Impact of Maternal Intrapartum Antibiotics, and Caesarean Section with and without Labour on Bifidobacterium and Other Infant Gut Microbiota
Abstract
:1. Introduction
2. Methods
2.1. Study Design
2.2. Faecal Microbiota Analysis
2.3. Quantitative PCR (qPCR) for Total Bifidobacteria and Bifidobacterium longum subsp. Infantis Detection
2.4. Statistical Analysis
3. Results
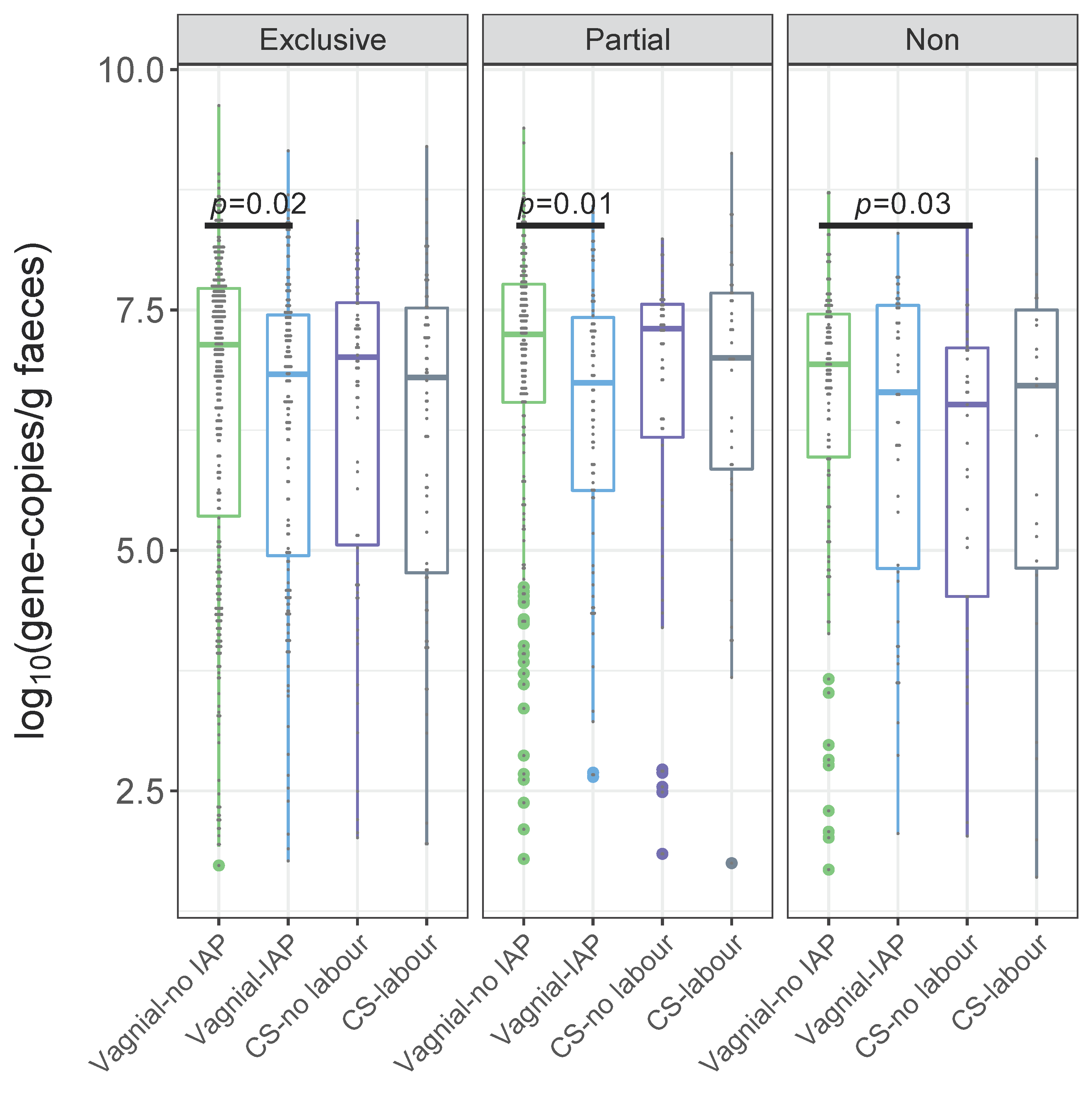
3.1. Quantification of Bifidobacteria and B. longum subsp. infantis According to a Birth Mode
Independent Associations between Bifidobacteria, B. longum subsp. infantis and Birth Mode
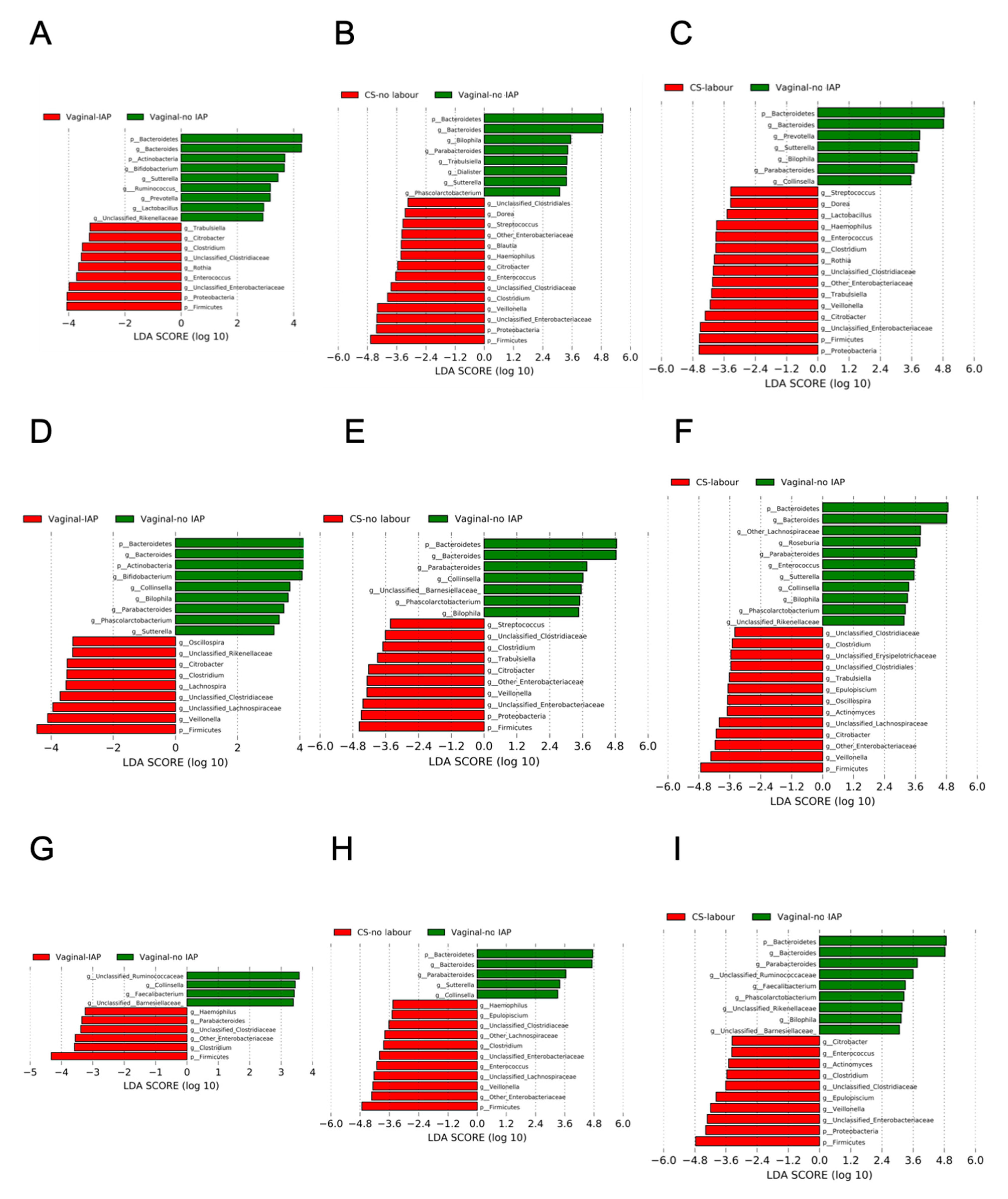
3.2. Alpha-Diversity, Beta-Diversity of Gut Microbial Community According to a Birth Mode
3.3. Overall Gut Microbiota According to a Birth Mode, Stratified by Breastfeeding Status
3.4. Co-Occurrence Network between Bifidobacterium and Other Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitchell, C.M.; Mazzoni, C.; Hogstrom, L.; Bryant, A.; Bergerat, A.; Cher, A.; Pochan, S.; Herman, P.; Carrigan, M.; Sharp, K.; et al. Delivery mode affects stability of early infant gut microbiota. Cell Rep. Med. 2020, 1, 100156. [Google Scholar] [CrossRef]
- Shaterian, N.; Abdi, F.; Ghavidel, N.; Alidost, F. Role of cesarean section in the development of neonatal gut microbiota: A systematic review. Open Med. 2021, 16, 624–639. [Google Scholar] [CrossRef]
- Wampach, L.; Heintz-Buschart, A.; Hogan, A.; Muller, E.E.L.; Narayanasamy, S.; Laczny, C.C.; Hugerth, L.W.; Bindl, L.; Bottu, J.; Andersson, A.F.; et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front. Microbiol. 2017, 8, 738. [Google Scholar] [CrossRef] [PubMed]
