Cold-Active, Heterotrophic Bacteria from the Highly Oligotrophic Waters of Lake Vanda, Antarctica
Abstract
:1. Introduction
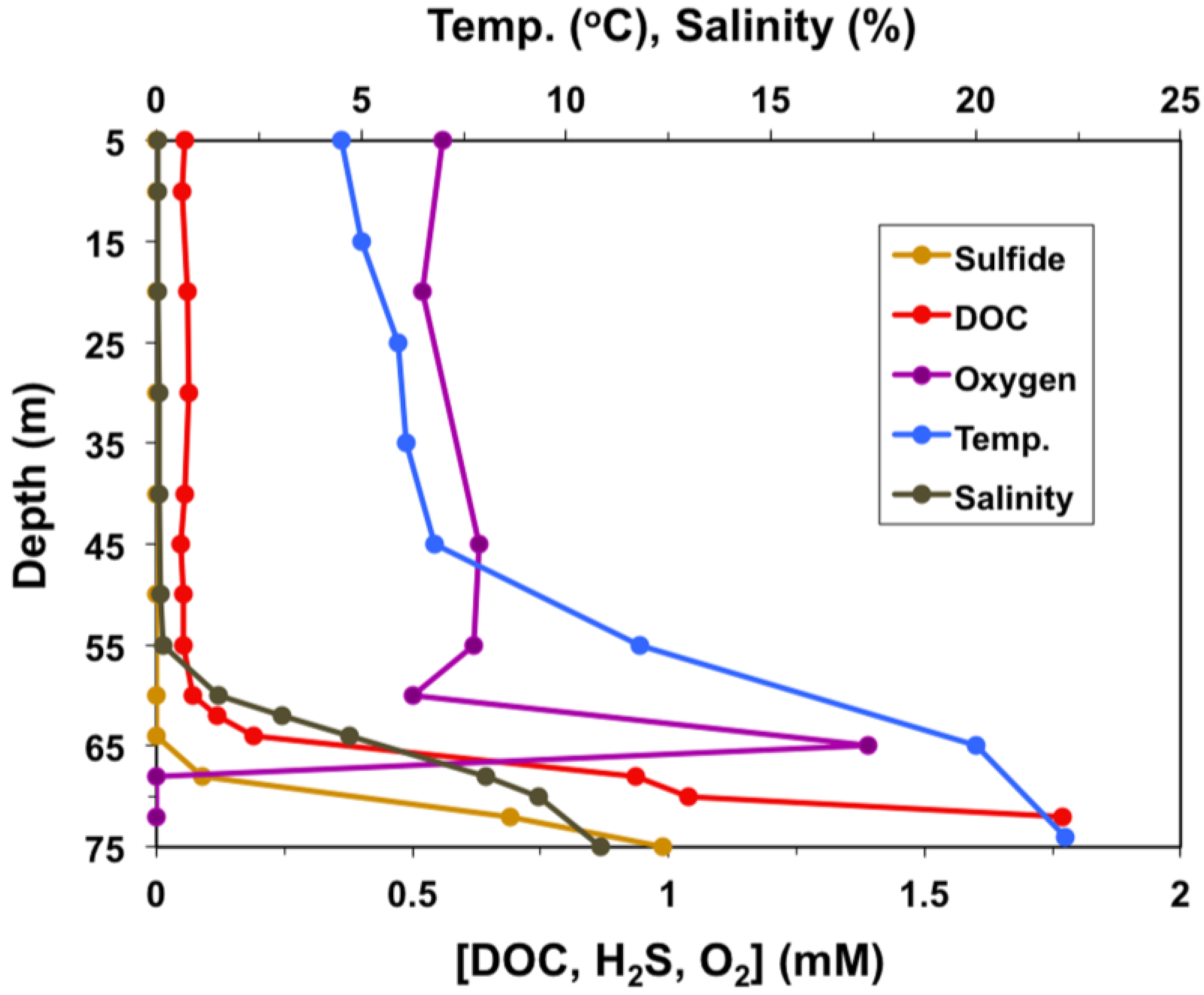
2. Materials and Methods
2.1. Sample Collection and Analysis, Enrichment, and Isolation
2.2. Morphology
2.3. Physiological Studies
2.4. Phylogenetic Analyses
3. Results
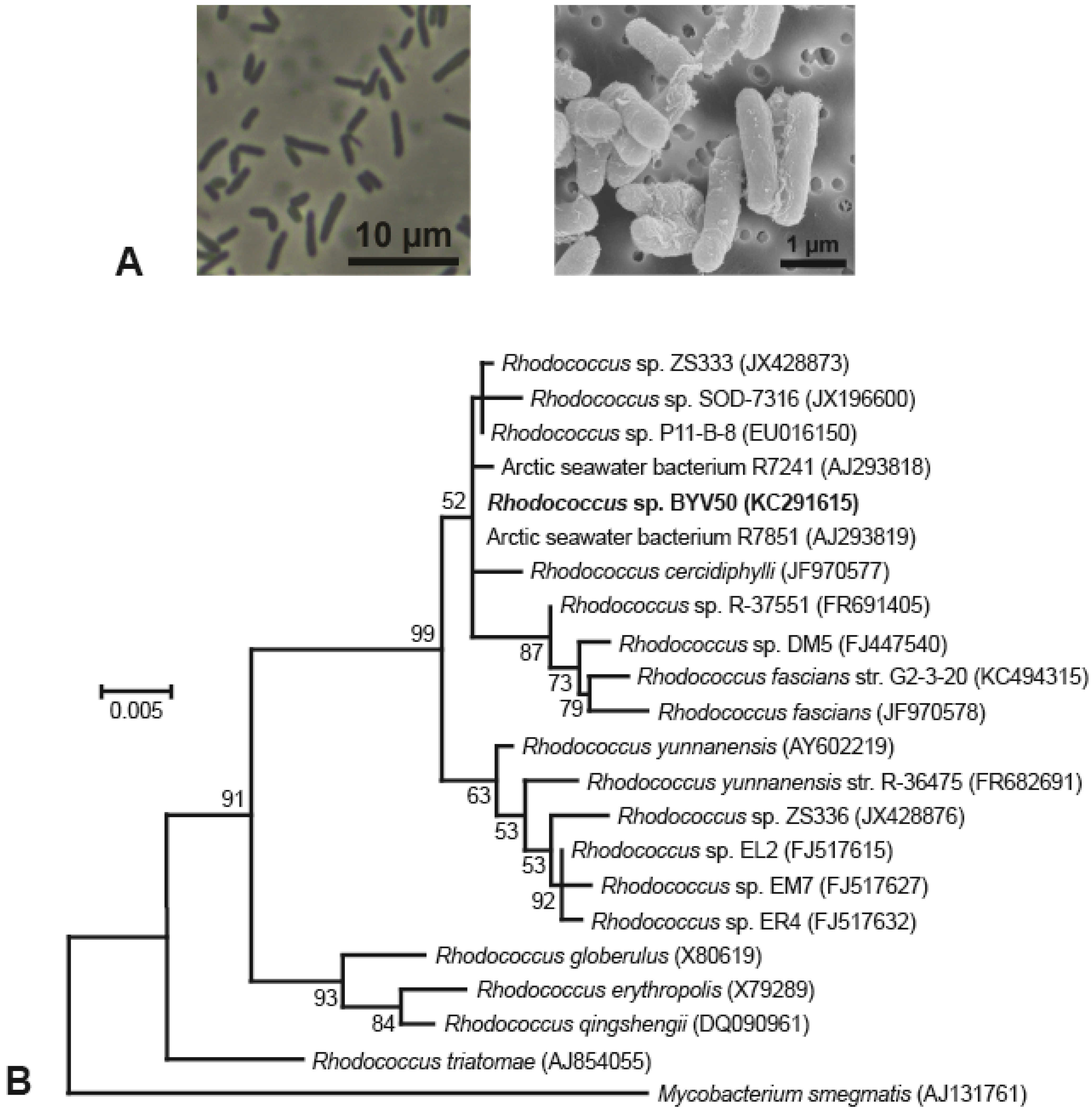
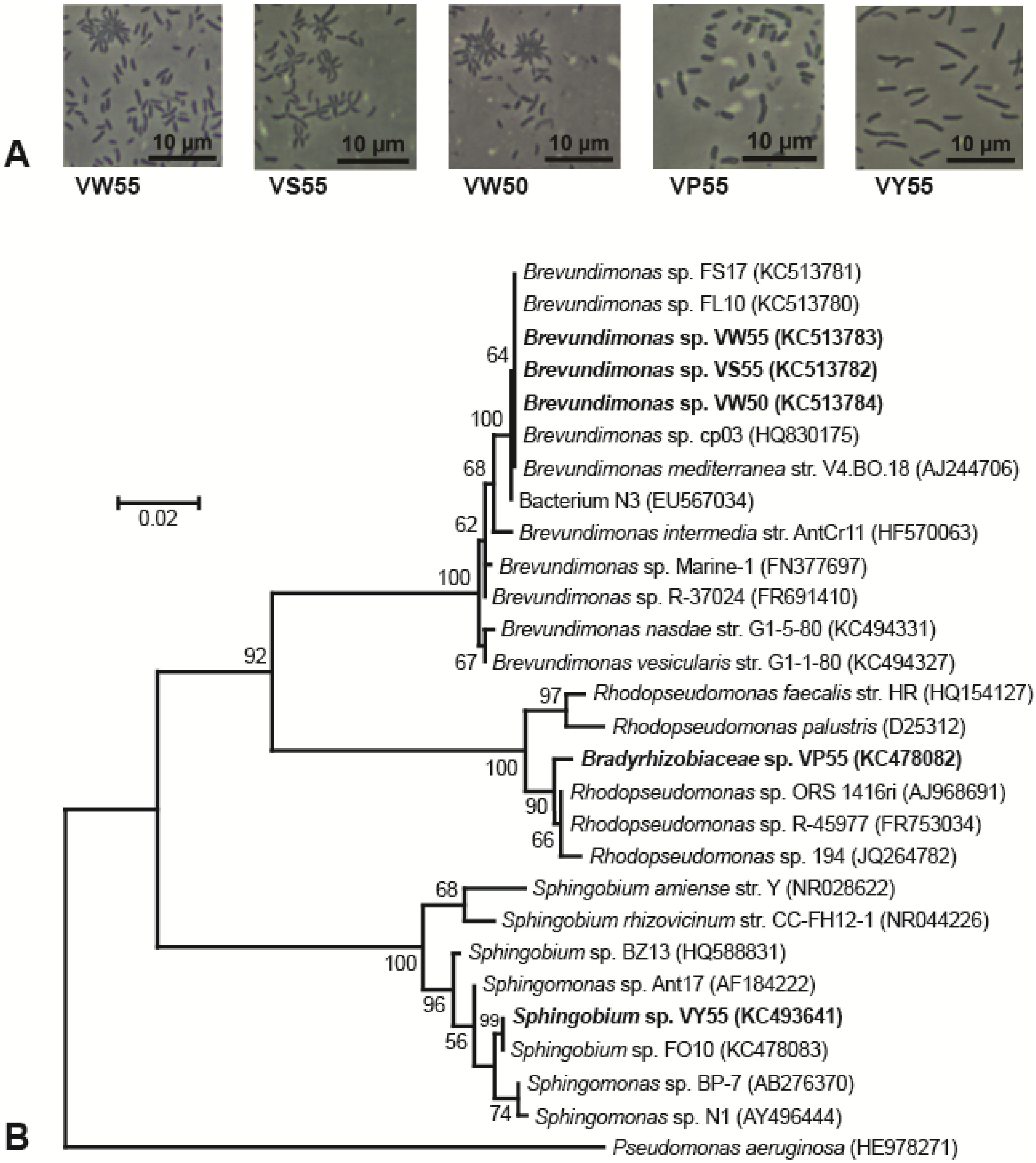
3.1. Isolation and Morphology of Lake Vanda Strains
| Property | Rhodococcus sp. BYV50 | Brevundimonas spp. VW50, VW55, and VS55 | Bradyrhizobiaceae sp. VP55 | Sphingobium sp. VY55 |
|---|---|---|---|---|
| Cell shape | Straight rod | Straight-to-curved rod | Straight rod, often tapered | Straight rod |
| Motility | − | + | − | + |
| Cell size (l × w, μm) | 2–5 × 1–1.2 | 1.4–3.2 × 0.7–0.8 | 1.5–3.5 × 0.9–1.1 | 1.8–8 × 0.9–1 |
| Gram stain reaction | + | − | − | − |
| Pigment | Bright yellow/Orange | None | Pink | Bright yellow |
| Temp. range (°C) | −2–32 | −2–35 | 2–29 | 0–30 |
| Temp. optimum (°C) | 18–24 | 20 | 20 | 20 |
| Salinity range (% NaCl) | 0–10 | 0–6 | 0–1 | 0–6 |
3.2. Phylogeny
3.3. Physiology
| Carbon Source | Rhodococcus sp. BYV50 | Brevundimonas spp. VW50, VW55, VS55 | Bradyrhizobiaceae sp. VP55 | Sphingobium sp. VY55 |
|---|---|---|---|---|
