Molecular Epidemiological Survey of Cryptosporidium in Ochotona curzoniae and Bos grunniens of Zoige County, Sichuan Province
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. PCR Amplification
2.4. Sequence Analysis and Phylogenetic Tree
2.5. Statistical Analysis
3. Results
3.1. Occurrence of Cryptosporidium spp.
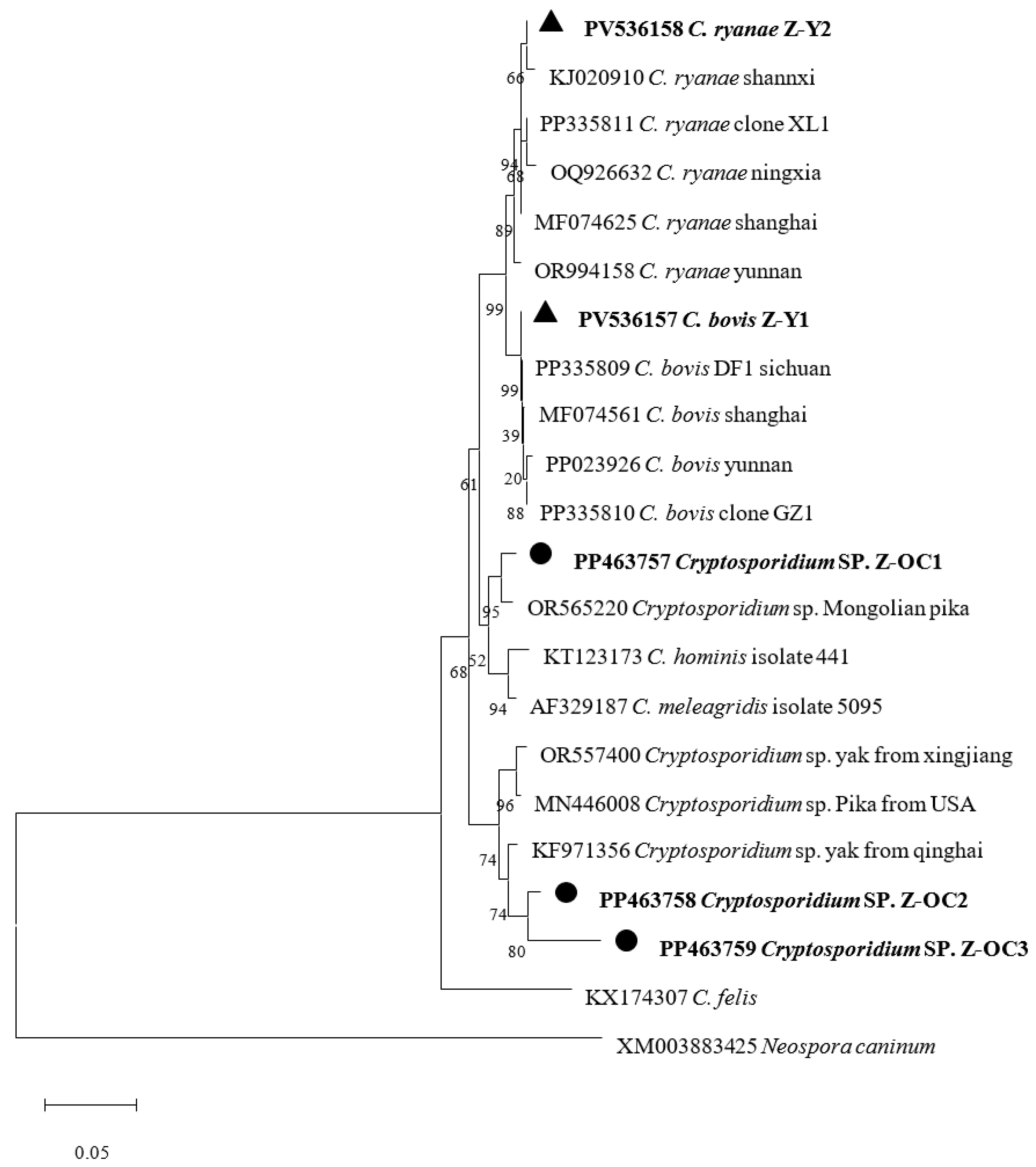
3.2. Phylogenetic Analysis Based on SSU rRNA Gene of Cryptosporidium
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SSU rRNA | Small Subunit Ribosomal RNA |
| NCBI | National Center for Biotechnology Information |
| PCR | Polymerase Chain Reaction |
| NJ | Neighbor-Joining |
References
- Xiao, L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010, 124, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Tyzzer, E.E. A sporozoan found in the peptic glands of the common mouse. Proc. Soc. Exp. Biol. Med. 1907, 5, 12–13. [Google Scholar] [CrossRef]
- Shahbaz, M.K.; William, H.W. Past, current, and potential treatments for cryptosporidiosis in humans and farm animals: A comprehensive review. Front. Cell. Infect. Microbiol. 2023, 13, 1115522. [Google Scholar] [CrossRef] [PubMed]
- Nigel, Y.; Mary, M.; Deborah, A.S.; Kevin, A.; Elizabeth, C.; Rodrigo, P.B.; Michael, W.R.; Jessica, C.K. Genomic and virulence analysis of in vitro cultured Cryptosporidium parvum. PLoS Pathog. 2024, 20, e1011992. [Google Scholar] [CrossRef]
- Egan, S.; Barbosa, A.; Feng, Y.Y.; Xiao, L.H.; Ryan, U. Critters and contamination: Zoonotic protozoans in urban rodents and water quality. Water Res. 2024, 251, 121165. [Google Scholar] [CrossRef] [PubMed]
- Ng-Hublin, J.S.Y.; Combs, B.; MacKenzie, B.; Ryan, U. Human Cryptosporidiosis Diagnosed in Western Australia: A Mixed Infection with Cryptosporidium meleagridis, the Cryptosporidium Mink Genotype, and an Unknown Cryptosporidium Species. J. Clin. Microbiol. 2013, 51, 2463–2465. [Google Scholar] [CrossRef] [PubMed]
- Koehler, A.V.; Whipp, M.J.; Haydon, S.R.; Gasser, R.B. Cryptosporidium cuniculus—New records in human and kangaroo in Australia. Parasites Vectors 2014, 7, 492. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, F.T.; Fonseca, A.B.M.; Oliveira Coelho, L.F.; Silva Barbosa, A.; Bastos, O.M.P.; Uchôa, C.M.A. Cryptosporidium species in non-human animal species in Latin America: Systematic review and meta-analysis. Veter Parasitol. Reg. Stud. Rep. 2022, 29, 100690. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Paparini, A. Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol. 2016, 38, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Danišová, O.; Valenčáková, A.; Stanko, M.; Luptáková, L.; Hatalová, E.; Čanády, A. Rodents as a Reservoir of Infection Caused by Multiple Zoonotic Species/Genotypes of C. Parvum, C. Hominis, C. Suis, C. Scrofarum, and the First Evidence of Cryptosporidium Muskrat Genotypes I and II of Rodents in Europe. Acta Trop. 2017, 172, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Bujila, I.; Troell, K.; Ögren, J.; Hansen, A.; Killander, G.; Agudelo, L.; Lebbad, M.; Beser, J. Cryptosporidium species and subtypes identified in human domestic cases through the national microbiological surveillance programme in Sweden from 2018 to 2022. BMC Infect. Dis. 2024, 24, 146. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.Q.; Gong, B.Y.; Liu, X.H.; Shen, Y.J.; Wu, Y.C.; Zhang, W.Z.; Cao, J.P. Aretrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987–2018). PLoS Negl. Trop. Dis. 2020, 14, e0008146. [Google Scholar] [CrossRef] [PubMed]
