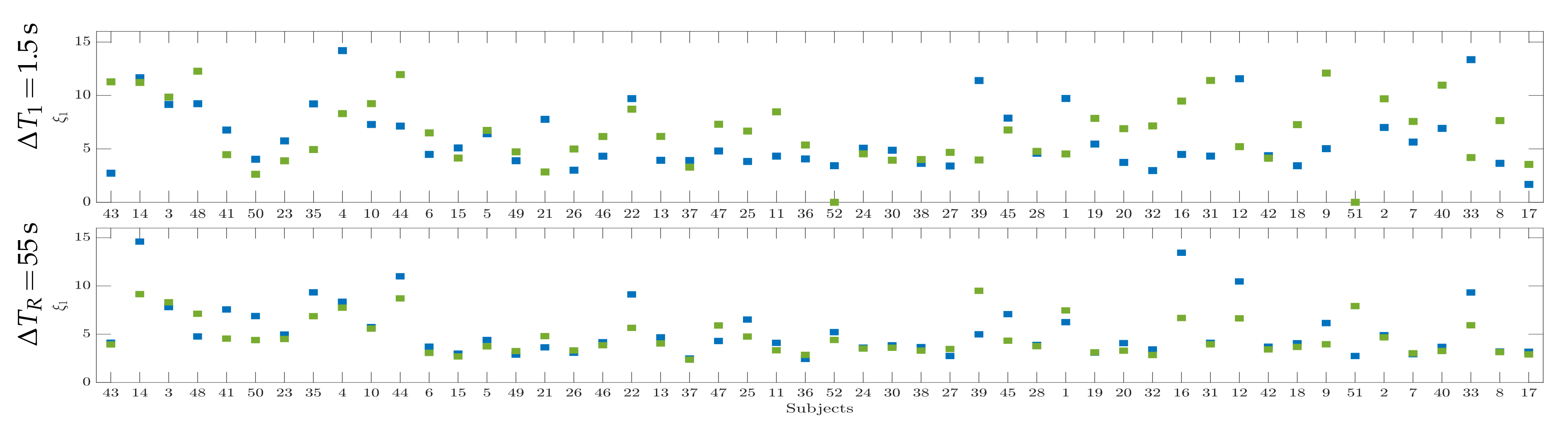
4.2. Initial Training Synchronization Assessment
4.3. Drn-Based Indicator Extraction and Regression
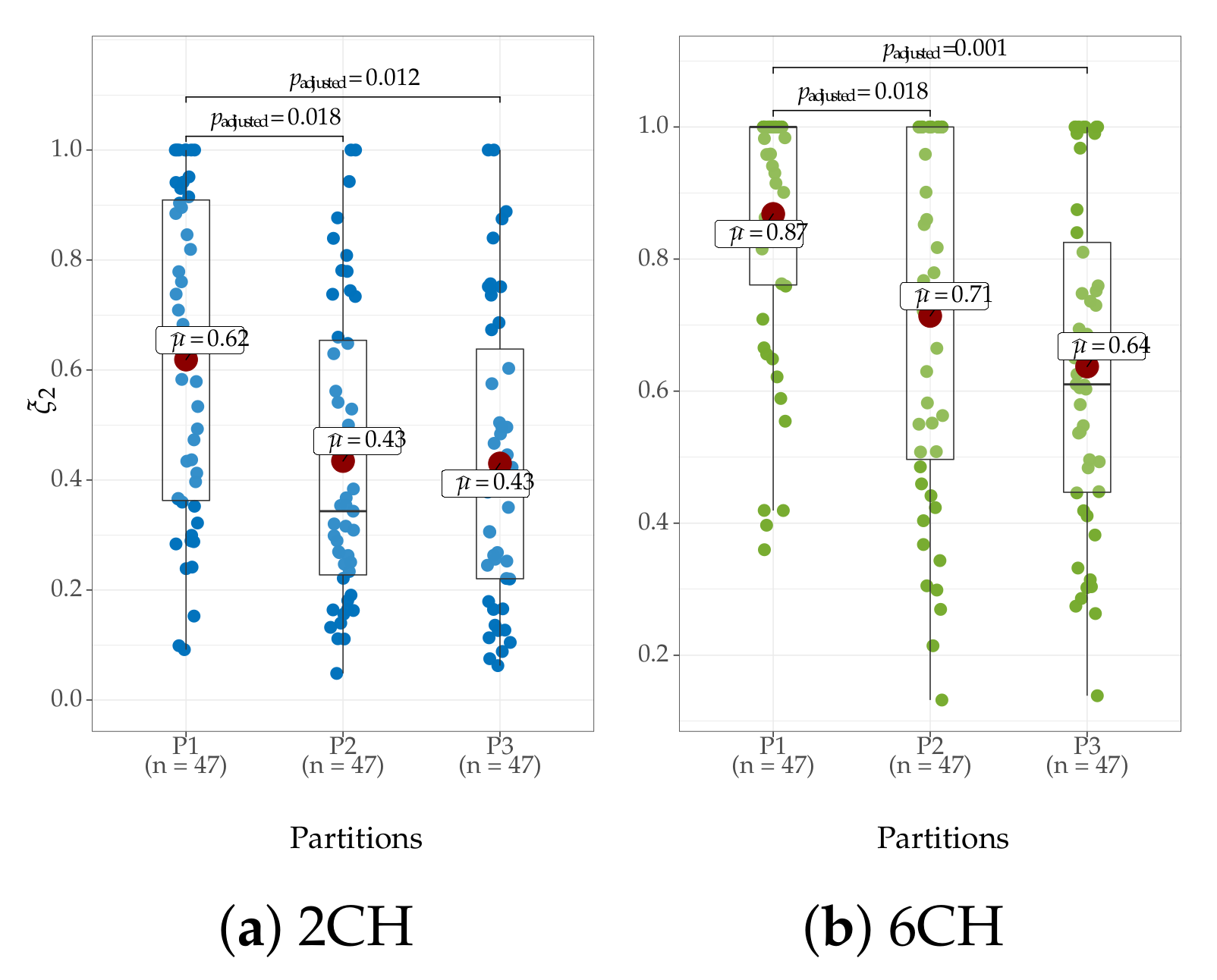
4.4. Clustering of Subject-Level Efficiency
- (i)
- A group that holds the individuals performing the best accuracy with very low variability (yellow color).
- (ii)
- A group that contains the subjects that reach important values of accuracy, but performing with some fluctuations.
- (iii)
- A group with modest accuracy performed with high unevenness.
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Eaves, D.; Riach, M.; Holmes, P.; Wright, D. Motor Imagery during Action Observation: A Brief Review of Evidence, Theory and Future Research Opportunities. Front. Neurosci. 2016, 10, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, E.; Richardson, M.; Nestel, D. Mental imagery and learning: A qualitative study in orthopaedic trauma surgery. Med. Educ. 2015, 49, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Agosti, V.; Sirico, M. Motor imagery as a tool for motor learning and improving sports performance: A mini review on the state of the art. Sport Sci. 2020, 13, 13–17. [Google Scholar]
- Lee, D.; Hwang, S. Motor imagery on upper extremity function for persons with stroke: A systematic review and meta-analysis. Phys. Ther. Rehabil. Sci. 2019, 8, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Yuan, Y.; Luo, L.; Su, W.; Zhao, K.; Xu, C.; Huang, J.; Pi, M. Hybrid brain/muscle signals powered wearable walking exoskeleton enhancing motor ability in climbing stairs activity. IEEE Trans. Med Robot. Bionics 2019, 1, 218–227. [Google Scholar] [CrossRef]
- Frau-Meigs, D. Media Education. A Kit for Teachers, Students, Parents and Professionals; UNESCO: Paris, France, 2007. [Google Scholar]
- Power, L.; Neyedli, H.; Boe, G.; Bardouille, T. Efficacy of low-cost wireless neurofeedback to modulate brain activity during motor imagery. Biomed. Phys. Eng. Express 2020, 6, 035024. [Google Scholar] [CrossRef]
- León, J.; Escobar, J.; Ortiz, A.; Ortega, J.; González, J.; Martín-Smith, P.; Gan, J.; Damas, M. Deep learning for EEG-based Motor Imagery classification: Accuracy-cost trade-off. PLoS ONE 2020, 15, e0234178. [Google Scholar] [CrossRef]
- Appriou, A.; Cichocki, A.; Lotte, F. Modern Machine-Learning Algorithms: For Classifying Cognitive and Affective States From Electroencephalography Signals. IEEE Syst. Man Cybern. Mag. 2020, 6, 29–38. [Google Scholar] [CrossRef]
