High Antibiotic Resistance of Helicobacter pylori and Its Associated Novel Gene Mutations among the Mongolian Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population and Sampling
2.2. H. pylori Isolation and DNA Sequencing
2.2.1. Isolation and Culture of H. pylori
2.2.2. DNA Extraction and WGS
2.3. Antibiotic Susceptibility Tests and Detection of Genotypic Determinants of Antibiotic Resistance
2.3.1. Antibiotic Susceptibility Test
2.3.2. Detection of Genotypic Determinants of Antibiotic Resistance
2.3.3. Statistical Analysis
2.3.4. Nucleotide Sequence Accession Number
2.4. Ethics
3. Results
3.1. Genotypic Determination of Antibiotic Resistance
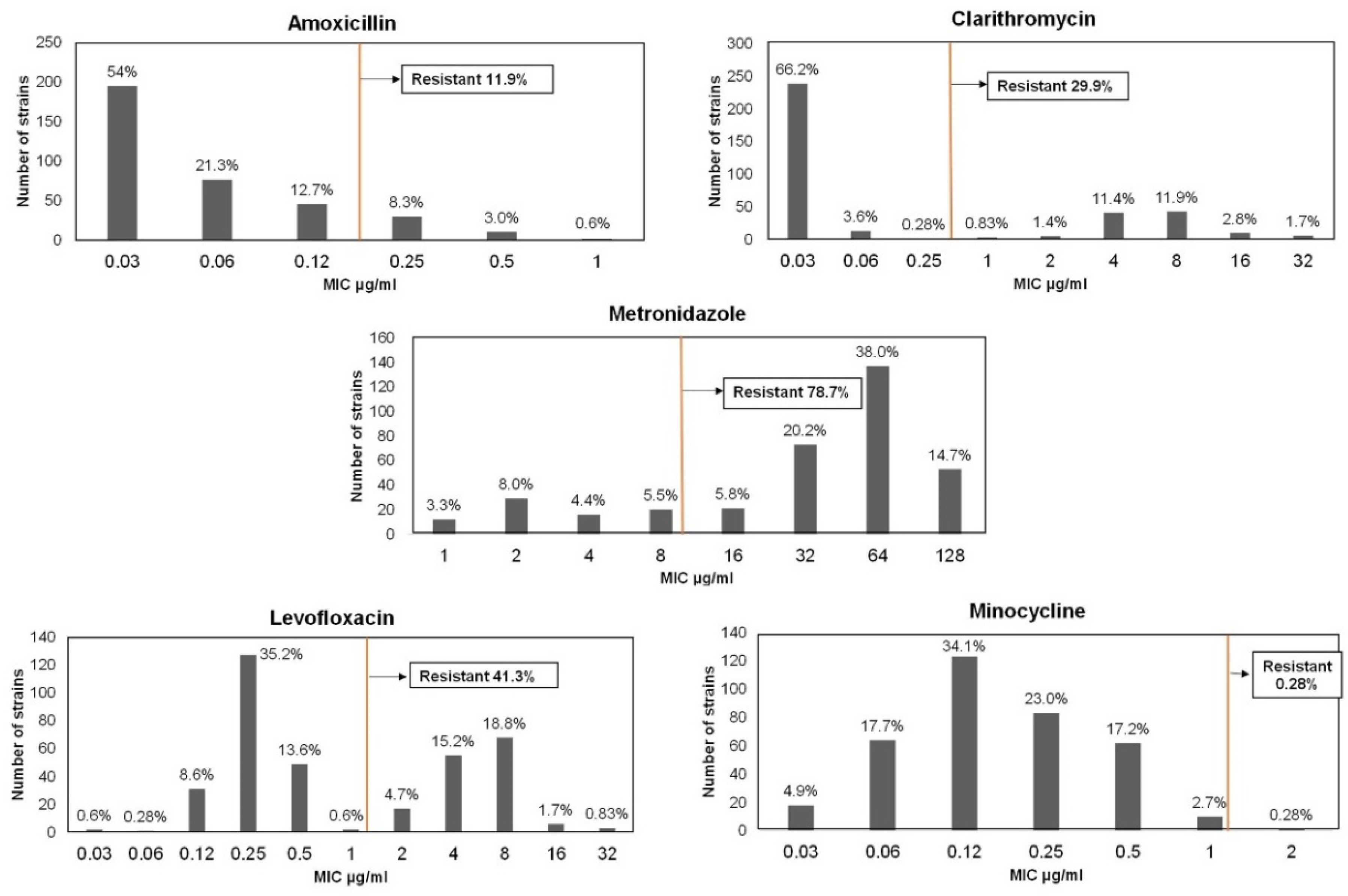
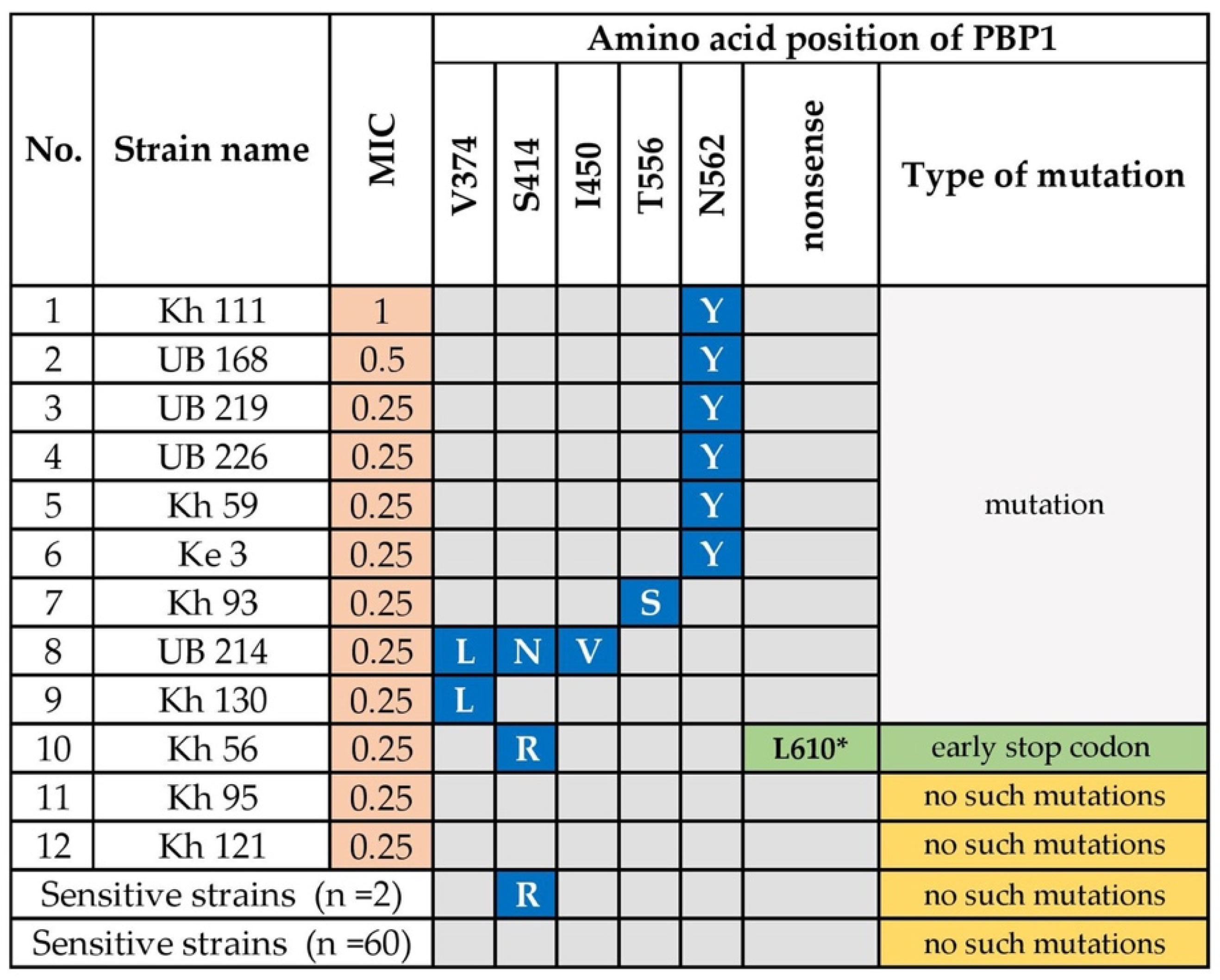
3.1.1. Amoxicillin Resistance
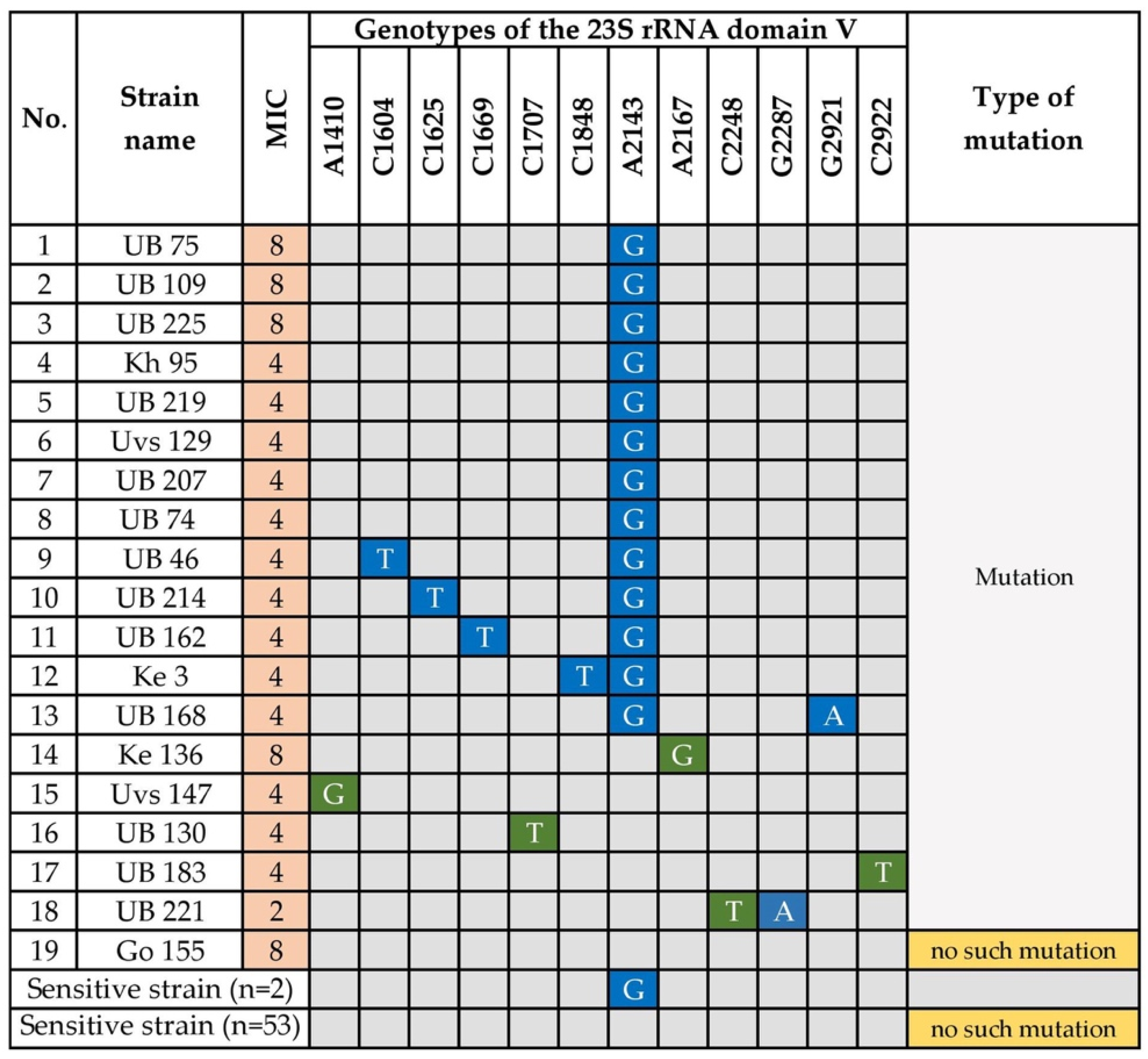
3.1.2. Clarithromycin Resistance
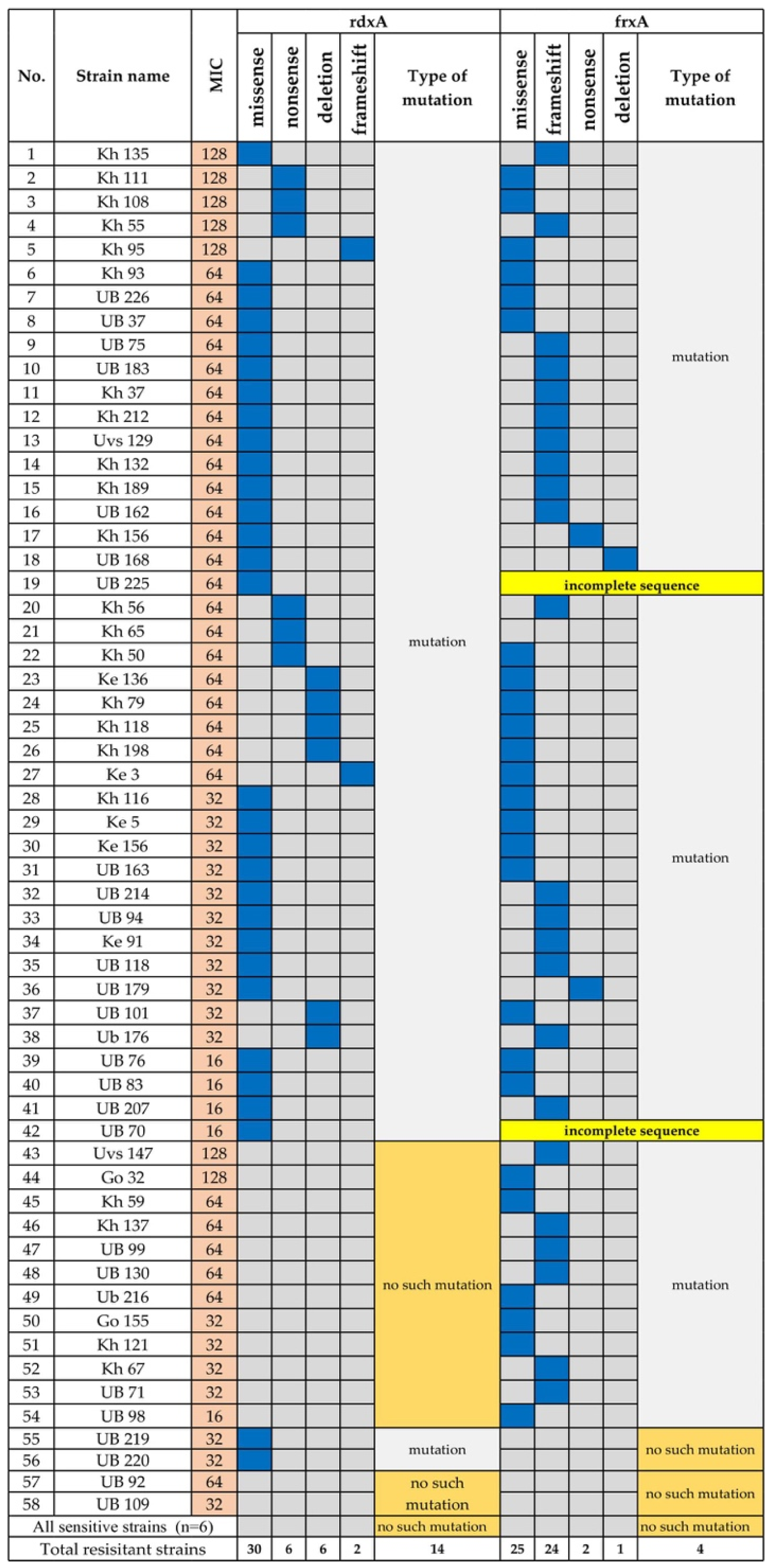