- King, P.; Tyler, K.M.; Hunter, P.R. Anthroponotic transmission of Cryptosporidium parvum predominates in countries with poorer sanitation: A systematic review and meta-analysis. Parasites Vectors 2019, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.T.; Foggin, J.M. The plateau pika (Ochotona curzoniae) is a keystone species for biodiversity on the Tibetan plateau. Anim. Conserv. 1999, 2, 235–240. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, M.Y.; Wang, L.F.; Wang, Y.; Zhang, S.; Zhang, Z.S.; Chai, H.L.; Fan, W.J.; Yi, C.; Ding, Y.L.; et al. Prevalence and molecular characterization of Cryptosporidium spp. in dairy and beef cattle in Shanxi, China. Parasitol. Res. 2023, 123, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zou, Y.; Wang, P.; Qu, M.R.; Zheng, W.B.; Wang, P.; Chen, X.Q.; Zhu, X.Q. Prevalence and multilocus genotyping of Cryptosporidium spp. in cattle in Jiangxi Province, southeastern China. Parasitol. Res. 2021, 120, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, H.Y.; Jing, B.; Wang, D.F.; Wang, R.J.; Zhang, L.X. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Vet. Parasitol. 2015, 212, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhang, S.; Zhang, Z.S.; Qian, X.Y.; Chai, H.L.; Wang, Y.; Fan, W.J.; Yi, C.; Ding, U.L.; Han, W.X.; et al. Prevalence and molecular characterization of Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. Vet. Res. Commun. 2024, 48, 2629–2643. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.L.; Heng, Z.J.; Li, L.J.; Yang, J.F.; He, J.J.; Zou, F.C.; Shu, F.F. Cryptosporidium spp. Infection and Genotype Identification in Pre-Weaned and Post-Weaned Calves in Yunnan Province, China. Animals 2024, 14, 1907. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chai, H.L.; Wang, M.Y.; Zhang, Z.S.; Han, W.X.; Yang, B.; Wang, Y.; Zhang, S.; Zhao, W.H.; Ma, Y.M.; et al. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle in Central Inner Mongolia, Northern China. BMC Vet. Res. 2023, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.N.; Zhang, N.Z.; Gong, Q.L.; Zhao, Q.; Zhang, X.X. Prevalence of Cryptosporidium in dairy cattle in China during 2008–2018: A systematic review and meta-analysis. Microb. Pathog. 2019, 132, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.L.; Ni, H.B.; Li, J.H.; Jiang, J.; Wang, W.; Wei, X.Y.; Zhang, Y.; Sun, H.T. Prevalence of Cryptosporidium spp. in Yaks (Bos grunniens) in China: A Systematic Review and Meta-Analysis. Front. Cell. Infect. Microbiol. 2021, 11, 770612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Jian, Y.J.; Li, X.P.; Ma, L.Q.; Karanis, G.; Karanis, P. The first report of Cryptosporidium spp. in Microtus fuscus (Qinghai vole) and Ochotona curzoniae (wild plateau pika) in the Qinghai-Tibetan Plateau area, China. Parasitol. Res. 2018, 117, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.Y.; Sun, H.T.; Chuang, L.; Zhu, J.H.; Wang, Z.J.; Ma, T.; Zhao, Q.; Lan, Y.G.; He, W.Q. Prevalence and Characterization of Cryptosporidium Species in Tibetan Antelope (Pantholops hodgsonii). Front. Cell. Infect. Microbiol. 2021, 11, 713873. [Google Scholar] [CrossRef] [PubMed]