| SUGARS | ||||
| Glucose | + | + | − | (+) |
| Fructose | ++ | (+) | − | − |
| Ribose | + | (+) | − | − |
| Galactose | + | + | − | (+) |
| Sucrose | ++ | − | − | (+) |
| Maltose | + | + | − | (+) |
| Lactose | + | (+) | − | (+) |
| Xylose | ++ | − | − | − |
| Mannose | + | (+) | − | − |
| ALCOHOLS | ||||
| Ethanol | + | + | − | (+) |
| Propanol | ++ | (+) | − | − |
| FATTY AND ORGANIC ACIDS | ||||
| Acetate | + | + | − | (+) |
| Pyruvate | + | + | + | (+) |
| Propionate | ++ | − | − | − |
| Butyrate | ++ | + | (+) | (+) |
| Lactate | ++ | + | + | − |
| Fumarate | + | + | − | − |
| Succinate | ++ | + | (+) | (+) |
| Benzoate | + | − | − | − |
| Yeast Extract | ++ | ++ | + | + |
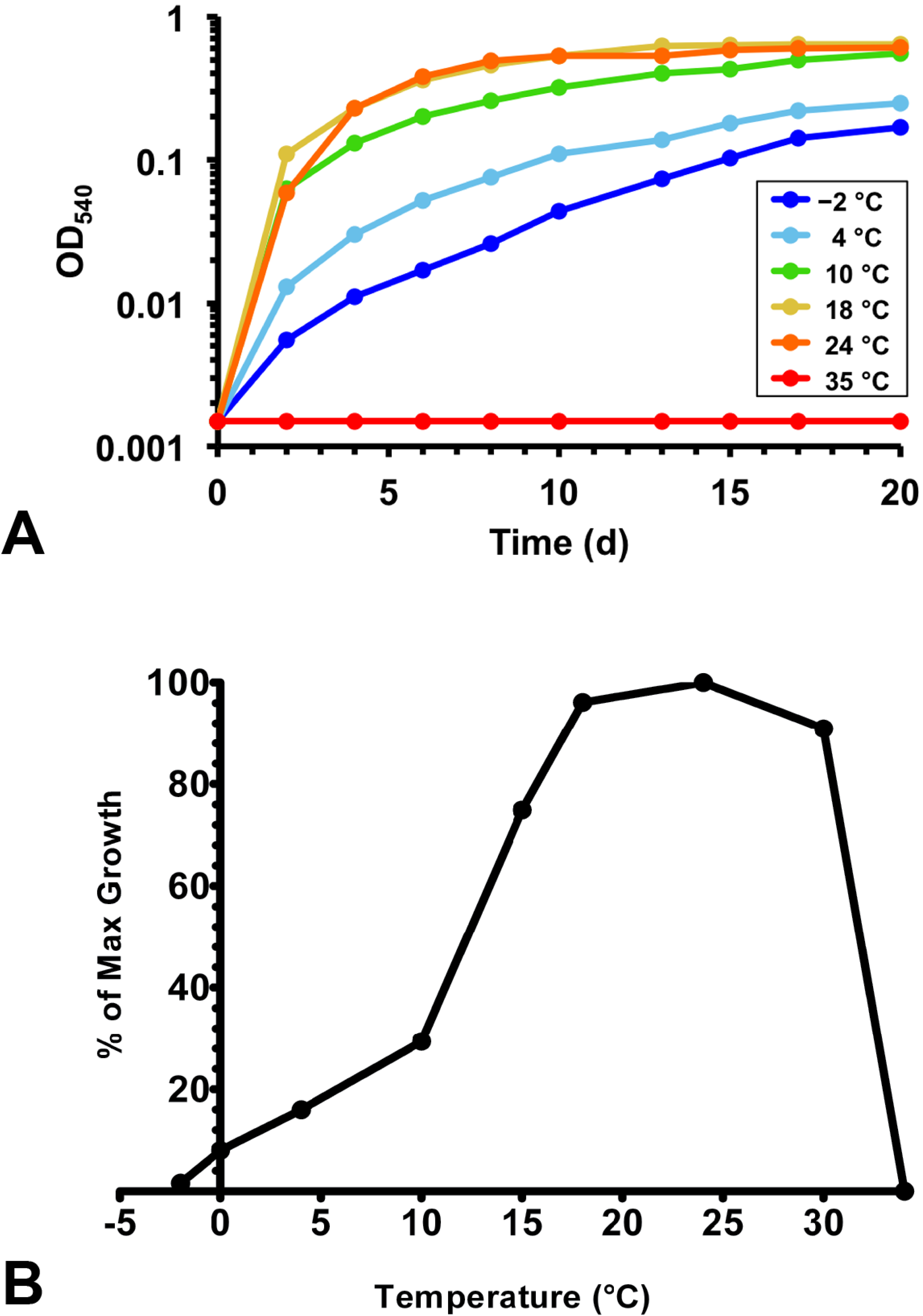
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Spigel, R.H.; Priscu, J.C. Physical limnology of McMurdo Dry Valley Lakes. In Ecosystem Dynamics in a Polar Desert; Priscu, J.C., Ed.; American Geophysical Union: Washington, DC, USA, 1998; Volume 72, pp. 153–187. [Google Scholar]
- Vincent, W.F.; Vincent, C.L. Factors controlling phytoplankton production in Lake Vanda (77 °C). Can. J. Fish. Aquat. Sci. 1982, 39, 1602–1609. [Google Scholar] [CrossRef]