- Princisval, L.; Rebelo, F.; Williams, B.L.; Coimbra, A.C.; Crovesy, L.; Ferreira, A.L.; Kac, G. Association between the mode of delivery and infant gut microbiota composition up to 6 months of age: A Systematic literature review considering the role of breastfeeding. Nutr. Rev. 2021, 1–15. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Mahony, J.; van Sinderen, D.; Ventura, M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2018, 26, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Dunn, A.B.; Jordan, S.; Baker, B.J.; Carlson, N.S. The maternal infant microbiome: Considerations for labor and birth. MCN Am. J. Matern. Child. Nurs. 2017, 42, 318–325. [Google Scholar] [CrossRef]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Shin, H.; Pizoni, A.; Werlang, I.C.; Matte, U.; Goldani, M.Z.; Goldani, H.A.S.; Dominguez-Bello, M.G. Delivery mode and the transition of pioneering gut-microbiota structure, composition and predicted metabolic function. Genes 2017, 8, 364. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.S.W.; Sabu, P.; Deopujari, V.; Levy, S.; Shah, A.A.; Clemency, N.; Provenzano, M.; Saadoon, R.; Munagala, A.; Baker, R.; et al. Prenatal and peripartum exposure to antibiotics and cesarean section delivery are associated with differences in diversity and composition of the infant meconium microbiome. Microorganisms 2020, 8, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, K.; Lou, W.; Tun, H.M.; Konya, T.B.; Morales-Lizcano, N.; Chari, R.S.; Field, C.J.; Guttman, D.S.; Mandal, R.; Wishart, D.S.; et al. From birth to overweight and atopic disease: Multiple and common pathways of the infant gut microbiome. Gastroenterology 2021, 160, 128–144.e110. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145.e135. [Google Scholar] [CrossRef]
- Van Schalkwyk, J.; Van Eyk, N. Antibiotic prophylaxis in obstetric procedures. J. Obstet. Gynaecol. Can. 2010, 32, 878–884. [Google Scholar] [CrossRef]
- Stearns, J.C.; Simioni, J.; Gunn, E.; McDonald, H.; Holloway, A.C.; Thabane, L.; Mousseau, A.; Schertzer, J.D.; Ratcliffe, E.M.; Rossi, L.; et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci. Rep. 2017, 7, 16527. [Google Scholar] [CrossRef]