- Koehler, A.V.; Wang, T.; Haydon, S.R.; Gasser, R.B. Cryptosporidium viatorum from the Native Australian Swamp Rat Rattus Lutreolus—An Emerging Zoonotic Pathogen? Int. J. Parasitol. Parasites Wildl. 2018, 7, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.B.; Wieger, V.; Johnstone, T.; Sandra, M.; Nienke, V.; Laura, M.; Daisy, B.d.J.; Merlin, V.L.; Petra, F.M.; James, A.B.; et al. Performance of three rapid diagnostic tests for the detection of Cryptosporidium spp. and Giardia duodenalis in children with severe acute malnutrition and diarrhoea. Infect. Dis. Poverty 2019, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liu, Y.Y.; Mei, J.J.; Zheng, W.B.; Liu, Q.; Gao, W.W.; Zhu, X.Q.; Xie, S.C. Molecular Identification and Genotyping of Cryptosporidium spp. and Blastocystis sp. in Cattle in Representative Areas of Shanxi Province, North China. Animals 2023, 13, 2929. [Google Scholar] [CrossRef] [PubMed]
- Angus, K. Cryptosporidiosis in Ruminants. In CRC Press eBooks; CRC Press: Boca Raton, FL, USA, 2018; pp. 83–104. [Google Scholar] [CrossRef]
- Kvác, M.; Sak, B.; Kvetonová, D.; Secor, W.E. Infectivity of gastric and intestinal Cryptosporidium species in immunocompetent Mongolian gerbils (Meriones unguiculatus). Veter Parasitol. 2009, 163, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Zhang, Q.Z.; Qi, R. Global prevalence of lagomorpha coccidiosis from 1951 to 2024: A systematic review and meta-analysis. Res. Veter Sci. 2025, 183, 105519. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Chen, Y.S. Prevalence of coccidia in lagomorphs in China between 1981 and 2023: A systematic review and meta-analysis. Veter Parasitol. 2024, 328, 110185. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.B.; Cai, J.Z.; Ma, J.W.; Feng, Y.Y.; Xiao, L.H. Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet. Parasitol. 2014, 202, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Li, P.L.; Cai, J.Z.; Cai, M.; Wu, W.X.; Li, C.H.; Lei, M.T.; Xu, H.L.; Feng, L.J.; Ma, J.W.; Feng, Y.Y.; et al. Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet. Parasitol. 2016, 215, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Mi, R.; Wang, X.; Li, C.; Huang, Y.; Zhou, P.; Li, Z.; Lei, M.; Cai, J.; Chen, Z. Prevalence and genetic characterization of Cryptosporidium in yaks in Qinghai Province of China. PLoS ONE 2013, 8, e74985. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.L.; Li, R.; Duan, L.; He, L. Molecular epidemiological Investigation on Cryptosporidium Infection fromYaks in Hongyuan County, Sichuan Province. Acta Agric. Zhejiangensis 2016, 28, 1842–1846. [Google Scholar]