- Muller, K. Analysing the Changing Brain: Immediate Brain Plasticity After One Hour of BCI. In Proceedings of the 2020 8th International Winter Conference on Brain-Computer Interface (BCI), Gangwon-do, Korea, 26–28 February 2020; pp. 1–2. [Google Scholar]
- Zhuang, M.; Wu, Q.; Wan, F.; Hu, Y. State-of-the-art non-invasive brain-computer interface for neural rehabilitation: A review. J. Neurorestoratol. 2020, 8, 12. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M. Critiquing the Concept of BCI Illiteracy. Sci. Eng. Ethics 2019, 25, 1217–12133. [Google Scholar] [CrossRef]
- Liu, T.; Huang, G.; Jiang, N.; Yao, L.; Zhang, Z. Reduce brain computer interface inefficiency by combining sensory motor rhythm and movement-related cortical potential features. J. Neural Eng. 2020, 17, 35003. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.; Vidaurre, C.; Schreuder, M.; Meinecke, F.; Bunau, P.; Muller, K. A mathematical model for the two-learners problem. J. Neural Eng. 2017, 14, 036005. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Lal, S.; Guesgen, H. Architectural Review of Co-Adaptive Brain Computer Interface. In Proceedings of the 4th Asia-Pacific World Congress on Computer Science and Engineering (APWC on CSE), Mana Island, Fiji, 11–13 December 2017; pp. 200–207. [Google Scholar]
- Abu-Rmileh, A.; Zakkay, E.; Shmuelof, L.; Shriki, O. Co-adaptive Training Improves Efficacy of a Multi-Day EEG-Based Motor Imagery BCI Training. Front. Hum. Neurosci. 2019, 13, 362. [Google Scholar] [CrossRef] [Green Version]
- Alonso-Valerdi, L. Python Executable Script for Estimating Two Effective Parameters to Individualize Brain-Computer Interfaces: Individual Alpha Frequency and Neurophysiological Predictor. Front. Neuroinform. 2016, 10, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noor, N.; Ibrahim, H. Machine Learning Algorithms and Quantitative Electroencephalography Predictors for Outcome Prediction in Traumatic Brain Injury: A Systematic Review. IEEE Access 2020, 8, 102075–102092. [Google Scholar] [CrossRef]
- Weber, L.; Ethofer, T.; Ehlis, A. Predictors of neurofeedback training outcome: A systematic review. NeuroImage Clin. 2020, 27, 102301. [Google Scholar] [CrossRef]
- Vukelic, M.; Gharabaghi, A. Self-regulation of circumscribed brain activity modulates spatially selective and frequency specific connectivity of distributed resting state networks. Front. Behav. Neurosci. 2015, 9, 181. [Google Scholar] [CrossRef] [Green Version]
- Ahn, M.; Cho, H.; Ahn, S.; Jun, S. High theta and low alpha powers may be indicative of BCI-illiteracy in motor imagery. PloS ONE 2013, 8, e80886. [Google Scholar] [CrossRef] [Green Version]