- Azad, M.B.; Konya, T.; Persaud, R.R.; Guttman, D.S.; Chari, R.S.; Field, C.J.; Sears, M.R.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: A prospective cohort study. BJOG 2016, 123, 983–993. [Google Scholar] [CrossRef]
- Alcon-Giner, C.; Caim, S.; Mitra, S.; Ketskemety, J.; Wegmann, U.; Wain, J.; Belteki, G.; Clarke, P.; Hall, L.J. Optimisation of 16S rRNA gut microbiota profiling of extremely low birth weight infants. BMC Genomics 2017, 18, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, A.W.; Martin, J.C.; Scott, P.; Parkhill, J.; Flint, H.J.; Scott, K.P. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef] [Green Version]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Li, Z.; Zhang, W.; Zhang, C.; Zhang, Y.; Mei, H.; Zhuo, N.; Wang, H.; Wang, L.; Wu, D. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: A study of 91 term infants. Sci. Rep. 2020, 10, 15792. [Google Scholar] [CrossRef] [PubMed]
- Smilowitz, J.T.; Lebrilla, C.B.; Mills, D.A.; German, J.B.; Freeman, S.L. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forbes, J.D.; Azad, M.B.; Vehling, L.; Tun, H.M.; Konya, T.B.; Guttman, D.S.; Field, C.J.; Lefebvre, D.; Sears, M.R.; Becker, A.B.; et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018, 172, e181161. [Google Scholar] [CrossRef]
- Ly, N.P.; Litonjua, A.; Gold, D.R.; Celedón, J.C. Gut microbiota, probiotics, and vitamin D: Interrelated exposures influencing allergy, asthma, and obesity? J. Allergy Clin. Immunol. 2011, 127, 1087–1094, quiz 1095–1086. [Google Scholar] [CrossRef]
- Roberts, C.L.; Bell, J.C.; Ford, J.B.; Morris, J.M. Monitoring the quality of maternity care: How well are labour and delivery events reported in population health data? Paediatr. Perinat. Epidemiol. 2009, 23, 144–152. [Google Scholar] [CrossRef]