- Li, K.; Li, Z.X.; Zeng, Z.B.; Li, A.Y.; Khalid, M.; Muhammad, S.; Gao, K.; Li, J.K. Prevalence and Molecular Characterization of Cryptosporidium spp. In Yaks (Bos Grunniens) in Naqu, China. Microb. Pathog. 2020, 144, 104190. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.Y.; Zhang, X.X.; Zhao, G.H.; Zhou, D.H.; Yin, M.Y.; Zhao, Q.; Zhu, X.Q. First report of Cryptosporidium spp. in white yaks in China. Parasites Vectors 2014, 7, 230. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.W.; Zhao, G.H.; Gong, Y.Y.; Zhang, L.X. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Front. Microbiol. 2017, 8, 1823. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gong, Q.L.; Zeng, A.; Li, M.H.; Zhao, Q.; Ni, H.B. Prevalence of Cryptosporidium in pigs in China: A systematic review and meta-analysis. Transbound. Emerg. Dis. 2021, 68, 1400–1413. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Gong, Q.L.; Zhao, B.; Cai, Y.N.; Zhao, Q. Prevalence of Cryptosporidium Infection in Sheep and Goat Flocks in China During 2010–2019: A Systematic Review and Meta-Analysis. Vector Borne Zoonotic Dis. 2021, 21, 692–706. [Google Scholar] [CrossRef] [PubMed]
- Paul, O.O.; Isaiah, O.A. Epidemiology of Cryptosporidium infection in different hosts in Nigeria: A meta-analysis. Parasitol. Int. 2019, 71, 194–206. [Google Scholar] [CrossRef]
- Zhong, Z.J.; Dan, J.M.; Yan, G.W.; Tu, R.; Tian, Y.N.; Cao, S.Z.; Shen, L.H.; Deng, J.L.; Yu, S.M.; Geng, Y.; et al. Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan province, China. Parasite 2018, 25, 45. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.X.; Wang, X.; Jian, J.H.; Zuo, Q.Q.; Liu, H.; Wang, Y.X.; Su, Y.X.; Cao, J.P.; Jiang, B.; Shen, Y.J. Investigation of Cryptosporidium spp. and Enterocytozoon bieneusi in free-ranged livestock on the southeastern Qinghai-Xizang Plateau, China. BMC Infect. Dis. 2025, 25, 356. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.L.; Li, N.; Huang, Y.J.; Chen, C.Y.; Wen, L.X.; Wang, W.J.; Una, M.R.; Xiao, L.H.; Feng, Y.Y.; Guo, Y.Q. Longitudinal follow-up reveals occurrence of successive Cryptosporidium bovis and Cryptosporidium ryanae infections by different subtype families in dairy cattle. Int. J. Parasitol. 2023, 53, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Cao, X.F.; Deng, L.; Li, W.; Huang, X.M.; Lan, J.C.; Xiao, Q.C.; Zhong, Z.J.; Feng, F.; Zhang, Y.; et al. Epidemiology of Cryptosporidium infection in cattle in China: A review. Parasite 2017, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santín, M.; Trout, J.M. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). J. Parasitol. 2005, 91, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, M.K.; Chanchal, D.; Amiya, K.P.; Xiao, L.H.; Tomoyoshi, N.; Sandipan, G. Molecular characterization and assessment of zoonotic transmission of Cryptosporidium from dairy cattle in West Bengal, India. Veter Parasitol. 2010, 171, 41–47. [Google Scholar] [CrossRef]
- Josephine, S.Y.N.; Keith, E.; Belinda, W.; David, N.D.; Peter, D.M.; Philippe, P.; Ross, K.; Bob, M.; Kate, L.; David, M.; et al. Evidence of Cryptosporidium transmission between cattle and humans in northern New South Wales. Exp. Parasitol. 2012, 130, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Yosra, A.H.; Jürgen, K.; Karsten, N.; Georgvon, S.H.; Karl, H.Z. Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Veter Parasitol. 2013, 193, 15–24. [Google Scholar] [CrossRef]