3.1.3. Metronidazole Resistance
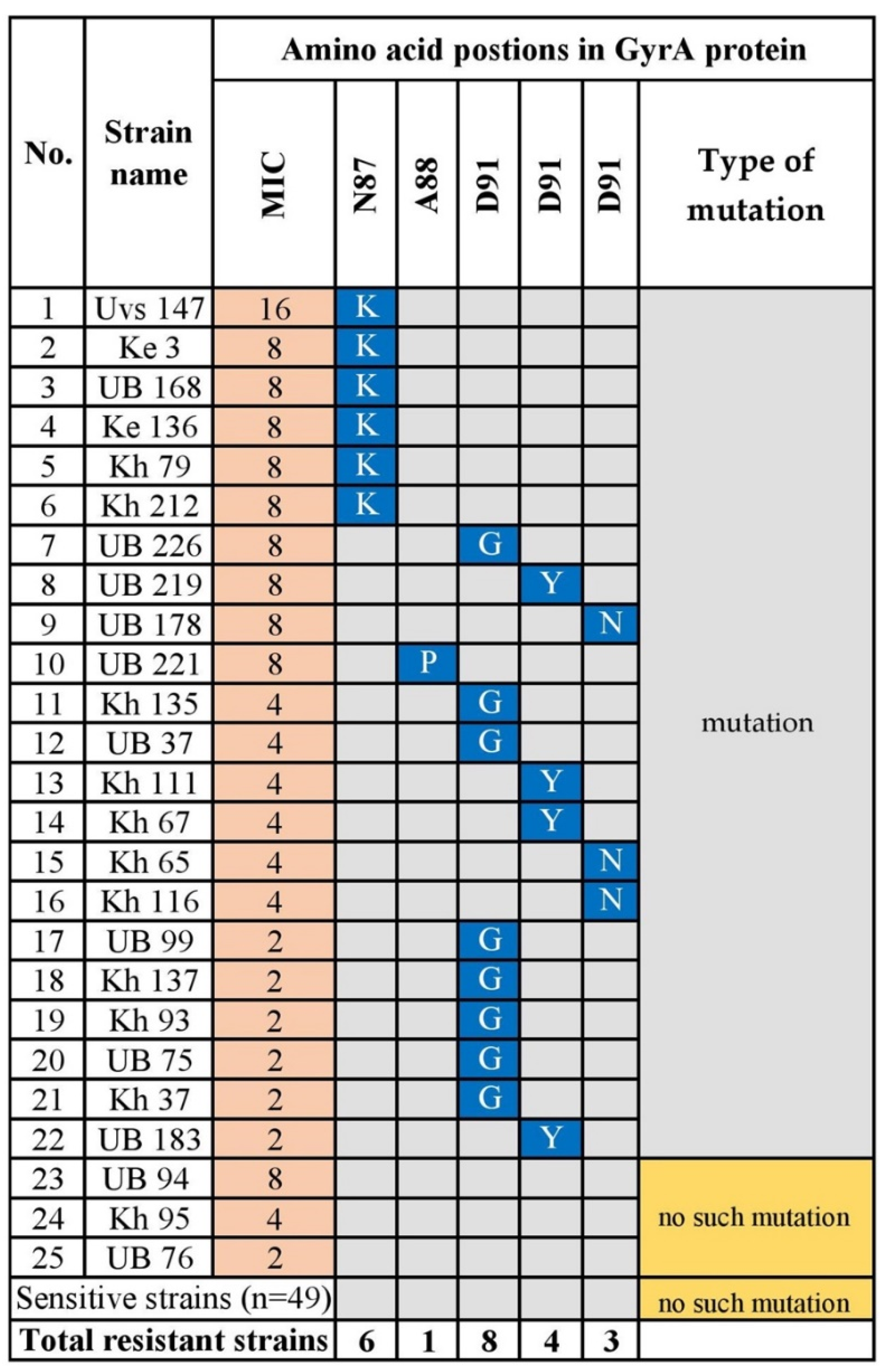
3.1.4. Levofloxacin Resistance
3.1.5. Minocycline Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global prevalence of Helicobacter pylori infection: Systematic review and meta-analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, J.R.; Marshall, B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983, 1, 1273–1275. [Google Scholar] [PubMed]
- IARC. Helicobacter Pylori Eradication as a Strategy for Preventing Gastric Cancer 2014; IARC: Lyon, France, 2014. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuhisa, T.; Yamaoka, Y.; Uchida, T.; Duger, D.; Adiyasuren, B.; Khasag, O.; Tegshee, T.; Tsogt-Ochir, B. Gastric mucosa in Mongolian and Japanese patients with gastric cancer and Helicobacter pylori infection. World J. Gastroenterol. 2015, 21, 8408–8417. [Google Scholar] [CrossRef]
- Khasag, O.; Boldbaatar, G.; Tegshee, T.; Duger, D.; Dashdorj, A.; Uchida, T.; Matsuhisa, T.; Yamaoka, Y. The prevalence of Helicobacter pylori infection and other risk factors among Mongolian dyspeptic patients who have a high incidence and mortality rate of gastric cancer. Gut Pathog. 2018, 10, 14. [Google Scholar] [CrossRef] [Green Version]
- Global Cancer Observatory (GCO). Available online: https://gco.iarc.fr/ (accessed on 1 June 2018).