- Bamdadian, A.; Guan, C.; Ang, K.; Xu, J. The predictive role of pre-cue EEG rhythms on MI-based BCI classification performance. J. Neurosci. Methods 2014, 235, 138–144. [Google Scholar] [CrossRef]
- Acqualagna, L.; Botrel, L.; Vidaurre, C.; Kubler, A.; Blankertz, B. Large-Scale Assessment of a Fully Automatic Co-Adaptive Motor Imagery-Based Brain Computer Interface. PLoS ONE 2016, 11, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Samek, W.; Blythe, D.; Curio, G.; Muller, K.; Blankertz, K.; Nikulin, V. Multiscale temporal neural dynamics predict performance in a complex sensorimotor task. NeuroImage 2016, 141, 291–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, M.; Cho, H.; Won, K.; Ahn, A.; Jun, S. Use of Both Eyes-Open and Eyes-Closed Resting States May Yield a More Robust Predictor of Motor Imagery BCI Performance. Electronics 2020, 9, 690. [Google Scholar] [CrossRef]
- Wan, F.; Nan, W.; Vai, M.; Rosa, A. Resting alpha activity predicts learning ability in alpha neurofeedback. Front. Hum. Neurosci. 2014, 8, 500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solesio-Jofre, E.; Beets, I.; Woolley, D.; Pauwels, L.; Chalavi, S.; Mantini, D.; Swinnen, S. Age-Dependent Modulations of Resting State Connectivity Following Motor Practice. Front. Aging Neurosci. 2018, 10, 25. [Google Scholar] [CrossRef]
- Corsi, M.; Chavez, M.; Schwartz, D.; George, N.; Hugueville, L.; Kahn, A.; Dupont, S.; Bassett, D.; De Vico Fallani, F. Functional disconnection of associative cortical areas predicts performance during BCI training. NeuroImage 2020, 209, 116500. [Google Scholar] [CrossRef]
- Filho, S.; Costa, T.; Uribe, L.; Rodrigues, P.; Soriano, D.; Attux, R.; Castellano, G. On the (in)efficacy of motor imagery training without feedback and event-related desynchronizations considerations. Biomed. Phys. Eng. Express 2020, 6, 035030. [Google Scholar] [CrossRef]
- Daeglau, M.; Wallhoff, F.; Debener, S.; Condro, I.; Kranczioch, C.; Zich, C. Challenge Accepted? Individual Performance Gains for Motor Imagery Practice with Humanoid Robotic EEG Neurofeedback. Sensors 2020, 20, 1620. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Wang, J.; Chen, L. Event-related desynchronization and synchronization quantification in motor-related EEG by Kolmogorov entropy. J. Neural Eng. 2013, 10, 036023. [Google Scholar] [CrossRef]
- Velasquez, L.; Caicedo, J.; Castellanos, G. Entropy-Based Estimation of Event-Related De/Synchronization in Motor Imagery Using Vector-Quantized Patterns. Entropy 2020, 22, 703. [Google Scholar] [CrossRef]