- Tun, H.M.; Bridgman, S.L.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Sears, M.R.; et al. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. JAMA Pediatr. 2018, 172, 368–377. [Google Scholar] [CrossRef]
- Shrier, I.; Platt, R.W. Reducing bias through directed acyclic graphs. BMC Med. Res. Methodol. 2008, 8, 70. [Google Scholar] [CrossRef] [Green Version]
- Evans, D.; Chaix, B.; Lobbedez, T.; Verger, C.; Flahault, A. Combining directed acyclic graphs and the change-in-estimate procedure as a novel approach to adjustment-variable selection in epidemiology. BMC Med. Res. Methodol. 2012, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Van der Leek, A.P.; Bahreinian, S.; Chartier, M.; Dahl, M.E.; Azad, M.B.; Brownell, M.D.; Kozyrskyj, A.L. Maternal distress during pregnancy and recurrence in early childhood predicts atopic dermatitis and asthma in childhood. Chest 2020, 158, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.J.; Sears, M.R.; HayGlass, K.T.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Becker, A.B.; et al. Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 2015, 45, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drall, K.M.; Tun, H.M.; Morales-Lizcano, N.P.; Konya, T.B.; Guttman, D.S.; Field, C.J.; Mandal, R.; Wishart, D.S.; Becker, A.B.; Azad, M.B.; et al. Clostridioides difficile colonization is differentially associated with gut microbiome profiles by infant feeding modality at 3-4 months of age. Front. Immunol. 2019, 10, 2866. [Google Scholar] [CrossRef]
- Frese, S.A.; Hutton, A.A.; Contreras, L.N.; Shaw, C.A.; Palumbo, M.C.; Casaburi, G.; Xu, G.; Davis, J.C.C.; Lebrilla, C.B.; Henrick, B.M.; et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere 2017, 2, e00501-17. [Google Scholar] [PubMed] [Green Version]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 1 May 2021).