- Graham, D.Y.; Lu, H.; Yamaoka, Y. A report card to grade Helicobacter pylori therapy. Helicobacter 2007, 12, 275–278. [Google Scholar] [CrossRef]
- Megraud, F. H pylori antibiotic resistance: Prevalence, importance, and advances in testing. Gut 2004, 53, 1374–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, Y.T.; Liou, J.M.; El-Omar, E.M.; Wu, J.Y.; Leow, A.H.R.; Goh, K.L.; Das, R.; Lu, H.; Lin, J.T.; Tu, Y.K.; et al. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 707–715. [Google Scholar] [CrossRef]
- World Health Organization. List of Bacteria for Which New Antibiotics Are Urgently Needed; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Wang, D.; Guo, Q.; Yuan, Y.; Gong, Y. The antibiotic resistance of Helicobacter pylori to five antibiotics and influencing factors in an area of China with a high risk of gastric cancer. BMC Microbiol. 2019, 19, 152. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Ota, H.; Okuda, M.; Kikuchi, S.; Satoh, K.; Shimoyama, T.; Suzuki, H.; Handa, O.; Furuta, T.; Mabe, K.; et al. Guidelines for the management of Helicobacter pylori infection in Japan: 2016 Revised Edition. Helicobacter 2019, 24, e12597. [Google Scholar] [CrossRef] [PubMed]
- Storskrubb, T.; Aro, P.; Ronkainen, J.; Wreiber, K.; Nyhlin, H.; Bolling-Sternevald, E.; Talley, N.J.; Engstrand, L.; Agreus, L. Antimicrobial susceptibility of Helicobacter pylori strains in a random adult Swedish population. Helicobacter 2006, 11, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.; Kersulyte, D.; Sisson, G.; Veldhuyzen van Zanten, S.J.; Berg, D.E.; Hoffman, P.S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 1998, 28, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; El-Zaatari, F.A.; Kato, M.; Osato, M.S.; Reddy, R.; Yamaoka, Y.; Graham, D.Y. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob. Agents Chemother. 2000, 44, 2133–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, Y.; Yuan, Y. Resistance mechanisms of Helicobacter pylori and its dual target precise therapy. Crit. Rev. Microbiol. 2018, 44, 371–392. [Google Scholar] [CrossRef]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of antibiotic resistance in Helicobacter pylori: A systematic review and meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [Green Version]
- Gerrits, M.M.; de Zoete, M.R.; Arents, N.L.; Kuipers, E.J.; Kusters, J.G. 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2002, 46, 2996–3000. [Google Scholar] [CrossRef] [Green Version]
- Chey, W.D.; Leontiadis, G.I.; Howden, C.W.; Moss, S.F. ACG clinical guideline: Treatment of Helicobacter pylori infection. Am. J. Gastroenterol. 2017, 112, 212–239. [Google Scholar] [CrossRef]
- Bolor-Erdene, M.; Namdag, B.; Yamaoka, Y.; Jav, S. Antibiotic resistance of Helicobacter pylori in Mongolia. J. Infect. Dev. Ctries. 2017, 11, 887–894. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gerrits, M.M.; Godoy, A.P.; Kuipers, E.J.; Ribeiro, M.L.; Stoof, J.; Mendonca, S.; van Vliet, A.H.; Pedrazzoli, J., Jr.; Kusters, J.G. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter 2006, 11, 181–187. [Google Scholar] [CrossRef]
- Kim, B.J.; Kim, J.G. Substitutions in penicillin-binding protein 1 in amoxicillin-resistant Helicobacter pylori strains isolated from Korean patients. Gut Liver 2013, 7, 655–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyachi, H.; Miki, I.; Aoyama, N.; Shirasaka, D.; Matsumoto, Y.; Toyoda, M.; Mitani, T.; Morita, Y.; Tamura, T.; Kinoshita, S.; et al. Primary levofloxacin resistance and gyrA/B mutations among Helicobacter pylori in Japan. Helicobacter 2006, 11, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Byambajav, T.O.; Bira, N.; Choijamts, G.; Davaadorj, D.; Gantuya, B.; Sarantuya, T.; Sarantuya, G.; Enkhtsetseg, A.; Erdenetsogt, D.; Battulga, A.; et al. Initial trials with susceptibility-based and empiric anti-H. pylori therapies in Mongolia. Front. Pharmacol. 2019, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Center for Health Development: Surveillance of Antibiotic Consumption in Mongolia; Ministry of Health and World Health Organization: 2019. Available online: https://www.who.int/medicines/areas/rational_use/who-amr-amc-report-20181109.pdf (accessed on 5 June 2020).