- Chholak, P.; Pisarchik, A.N.; Kurkin, S.A.; Maksimenko, V.A.; Hramov, A.E. Phase-amplitude coupling between mu-and gamma-waves to carry motor commands. In Proceedings of the 2019 3rd School on Dynamics of Complex Networks and their Application in Intellectual Robotics (DCNAIR), Innopolis, Russia, 9–11 September 2019; pp. 39–45. [Google Scholar]
- Kim, B.; Winstein, C. Can neurological biomarkers of brain impairment be used to predict poststroke motor recovery? A systematic review. Neurorehabilit. Neural Repair 2017, 31, 3–24. [Google Scholar] [CrossRef]
- Yperman, J.; Becker, T.; Valkenborg, D.; Popescu, V.; Hellings, N.; Van Wijmeersch, B.; Peeters, L.M. Machine learning analysis of motor evoked potential time series to predict disability progression in multiple sclerosis. BMC Neurol. 2020, 20, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiederer, L.; Volker, M.; Schirrmeister, R.; Burgard, W.; Boedecker, J.; Ball, T. Hybrid Brain-Computer-Interfacing for Human-Compliant Robots: Inferring Continuous Subjective Ratings with Deep Regression. Front. Neurorobotics 2019, 13, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahid, A.; Singh, M. A deep learning approach for prediction of Parkinson’s disease progression. Biomed. Eng. Lett. 2020, 10, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.; Janghel, R. Recent Deep Learning Techniques, Challenges and Its Applications for Medical Healthcare System: A Review. Neural Process. Lett. 2019, 50, 1907–1935. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, Z.; Cheng, M.; Liu, Y.; Bian, J.; Zheng, G.; Zeng, Z. Nonlinear Regression via Deep Negative Correlation Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2019. [Google Scholar] [CrossRef]
- Blankertz, B.; Sannelli, C.; Halder, S.; Hammer, E.; Kubler, A.; Muller, K.; Curio, G.; Dickhaus, T. Neurophysiological predictor of SMR-based BCI performance. NeuroImage 2010, 51, 1303–1309. [Google Scholar] [CrossRef] [Green Version]
- Molla, M.; Shiam, A.; Islam, M.; Tanaka, T. Discriminative Feature Selection-Based Motor Imagery Classification Using EEG Signal. IEEE Access 2020, 8, 98255–98265. [Google Scholar] [CrossRef]
- Pfurtscheller, G.; Lopes-da Silva, F. Event-related EEG/MEG synchronization and desynchronization: Basic principles. Clin. Neurophysiol. 1999, 110, 1842–1857. [Google Scholar] [CrossRef]
- Cheng, H.T.; Koc, L.; Harmsen, J.; Shaked, T.; Chandra, T.; Aradhye, H.; Anderson, G.; Chai, W.; Ispir, M.; et al. Shaked. Wide & deep learning for recommender systems. In Proceedings of the 1st Workshop on Deep Learning for Recommender Systems, Boston, FL, USA, 15 September 2016; 2016; pp. 7–10. [Google Scholar]