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 1 May 2021).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ogle, D.H.; Doll, J.C.; Wheeler, P.; Dinno, A. FSA: Fisheries stock analysis. R Package Version 0.9.1.9000. 2021. Available online: https://github.com/droglenc/FSA (accessed on 1 May 2021).
- Osborne, J. Improving your data transformations: Applying the Box-Cox transformation. Pract. Assess. Res. Eval. 2010, 15, 12. [Google Scholar]
- Grech, A.; Collins, C.E.; Holmes, A.; Lal, R.; Duncanson, K.; Taylor, R.; Gordon, A. Maternal exposures and the infant gut microbiome: A systematic review with meta-analysis. Gut Microbes 2021, 13, 1–30. [Google Scholar] [CrossRef]
- Cortés-Macías, E.; Selma-Royo, M.; Martínez-Costa, C.; Collado, M.C. Breastfeeding practices influence the breast milk microbiota depending on pre-gestational maternal BMI and weight gain over pregnancy. Nutrients 2021, 13, 1518. [Google Scholar] [CrossRef] [PubMed]
- Persaud, R.R.; Azad, M.B.; Chari, R.S.; Sears, M.R.; Becker, A.B.; Kozyrskyj, A.L. Perinatal antibiotic exposure of neonates in Canada and associated risk factors: A population-based study. J. Matern. Fetal Neonatal. Med. 2015, 28, 1190–1195. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.; Gavini, F.; Vaugien, L.; Butel, M.J.; Doucet-Populaire, F. Antimicrobial susceptibility of bifidobacteria. J. Antimicrob. Chemother. 2005, 55, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, M.A.E.; O’Neill, I.J.; Kujawska, M.; Gowrinadh Javvadi, S.; Wijeyesekera, A.; Flegg, Z.; Chalklen, L.; Hall, L.J. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J 2020, 14, 635–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcobal, A.; Sonnenburg, J.L. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 2012, 18 (Suppl. 4), 12–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakwinska, O.; Foata, F.; Berger, B.; Brüssow, H.; Combremont, S.; Mercenier, A.; Dogra, S.; Soh, S.E.; Yen, J.C.K.; Heong, G.Y.S.; et al. Does the maternal vaginal microbiota play a role in seeding the microbiota of neonatal gut and nose? Benef. Microbes 2017, 8, 763–778. [Google Scholar] [CrossRef] [Green Version]
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Viappiani, A.; Lugli, G.A.; Ferrario, C.; Gioiosa, L.; Ferrarini, A.; et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 2016, 10, 1656–1668. [Google Scholar] [CrossRef]
- Chassard, C.; de Wouters, T.; Lacroix, C. Probiotics tailored to the infant: A window of opportunity. Curr. Opin. Biotechnol. 2014, 26, 141–147. [Google Scholar] [CrossRef]
- Fehr, K.; Moossavi, S.; Sbihi, H.; Boutin, R.C.T.; Bode, L.; Robertson, B.; Yonemitsu, C.; Field, C.J.; Becker, A.B.; Mandhane, P.J.; et al. Breastmilk Feeding practices are associated with the co-occurrence of bacteria in mothers’ milk and the infant gut: The CHILD cohort study. Cell Host Microbe 2020, 28, 285–297.e284. [Google Scholar] [CrossRef]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.S.; Ivanov, I.; Donovan, S.M. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef] [Green Version]