- Rimbara, E.; Noguchi, N.; Kawai, T.; Sasatsu, M. Correlation between substitutions in penicillin-binding protein 1 and amoxicillin resistance in Helicobacter pylori. Microbiol. Immunol. 2007, 51, 939–944. [Google Scholar] [CrossRef]
- Kwon, Y.H.; Kim, J.Y.; Kim, N.; Park, J.H.; Nam, R.H.; Lee, S.M.; Kim, J.W.; Kim, J.M.; Park, J.Y.; Lee, D.H. Specific mutations of penicillin-binding protein 1A in 77 clinically acquired amoxicillin-resistant Helicobacter pylori strains in comparison with 77 amoxicillin-susceptible strains. Helicobacter 2017, 22, e12437. [Google Scholar] [CrossRef]
- Gerrits, M.M.; Schuijffel, D.; van Zwet, A.A.; Kuipers, E.J.; Vandenbroucke-Grauls, C.M.; Kusters, J.G. Alterations in penicillin-binding protein 1A confer resistance to beta-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 2002, 46, 2229–2233. [Google Scholar] [CrossRef] [Green Version]
- Rimbara, E.; Noguchi, N.; Kawai, T.; Sasatsu, M. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J. Antimicrob. Chemother. 2008, 61, 995–998. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.M.; Kim, J.S.; Kim, N.; Kim, Y.J.; Kim, I.Y.; Chee, Y.J.; Lee, C.H.; Jung, H.C. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J. Microbiol. Biotechnol. 2008, 18, 1584–1589. [Google Scholar]
- Tran, V.H.; Ha, T.M.T.; Le, P.T.Q.; Phan, T.N.; Tran, T.N.H. Characterisation of point mutations in domain V of the 23S rRNA gene of clinical Helicobacter pylori strains and clarithromycin-resistant phenotype in central Vietnam. J. Glob. Antimicrob. Resist. 2019, 16, 87–91. [Google Scholar] [CrossRef]
- Kwon, D.H.; Kato, M.; El-Zaatari, F.A.; Osato, M.S.; Graham, D.Y. Frame-shift mutations in NAD(P)H flavin oxidoreductase encoding gene (frxA) from metronidazole resistant Helicobacter pylori ATCC43504 and its involvement in metronidazole resistance. FEMS Microbiol. Lett. 2000, 188, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Sugimoto, M.; Mizuno, H.; Kanno, T.; Satoh, K. Clarithromycin versus metronidazole in first-line Helicobacter pylori triple eradication therapy based on resistance to antimicrobial agents: Meta-analysis. J. Clin. Med. 2020, 9, 543. [Google Scholar] [CrossRef] [Green Version]
- Gisbert, J.P.; Morena, F. Systematic review and meta-analysis: Levofloxacin-based rescue regimens after Helicobacter pylori treatment failure. Aliment. Pharmacol. Ther. 2006, 23, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Y.; Lu, N.H. Primary antibiotic resistance of Helicobacter pylori in China. Dig. Dis. Sci. 2017, 62, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Tuan, V.P.; Narith, D.; Tshibangu-Kabamba, E.; Dung, H.D.Q.; Viet, P.T.; Sokomoth, S.; Binh, T.T.; Sokhem, S.; Tri, T.D.; Ngov, S.; et al. A next-generation sequencing-based approach to identify genetic determinants of antibiotic resistance in Cambodian Helicobacter pylori clinical isolates. J. Clin. Med. 2019, 8, 858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.; Raymond, J.; Garnier, M.; Cremniter, J.; Burucoa, C. Distribution of spontaneous gyrA mutations in 97 fluoroquinolone-resistant Helicobacter pylori isolates collected in France. Antimicrob. Agents. Chemother. 2012, 56, 550–551. [Google Scholar] [CrossRef] [Green Version]
- Mori, H.; Suzuki, H.; Matsuzaki, J.; Masaoka, T.; Kanai, T. Acquisition of double mutation in gyrA caused high resistance to sitafloxacin in Helicobacter pylori after unsuccessful eradication with sitafloxacin-containing regimens. United Eur. Gastroenterol. J. 2018, 6, 391–397. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Suo, B.; Zhang, L.; Zhou, L. Rabeprazole, minocycline, amoxicillin, and bismuth as first-line and second-line regimens for Helicobacter pylori eradication. Helicobacter 2016, 21, 462–470. [Google Scholar] [CrossRef]