- Nagabushanam, P.; George, S.T.; Radha, S. EEG signal classification using LSTM and improved neural network algorithms. Soft Comput. 2019, 24, 9981–10003. [Google Scholar] [CrossRef]
- Baig, M.; Aslam, N.; Shum, H. Filtering techniques for channel selection in motor imagery EEG applications: A survey. Artif. Intell. Rev. 2020, 53, 1207–1232. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Ahn, M.; Ahn, S.; Kwon, M.; Jun, S.C. EEG datasets for motor imagery brain-computer interface. GigaScience 2017, 6, 1–8. [Google Scholar] [CrossRef]
- Vidaurre, C.; Sander, T.; A, S. BioSig: The free and open source software library for biomedical signal processing. Comput. Intell. Neurosci. 2011, 2011, 935364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Yao, D.; Valdes-Sosa, P.; Li, F.; Li, P.; Zhang, T.; Ma, T.; Li, Y.; Xu, P. Efficient resting-state EEG network facilitates motor imagery performance. J. Neural Eng. 2015, 12, 66024. [Google Scholar] [CrossRef] [PubMed]
- Miao, M.; Zeng, H.; Wang, A.; Zhao, C.; Liu, F. Discriminative spatial-frequency-temporal feature extraction and classification of motor imagery EEG: An sparse regression and Weighted Naïve Bayesian Classifier-based approach. J. Neurosci. Methods 2017, 278, 13–24. [Google Scholar] [CrossRef]
- Miao, M.; Wang, A.; Liu, F. A spatial-frequency-temporal optimized feature sparse representation-based classification method for motor imagery EEG pattern recognition. Med. Biol. Eng. Comput. 2017, 55, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Scherer, R.; Vidaurre, C. Motor imagery based brain–computer interfaces. In Smart Wheelchairs and Brain-Computer Interfaces; Elsevier: Amsterdam, The Netherlands, 2018; pp. 171–195. [Google Scholar]
- Meng, J.; He, B. Exploring Training Effect in 42 Human Subjects Using a Non-invasive Sensorimotor Rhythm Based Online BCI. Front. Hum. Neurosci. 2019, 13, 128. [Google Scholar] [CrossRef] [Green Version]
- Shu, X.; Chen, S.; Yao, L.; Sheng, X.; Zhang, D.; Jiang, N.; Jia, J.; Zhu, X. Fast recognition of BCI-inefficient users using physiological features from EEG signals: A screening study of stroke patients. Front. Neurosci. 2018, 12, 93. [Google Scholar] [CrossRef] [Green Version]







| Resting Data | Electrode | [s] | |||||
|---|---|---|---|---|---|---|---|
| Configuration | Mean | PCA | |||||
| Baseline inverval | 2Ch(LC) | 0.15 | 0.15 | 0.17 | 0.16 | 0.13 | 0.15 |
| 6Ch(LC) | 0.07 | 0.04 | 0.11 | 0.13 | 0.05 | 0.07 | |
| 2Ch(DRN ) | 0.15 | 0.16 | 0.18 | 0.16 | 0.14 | 0.15 | |
| 6Ch(DRN ) | 0.07 | 0.04 | 0.12 | 0.14 | 0.06 | 0.08 | |
| 2Ch(DRN ) | 0.86 | 0.85 | 0.96 | 0.97 | 0.83 | 0.87 | |