- Marcobal, A.; Barboza, M.; Froehlich, J.W.; Block, D.E.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010, 58, 5334–5340. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Fanning, S.; Hall, L.J.; Cronin, M.; Zomer, A.; MacSharry, J.; Goulding, D.; Motherway, M.O.; Shanahan, F.; Nally, K.; Dougan, G.; et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. USA 2012, 109, 2108–2113. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Stokholm, J.; Brejnrod, A.; Vestergaard, G.A.; Russel, J.; Trivedi, U.; Thorsen, J.; Gupta, S.; Hjelmsø, M.H.; Shah, S.A.; et al. The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host Microbe 2021, 29, 975–987.e974. [Google Scholar] [CrossRef] [PubMed]
- Podlesny, D.; Fricke, W.F. Strain inheritance and neonatal gut microbiota development: A meta-analysis. Int. J. Med. Microbiol. 2021, 311, 151483. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.M.; Sitarik, A.R.; Havstad, S.L.; Fujimura, K.E.; Wegienka, G.; Cassidy-Bushrow, A.E.; Kim, H.; Zoratti, E.M.; Lukacs, N.W.; Boushey, H.A.; et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci. Rep. 2016, 6, 31775. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.M.; Meng, L.; Liu, H.; Bao, D. Changes in intestinal flora in preeclampsia rats and effects of probiotics on their inflammation and blood pressure. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10155–10161. [Google Scholar] [PubMed]


| Target Microorganism | Primer Set/Probe Name | Sequence (5′–3′) | References |
|---|---|---|---|
| Total bacteria | Forward (Total F) | TCCTACGGGAGGCAGCAGT | [34] |
| Reverse (Total R) | GGACTACCAGGGTATCTAATCCTGTT | ||
| Probe (Total Probe) | 5′6-FAM /CGTATTACCGCGGCTGCTGGCAC/3′TAMRA | ||
| genus Bifidobacterium | Forward (Bif F) | GCGTGCTTAACACATGCAAGTC | [33] |
| Reverse (Bif R) | CACCCGTTTCCAGGAGCTATT | ||
| Probe (Bif probe) | 5′TexRd-XN/TCACGCATTACTCACCCGTTCGCC/3′IAbRQSp | ||
| B. longum subsp. infantis | Forward (Blon0915-F) | CGTATTGGCTTTGTACGCATTT | [33] |
| Reverse (Blon0915R) | ATCGTGCCGGTGAGATTTAC | ||
| Probe (Blon0915 Probe) | 5′6-FAM/CCAGTATGG/ZEN/CTGGTAAAGTTCACTGCA/3′IABkFQ |
| CS Groups Defined in This Study | |||||||
| Breastfeeding Status | Linear Regression Model | Vaginal-IAP | CS-No Labour | CS-Labour | |||
| Beta-Coefficient (95% CI) | p Value | Beta-Coefficient (95% CI) | p Value | Beta-Coefficient (95% CI) | p Value | ||
| All | Crude | −1.12 (−1.75, −0.49) | 0.0005 | −0.98 (−1.78, −0.19) | 0.02 | −0.88 (−1.75, −0.01) | 0.047 |
| Adjusted ‡ | −1.18 (−1.83, −0.52) | 0.0004 | −1.05 (−1.87, −0.22) | 0.01 | −1.29 (−2.19, −0.39) | 0.005 | |