- Lee, P.A.; Mikucki, J.A.; Foreman, C.M.; Priscu, J.C.; DiTullio, G.R.; Riseman, S.F.; de Mora, S.J.; Wolf, C.F.; Kester, L. Thermodynamic constraints on microbially mediated processes in lakes of the McMurdo Dry Valleys, Antarctica. Geomicrobiol. J. 2004, 21, 221–237. [Google Scholar] [CrossRef]
- Matsumoto, G.; Torii, T.; Hanya, T. Vertical distribution of organic constituents in an Antarctic Lake: Lake Vanda. Hydrobiologia 1984, 111, 119–126. [Google Scholar] [CrossRef]
- McKnight, D.M.; Aiken, G.R.; Andrews, E.D.; Bowles, E.C.; Harnish, R.A. Dissolved organic material in Dry Valley Lakes: A comparison of Lake Fryxell, Lake Hoare, and Lake Vanda. In Physical and Biogeochemical Processes in Antarctic Lakes, Antarctic Research Series; Green, W., Friedmann, E.I., Eds.; American Geophysical Union: Washington, DC, USA, 1993; Volume 59, pp. 119–133. [Google Scholar]
- Vincent, W.F.; Downes, M.T.; Vincent, C.L. Nitrous oxide cycling in Lake Vanda, Antarctica. Nature 1981, 292, 618–620. [Google Scholar] [CrossRef]
- Canfield, D.E.; Green, W.J. The cycling of nutrients in a closed-basin Antarctic Lake: Lake Vanda. Biogeochemistry 1985, 1, 233–256. [Google Scholar] [CrossRef]
- Howes, B.L.; Schlezinger, D.R.; Goehringer, D.D.; Brown-Leger, S. Carbon cycling in a redox-stratified Antarctic Lake, Lake Fryxell. Antarct. J. US 1992, 27, 263–265. [Google Scholar]