- Adriana, H.; Ximena, V.; Giovanny, H.; Julio, C.G.; Luis, R.V.; Plutarco, U.; Oswaldo, V.; Catalina, T.; Juan, D.R. Molecular detection and genotyping of intestinal protozoa from different biogeographical regions of Colombia. PeerJ 2020, 8, e8554. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santín, M.; Trout, J.M. Cryptosporidium ryanaen. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). Veter Parasitol. 2008, 156, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ma, L.; Gou, J.M.; Yao, H.Z.; Ren, M.; Yang, B.K.; Lin, Q. Seasonal distribution of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in Tibetan sheep in Qinghai, China. Parasites Vectors 2022, 15, 394. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Tamara, V.; Edward, R.A. Diverse Genotypes of Cryptosporidium in Sheep in California, USA. Pathogens 2022, 11, 1023. [Google Scholar] [CrossRef] [PubMed]
- Kaupke, A.; Michalski, M.M.; Rzeżutka, A. Diversity of Cryptosporidium species occurring in sheep and goat breeds reared in Poland. Parasitol. Res. 2017, 116, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Firoozi, Z.; Sazmand, A.; Zahedi, A.; Astani, A.; Bafghi, A.; Kiani-Salmi, N.; Ebrahimi, B.; Tafti, A.; Ryan, U.; Mohajeri, F. Prevalence and genotyping identification of Cryptosporidium in adult ruminants in central Iran. Parasites Vectors 2019, 12, 510. [Google Scholar] [CrossRef] [PubMed]
- Soltane, R.; Guyot, K.; Dei-Cas, E.; Àyadi, A. Prevalence of Cryptosporidium spp. (Eucoccidiorida: Cryptosporiidae) in seven species of farm animals in Tunisia. Parasite 2007, 14, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Zhang, Z.C.; Ai, S.T.; Wang, X.Q.; Zhang, R.Y.; Duan, Z.Y. Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis from animal sources in the Qinghai-Tibetan Plateau Area (QTPA) in China. Comp. Immunol. Microbiol. Infect. Dis. 2019, 67, 101346. [Google Scholar] [CrossRef] [PubMed]
- Mirhashemi, M.E.; Zintl, A.; Grant, T.; Lucy, F.; Mulcahy, G.; Waal, T.D. Molecular epidemiology of Cryptosporidium species in livestock in Ireland. Veter Parasitol. 2016, 216, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.M.; Köster, P.C.; Dashti, A.; Torres, R.T.; Fonseca, C.; Mysterud, A.; Bailo, B.; Carvalho, J.; Ferreira, E.; Hipólito, D.; et al. Molecular Detection and Distribution of Giardia duodenalis and Cryptosporidium spp. Infections in Wild and Domestic Animals in Portugal. Transbound. Emerg. Dis. 2023, 2023, 5849842. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.C.; Joyce, L.J.Y.; Monis, P.; Ryan, U. Molecular characterisation of Cryptosporidium and Giardia in cats (Felis catus) in Western Australia. Exp. Parasitol. 2015, 155, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, W.; Oliveira, M.; Peres, P.; Oliveira, B.; Nagata, W.; Vieira, D.; Andrade, A.; Ferrari, E.; Duarte, J.; Meireles, M.; et al. First report of genus Cryptosporidium in cervids species: Mazama americana, Mazama nana and Blastocerus dichotomus. Vet. Res. Commun. 