| Exclusive | Crude | −1.01 (−1.88, −0.13) | 0.02 | −0.66 (−1.89, 0.57) | 0.29 | −1.03 (−2.27, 0.21) | 0.10 |
| Adjusted | −1.03 (−1.93, −0.13) | 0.03 | −0.91 (−2.19, 0.38) | 0.17 | −1.31 (−2.58, −0.04) | 0.04 | |
| Partial | Crude | −5.73 (−10.00, −1.46) | 0.01 | −3.96 (−8.79, 0.87) | 0.11 | −2.09 (−7.74, 3.55) | 0.47 |
| Adjusted | −6.24 (−10.72, −1.77) | 0.01 | −3.94 (−8.91, 1.03) | 0.12 | −2.86 (−8.66, 2.94) | 0.33 | |
| Non | Crude | −0.86 (−2.26, 0.55) | 0.23 | −1.77 (−3.40,−0.15) | 0.03 | −1.02 (−2.95, 0.91) | 0.30 |
| Adjusted | −0.91 (−2.32, 0.50) | 0.20 | −1.76 (−3.43, −0.09) | 0.04 | −2.59 (−4.61, −0.56) | 0.01 | |
| CS Groups Defined Clinically | |||||||
| Breastfeeding Status | Linear Regression Model | Vaginal-IAP | CS-Elective | CS-Emergency | |||
| Beta-Coefficient (95% CI) | p Value | Beta-Coefficient (95% CI) | p Value | Beta-Coefficient (95% CI) | p Value | ||
| All | Crude | −1.12 (−1.75, −0.49) | 0.0005 | −0.79 (−1.71, 0.13) | 0.09 | −1.04 (−1.80, −0.27) | 0.01 |
| Adjusted | −1.18 (−1.83, −0.52) | 0.0004 | −0.88 (−1.84, 0.08) | 0.07 | −1.33 (−2.13, −0.54) | 0.001 | |
| Exclusive | Crude | −1.01 (−1.88, −0.13) | 0.02 | 0.18 (−1.30, 1.65) | 0.82 | −1.34 (−2.43, −0.25) | 0.02 |
| Adjusted | −1.03 (−1.93, −0.13) | 0.03 | −0.22 (−1.78, 1.33) | 0.78 | −1.52 (−2.65, −0.40) | 0.01 | |
| Partial | Crude | −5.73 (−9.99, −1.47) | 0.01 | −5.74 (−11.01, −0.47) | 0.03 | −0.92 (−5.98, 4.14) | 0.72 |
| Adjusted | −6.24 (−10.71, −1.77) | 0.01 | −5.72 (−11.17, −0.27) | 0.04 | −1.54 (−6.73, 3.66) | 0.56 | |
| Non | Crude | −0.86 (−2.26, 0.55) | 0.23 | −1.45 (−3.31, 0.42) | 0.13 | −1.50 (−3.16, 0.17) | 0.08 |
| Adjusted | −0.91 (−2.32, 0.49) | 0.20 | −1.25 (−3.17, 0.68) | 0.20 | −2.71 (−4.42, −0.99) | 0.002 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.Y.; Zhao, X.; Moeder, W.; Tun, H.M.; Simons, E.; Mandhane, P.J.; Moraes, T.J.; Turvey, S.E.; Subbarao, P.; Scott, J.A.; et al. Impact of Maternal Intrapartum Antibiotics, and Caesarean Section with and without Labour on Bifidobacterium and Other Infant Gut Microbiota. Microorganisms 2021, 9, 1847. https://doi.org/10.3390/microorganisms9091847
Chen YY, Zhao X, Moeder W, Tun HM, Simons E, Mandhane PJ, Moraes TJ, Turvey SE, Subbarao P, Scott JA, et al. Impact of Maternal Intrapartum Antibiotics, and Caesarean Section with and without Labour on Bifidobacterium and Other Infant Gut Microbiota. Microorganisms. 2021; 9(9):1847. https://doi.org/10.3390/microorganisms9091847
Chicago/Turabian StyleChen, Yuan Yao, Xin Zhao, Wolfgang Moeder, Hein M. Tun, Elinor Simons, Piushkumar J. Mandhane, Theo J. Moraes, Stuart E. Turvey, Padmaja Subbarao, James A. Scott, and et al. 2021. "Impact of Maternal Intrapartum Antibiotics, and Caesarean Section with and without Labour on Bifidobacterium and Other Infant Gut Microbiota" Microorganisms 9, no. 9: 1847. https://doi.org/10.3390/microorganisms9091847