- Takacs, C.D.; Priscu, J.C. Bacterioplankton dynamics in the McMurdo Dry Valley Lakes, Antarctica: Production and biomass loss over four seasons. Microbial Ecol. 1998, 36, 239–250. [Google Scholar] [CrossRef]
- Voytek, M.A.; Priscu, J.C.; Ward, B.B. The distribution and relative abundance of ammonia-oxidizing bacteria in lakes of the McMurdo Dry Valley, Antarctica. Hydrobiologia 1999, 401, 113–130. [Google Scholar] [CrossRef]
- Karr, E.A.; Sattley, W.M.; Jung, D.O.; Madigan, M.T.; Achenbach, L.A. Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic Lake. Appl. Environ. Microbiol. 2003, 69, 4910–4914. [Google Scholar] [CrossRef] [PubMed]
- Karr, E.A.; Sattley, W.M.; Rice, M.R.; Jung, D.O.; Madigan, M.T.; Achenbach, L.A. Diversity and distribution of sulfate-reducing bacteria in permanently frozen Lake Fryxell, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 2005, 71, 6353–6359. [Google Scholar] [CrossRef] [PubMed]
- Karr, E.A.; Ng, J.M.; Belchik, S.M.; Sattley, W.M.; Madigan, M.T.; Achenbach, L.A. Biodiversity of methanogenic and other Archaea in the permanently frozen Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 2006, 72, 1663–1666. [Google Scholar] [CrossRef] [PubMed]
- Glatz, R.E.; Lepp, P.W.; Ward, B.B.; Francis, C.A. Planktonic microbial community composition across steep physical/chemical gradients in permanently ice-covered Lake Bonney, Antarctica. Geobiology 2006, 4, 53–67. [Google Scholar] [CrossRef]