- Murakami, K.; Sato, R.; Okimoto, T.; Watanabe, K.; Nasu, M.; Fujioka, T.; Kodama, M.; Abe, T.; Sato, S.; Arita, T. Effectiveness of minocycline-based triple therapy for eradication of Helicobacter pylori infection. J. Gastroenterol. Hepatol. 2006, 21 Pt 2, 262–267. [Google Scholar] [CrossRef]
| Group | Total, n (%) | Antibiotic-Resistant Strains, n (%) | ||||
|---|---|---|---|---|---|---|
| Amoxicillin | Clarithromycin | Metronidazole | Levofloxacin | Minocycline | ||
| Total, n | 361 | 43 (11.9) | 108 (29.9) | 284 (78.7) | 149 (41.3) | 1 (0.28) |
| Sex | ||||||
| Female | 264 (73.1) | 31 (11.7) | 85 (32.2) | 218 (82.6) ** | 113 (42.8) | 0 (0) |
| Male | 97 (26.9) | 12 (12.4) | 23 (23.7) | 66 (68.0) | 36 (37.1) | 1 (1.0) |
| Age # | ||||||
| 19–29 | 65 | 9 (13.8) | 20 (30.8) | 44 (67.7) | 21 (32.3) | 0 (0) |
| 30–39 | 77 | 7 (9.1) | 27 (35.1) | 66 (85.7) | 34 (44.2) | 1 (1.3) |
| 40–49 | 78 | 6 (7.7) | 23 (29.5) | 63 (80.8) | 34 (43.6) | 0 (0) |
| 50–59 | 91 | 13 (14.3) | 25 (27.5) | 75 (82.4) | 40 (44.0) | 0 (0) |
| 60< | 50 | 8 (16.0) | 13 (26.0) | 36 (72.0) | 20 (40.0) | 0 (0) |
| Areas | Total, n | Amoxicillin | Clarithromycin | Metronidazole | Levofloxacin | Minocycline | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % (95% CI) | n | % (95% CI) | n | % (95% CI) | n | % (95% CI) | n | % (95% CI) | ||
| Khentii | 90 | 12 | 13.3 (7.5–21.5) | 22 | 24.4 (16.5–34) | 75 | 83.3 (74.6–89.9) | 42 | 46.7 (36.6–56.9) | 0 | 0.0 |
| Khuvsgul | 35 | 7 | 20.0 (9.4–35.3) | 6 | 17.1 (7.5–32) | 29 | 82.9 (68–92.5) | 14 | 40.0 (25.1–56.5) | 0 | 0.0 |
| Ulaanbaatar | 124 | 11 | 8.9 (4.8–14.8) | 42 | 33.9 (26–42.5) | 91 | 73.4 (65.1–80.6) | 47 | 37.9 (29.7–46.6) | 1 | 0.8 (0.1–3.7) |
| Umnugovi | 84 | 6 | 7.1 (3.0–14.1) | 24 | 28.6 (19.8–38.8) | 68 | 81.0 (71.6–88.2) | 35 | 41.7 (31.6–52.3) | 0 | 0.0 |
| Uvs | 28 | 7 | 25.0 * (11.9–42.9) | 14 | 50.0 * (32.2–67.8) | 21 | 75.0 (57.1–88.1) | 11 | 39.3 (23–57.7) | 0 | 0.0 |
| * p = 0.043 | * p = 0.034 | NS | NS | NS | |||||||
| Resistance Pattern | Number of Strains | Percentage (95% CI) |
|---|---|---|
| Sensitive to All Antibiotics | 43 | 11.9% (8.9–15.6) |
| Single drug | 133 | |
| MNZ | 106 | 29.4% (24.8–34.2) |
| LEV | 11 | 3.0% (1.6–5.2) |
| CLR | 10 | 2.8% (1.4–4.9) |
| AMX | 5 | 1.4% (0.5–3.0) |
| MNO | 1 | 0.28% (0.0–1.3) |
| Two drugs | 113 | |
| MNZ + LEV | 66 | 18.3% (14.6–22.5) |
| CLR + MNZ | 33 | 9.1% (6.5–12.4) |
| AMX + MNZ | 9 | 2.5% (1.2–4.5) |
| CLR + LEV | 5 | 1.4% (0.5–3.0) |
| Three drugs | 62 | |
| CLR + MNZ + LEV | 43 | 11.9% (8.9–15.6) |
| AMX + MNZ + LEV | 12 | 3.3% (1.8–5.6) |
| AMX + CLR + MNZ | 5 | 1.4% (0.5–3.0) |
| AMX + CLR + LEV | 2 | 0.56% (0.1–1.8) |
| Four drugs | 10 | |
| AMX+CLR+MNZ+LEV | 10 | 2.8% (1.4–4.9) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzaya, D.; Gantuya, B.; Oyuntsetseg, K.; Davaadorj, D.; Matsumoto, T.; Akada, J.; Yamaoka, Y. High Antibiotic Resistance of Helicobacter pylori and Its Associated Novel Gene Mutations among the Mongolian Population. Microorganisms 2020, 8, 1062. https://doi.org/10.3390/microorganisms8071062
Azzaya D, Gantuya B, Oyuntsetseg K, Davaadorj D, Matsumoto T, Akada J, Yamaoka Y. High Antibiotic Resistance of Helicobacter pylori and Its Associated Novel Gene Mutations among the Mongolian Population. Microorganisms. 2020; 8(7):1062. https://doi.org/10.3390/microorganisms8071062
Chicago/Turabian StyleAzzaya, Dashdorj, Boldbaatar Gantuya, Khasag Oyuntsetseg, Duger Davaadorj, Takashi Matsumoto, Junko Akada, and Yoshio Yamaoka. 2020. "High Antibiotic Resistance of Helicobacter pylori and Its Associated Novel Gene Mutations among the Mongolian Population" Microorganisms 8, no. 7: 1062. https://doi.org/10.3390/microorganisms8071062