| 2Ch(DRN ) LOO | 0.76 | 0.79 | 0.82 | 0.80 | 0.78 | 0.86 | |
| 6Ch(DRN ) | 0.92 | 0.86 | 0.95 | 0.97 | 0.83 | 0.88 | |
| 6Ch(DRN ) LOO | 0.83 | 0.87 | 0.85 | 0.87 | 0.89 | 0.91 | |
| Resting-state | 2Ch(LC) | 0.30 | 0.31 | 0.31 | 0.27 | 0.29 | 0.31 |
| 6Ch(LC) | 0.25 | 0.31 | 0.26 | 0.26 | 0.28 | 0.28 | |
| 2Ch(DRN ) | 0.31 | 0.31 | 0.31 | 0.28 | 0.30 | 0.32 | |
| 6Ch(DRN ) | 0.25 | 0.31 | 0.26 | 0.27 | 0.30 | 0.30 | |
| 2Ch(DRN ) | 0.79 | 0.80 | 0.92 | 0.94 | 0.78 | 0.82 | |
| 2Ch(DRN ) LOO | 0.85 | 0.87 | 0.83 | 0.82 | 0.79 | 0.84 | |
| 6Ch(DRN ) | 0.86 | 0.77 | 0.91 | 0.93 | 0.75 | 0.80 | |
| 6Ch(DRN ) LOO | 0.85 | 0.83 | 0.88 | 0.86 | 0.80 | 0.77 | |
| Rhythm | Electrode | [s] | |||||
|---|---|---|---|---|---|---|---|
| Subband | Configuration | Mean | PCA | ||||
| 2Ch(LC) | 0.12 | 0.064 | 0.04 | 0.003 | 0.6 | 0.05 | |
| 6Ch(LC) | 0.23 | 0.08 | 0.10 | 0.04 | 0.11 | 0.11 | |
| 2Ch(DRN ) | 0.13 | 0.064 | 0.13 | 0.17 | 0.06 | 0.17 | |
| 6Ch(DRN ) | 0.23 | 0.12 | 0.10 | 0.04 | 0.11 | 0.11 | |
| 2Ch(LC) | 0.11 | 0.06 | 0.08 | 0.02 | 0.07 | 0.06 | |
| 6Ch(LC) | 0.14 | 0.04 | 0.006 | 0.016 | 0.11 | 0.07 | |
| 2Ch(DRN ) | 0.16 | 0.15 | 0.20 | 0.23 | 0.16 | 0.20 | |
| 6Ch(DRN ) | 0.19 | 0.05 | 0.23 | 0.25 | 0.21 | 0.20 | |
| 2Ch(LC) | 0.06 | 0.05 | 0.05 | 0.01 | 0.04 | 0.04 | |
| 6Ch(LC) | 0.11 | 0.07 | 0.03 | 0.04 | 0.11 | 0.08 | |
| 2Ch(DRN ) | 0.08 | 0.06 | 0.10 | 0.18 | 0.11 | 0.09 | |
| 6Ch(DRN ) | 0.11 | 0.11 | 0.19 | 0.21 | 0.15 | 0.21 | |
| 2Ch(DRN ) | 0.84 | 0.80 | 0.94 | 0.91 | 0.78 | 0.83 | |
| 2Ch(DRN ) LOO | 0.15 | 0.17 | 0.24 | 0.19 | 0.18 | 0.21 | |
| 6Ch(DRN ) | 0.87 | 0.77 | 0.93 | 0.95 | 0.82 | 0.82 | |
| 6Ch(DRN ) LOO | 0.20 | 0.44 | 0.40 | 0.28 | 0.26 | 0.40 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velasquez-Martinez, L.; Caicedo-Acosta, J.; Acosta-Medina, C.; Alvarez-Meza, A.; Castellanos-Dominguez, G. Regression Networks for Neurophysiological Indicator Evaluation in Practicing Motor Imagery Tasks. Brain Sci. 2020, 10, 707. https://doi.org/10.3390/brainsci10100707
Velasquez-Martinez L, Caicedo-Acosta J, Acosta-Medina C, Alvarez-Meza A, Castellanos-Dominguez G. Regression Networks for Neurophysiological Indicator Evaluation in Practicing Motor Imagery Tasks. Brain Sciences. 2020; 10(10):707. https://doi.org/10.3390/brainsci10100707
Chicago/Turabian StyleVelasquez-Martinez, Luisa, Julian Caicedo-Acosta, Carlos Acosta-Medina, Andres Alvarez-Meza, and German Castellanos-Dominguez. 2020. "Regression Networks for Neurophysiological Indicator Evaluation in Practicing Motor Imagery Tasks" Brain Sciences 10, no. 10: 707. https://doi.org/10.3390/brainsci10100707