- Kong, W.; Dolhi, J.M.; Chiuchiolo, A.; Priscu, J.; Morgan-Kiss, R.M. Evidence of form II RubisCO (cbbM) in a permanently ice-covered Antarctic Lake. FEMS Microbiol. Ecol. 2012, 82, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Sattley, W.M.; Madigan, M.T. Isolation, characterization, and ecology of cold-active, chemolithotrophic, sulfur-oxidizing bacteria from perennially ice-covered Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 2006, 72, 5562–5568. [Google Scholar] [CrossRef] [PubMed]
- Sattley, W.M.; Madigan, M.T. Cold-active acetogenic bacteria from surficial sediments of perennially ice-covered Lake Fryxell, Antarctica. FEMS Microbiol. Lett. 2007, 272, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Sattley, W.M.; Madigan, M.T. Temperature and nutrient induced responses of Lake Fryxell sulfate-reducing prokaryotes and description of Desulfovibrio lacusfryxellense sp. nov., a pervasive, cold-active, sulfate-reducing bacterium from Lake Fryxell, Antarctica. Extremophiles 2010, 14, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Clocksin, K.M.; Jung, D.O.; Madigan, M.T. Cold-active chemoorganotrophic bacteria from permanently ice-covered Lake Hoare, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 2007, 73, 3077–3083. [Google Scholar] [CrossRef] [PubMed]