2022, 46, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Ježková, J.; Prediger, J.; Holubová, N.; Sak, B.; Konečný, R.; Feng, Y.; Xiao, L.; Rost, M.; McEvoy, J.; Kváč, M. Cryptosporidium rattin. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology 2021, 148, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Jian, Y.N.; Li, X.P.; Ma, L.Q.; Gabriele, K.; Cai, Q.G.; Panagiotis, K. Molecular detection and prevalence of Cryptosporidium spp. infections in two types of domestic farm animals in the Qinghai-Tibetan Plateau Area (QTPA) in China. Parasitol. Res. 2018, 117, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Taghipour, A.; Olfatifar, M.; Foroutan, M.; Bahadory, S.; Malih, N.; Norouzi, M. Global prevalence of Cryptosporidium infection in rodents: A systematic review and meta-analysis. Prev. Vet. Med. 2020, 182, 105119. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Qin, H.K.; Huang, J.Y.; Li, J.Q.; Zhang, L.X. The global prevalence of Cryptosporidium in sheep: A systematic review and meta-analysis. Parasitology 2022, 149, 1652–1665. [Google Scholar] [CrossRef] [PubMed]
| Location | Host | No. Positive/No. of Samples | Prevalence% (95% CI) | OR (95% CI) | Cryptosporidium spp. | Location Subtotal % (95% CI), OR |
|---|---|---|---|---|---|---|
| Dazhasi | O. curzoniae | 0/24 | 0 | |||
| B. grunniens | 0/18 | 0 | ||||
| Axi | O. curzoniae | 0/20 | 0 | |||
| B. grunniens | 0/40 | 0 | ||||
| Hongxing | O. curzoniae | 0/20 | 0 | 15.2 (6.6–28.0), 3.8 | ||
| B. grunniens | 7/26 | 27.9 (12.3–47.0) | C. ryanae (n = 2), C. bovis (n = 5) | |||
| Maixi | O. curzoniae | 8/20 | 40.0 (19.1–63.9) | 2.4 (0.5–11.6) | Cryptosporidium sp. (n = 8) | 32.4 (17.2–51.2), 10.0 |
| B. grunniens | 3/14 | 21.4 (4.8–46.6) | Reference | C. bovis (n = 3) | ||
| Tangke | O. curzoniae | 0/30 | 0 | 4.6% (0.6–14.8), Reference | ||
| B. grunniens | 2/14 | 14.3 (1.8–37.8) | C. bovis (n = 2) | |||
| Host subtotal | O. curzoniae | 8/114 | 7.0 (2.3–11.7) | Reference | ||
| B. grunniens | 12/128 | 9.4 (4.9–15.7) | 1.4 (0.5–3.5) | |||
| Total | 20/242 | 8.3 (5.1–12.5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, T.-C.; Jike, R.-H.; Zhu, L.-Q.; Chen, C.-X.; Hao, L.-L. Molecular Epidemiological Survey of Cryptosporidium in Ochotona curzoniae and Bos grunniens of Zoige County, Sichuan Province. Animals 2025, 15, 2140. https://doi.org/10.3390/ani15142140
Tang T-C, Jike R-H, Zhu L-Q, Chen C-X, Hao L-L. Molecular Epidemiological Survey of Cryptosporidium in Ochotona curzoniae and Bos grunniens of Zoige County, Sichuan Province. Animals. 2025; 15(14):2140. https://doi.org/10.3390/ani15142140
Chicago/Turabian StyleTang, Tian-Cai, Ri-Hong Jike, Liang-Quan Zhu, Chao-Xi Chen, and Li-Li Hao. 2025. "Molecular Epidemiological Survey of Cryptosporidium in Ochotona curzoniae and Bos grunniens of Zoige County, Sichuan Province" Animals 15, no. 14: 2140. https://doi.org/10.3390/ani15142140
APA StyleTang, T.-C., Jike, R.-H., Zhu, L.-Q., Chen, C.-X., & Hao, L.-L. (2025). Molecular Epidemiological Survey of Cryptosporidium in Ochotona curzoniae and Bos grunniens of Zoige County, Sichuan Province. Animals, 15(14), 2140. https://doi.org/10.3390/ani15142140