- Sattley, W.M.; Jung, D.O.; Madigan, M.T. Psychrosinus fermentans gen. nov., sp. nov., a lactate-fermenting bacterium from near-freezing oxycline waters of a meromictic Antarctic Lake. FEMS Microbiol. Lett. 2008, 287, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Stingl, U.; Cho, J.C.; Foo, W.; Vergin, K.L.; Lanoil, B.; Giovannoni, S.J. Dilution-to-extinction culturing of psychrotolerant planktonic bacteria from permanently ice-covered lakes in the McMurdo Dry Valleys, Antarctica. Microbial Ecol. 2008, 55, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Mondino, L.J.; Asao, M.; Madigan, M.T. Cold-active halophilic bacteria from the ice-sealed Lake Vida, Antarctica. Arch. Microbiol. 2009, 191, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Meyer, G.H.; Morrow, M.B.; Wyss, O.; Berg, T.E.; Littlepage, J.L. Antarctica: The microbiology of an unfrozen saline pond. Science 1962, 138, 1103–1104. [Google Scholar] [CrossRef] [PubMed]
- Kriss, A.E.; Mitskevich, I.N.; Rozanova, E.P.; Osnitskaya, L.K. Microbiological investigations of Lake Vanda (Antarctica). Microbiology 1976, 45, 917–922. [Google Scholar]
- Takii, S.; Konda, T.; Hiraishi, A.; Matsumoto, G.I.; Kawano, T.; Torii, T. Vertical distribution in and isolation of bacteria from Lake Vanda: An Antarctic Lake. Hydrobiologia 1986, 135, 15–21. [Google Scholar] [CrossRef]
- Nagashima, H.; Nishikawa, J.; Matsumoto, G.I.; Iizuka, H. Characterization and habitats of bacteria and yeasts isolated from Lake Vanda in Antarctica. Proc. NIPR Symp. Polar Biol. 1990, 3, 190–200. [Google Scholar]
- Bratina, B.J.; Stevenson, B.S.; Green, W.J.; Schmidt, T.M. Manganese reduction by microbes from oxic regions of the Lake Vanda (Antarctica) water column. Appl. Environ. Microbiol. 1998, 64, 3791–3797. [Google Scholar] [PubMed]
- Tregoning, G.S.; Kempher, M.L.; Jung, D.O.; Samarkin, V.A.; Joye, S.B.; Madigan, M.T. A halophilic bacterium inhabiting the warm, CaCl2-rich brine of the perennially ice-covered Lake Vanda, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 2015, 81, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Trüper, H.G.; Schlegel, H.G. Sulphur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek 1964, 30, 225–238. [Google Scholar] [CrossRef]
- Lyons, W.B.; Welch, K.A. McMurdo Long-Term Ecological Research Database. McMurdo Dry Valleys Long-Term Ecological Research, Albuquerque, NM. Available online: http://www.mcmlter.org (assessed on 14 July 2015).
- Reasoner, D.J.; Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [PubMed]
- Zimbro, M.J.; Power, D.A.; Miller, S.M.; Wilson, G.E.; Johnson, J.A. Difco & BBL Manual: Microbiological Culture Media, 2nd ed.; Becton, Dickinson and Company: Sparks, MD, USA, 2009; Volume 520, pp. 441–442. [Google Scholar]
- Maneval, W.E. Staining bacteria and yeasts with acid dyes. Biotech. Histochem. 1941, 16, 13–19. [Google Scholar] [CrossRef]
- Wahlund, T.M.; Woese, C.R.; Castenholz, R.W.; Madigan, M.T. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch. Microbiol. 1991, 156, 81–90. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; Tiedje, J.M. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Euzéby, J.P. List of Bacterial Names with Standing in Nomenclature: A folder available on the internet. Int. J. Syst. Bacteriol. 1997, 47, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Buck, J.D. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol. 1982, 44, 992–993. [Google Scholar] [PubMed]
- Mergaert, J.; Verhelst, A.; Cnockaert, M.C.; Tan, T.-L.; Swings, J. Characterization of facultative oligotrophic bacteria from polar seas by analysis of their fatty acids and 16S rDNA sequences. Syst. Appl. Microbiol. 2001, 24, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Shivaji, S.; Begum, Z.; Shiva Nageswara Rao, S.S.; Vishnu Vardhan Reddy, P.V.; Manasa, P.; Sailaja, B.; Prathiba, M.S.; Thamban, M.; Krishnan, K.P.; Singh, S.M.; et al. Antarctic ice core samples: Culturable bacterial diversity. Res. Microbiol. 2013, 164, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Fritz, I.; Strömpl, C.; Nikitin, D.I.; Lysenko, A.M.; Abraham, W.-R. Brevundimonas mediterranea sp. nov., a non-stalked species from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol. 2005, 55, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; An, X.; Zhang, W. Isolation and phylogenetic analysis of chromium(VI) reducing bacteria of a magnetite mine drainage from Hebei China. Modern Appl. Sci. 2011, 5, 113–118. [Google Scholar] [CrossRef]
- Zakhia, F.; Jeder, H.; Willems, A.; Gillis, M.; Dreyfus, B.; de Lajudie, P. Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microbial Ecol. 2006, 51, 375–393. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.E.; Van Hoorde, K.; Vekeman, B.; Braeckman, T.; Willems, A. Genetic diversity of rhizobia associated with indigenous legumes in different regions of Flanders (Belgium). Soil Biol. Biochem. 2011, 43, 2384–2396. [Google Scholar] [CrossRef]
- Madigan, M.T.; Jung, D.O. An overview of purple bacteria: Systematics, physiology, and habitat. In The Purple Phototrophic Bacteria; Hunter, C.N., Daldal, F., Thurnauer, M.C., Beatty, J.T., Eds.; Springer: Dordrecht, the Netherlands, 2009; pp. 1–15. [Google Scholar]
- Young, C.-C.; Arun, A.B.; Kӓmpfer, P.; Busse, H.-J.; Lai, W.-A.; Chen, W.-M.; Shen, F.-T.; Rekha, P.D. Sphingobium rhizovicinum sp. nov., isolated from rhizosphere soil of Fortunella hindsii (Champ. ex Benth.) Swingle. Int. J. Syst. Evol. Microbiol. 2008, 58, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Yamanaka, H.; Moriyoshi, K.; Oshmoto, T.; Ohe, T. Biodegradation of bisphenol A and related compounds by Sphingomonas sp. strain BP-7 isolated from seawater. Biosci. Biotech. Bioch. 2007, 71, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Helmke, E.; Weyland, H. Psychrophilic versus psychrotolerant bacteria—Occurrence and significance in polar and temperate marine habitats. Mol. Cell. Biol. 2004, 50, 553–561. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vander Schaaf, N.A.; Cunningham, A.M.G.; Cluff, B.P.; Kraemer, C.K.; Reeves, C.L.; Riester, C.J.; Slater, L.K.; Madigan, M.T.; Sattley, W.M. Cold-Active, Heterotrophic Bacteria from the Highly Oligotrophic Waters of Lake Vanda, Antarctica. Microorganisms 2015, 3, 391-406. https://doi.org/10.3390/microorganisms3030391
Vander Schaaf NA, Cunningham AMG, Cluff BP, Kraemer CK, Reeves CL, Riester CJ, Slater LK, Madigan MT, Sattley WM. Cold-Active, Heterotrophic Bacteria from the Highly Oligotrophic Waters of Lake Vanda, Antarctica. Microorganisms. 2015; 3(3):391-406. https://doi.org/10.3390/microorganisms3030391
Chicago/Turabian StyleVander Schaaf, Nicole A., Anna M. G. Cunningham, Brandon P. Cluff, CodyJo K. Kraemer, Chelsea L. Reeves, Carli J. Riester, Lauren K. Slater, Michael T. Madigan, and W. Matthew Sattley. 2015. "Cold-Active, Heterotrophic Bacteria from the Highly Oligotrophic Waters of Lake Vanda, Antarctica" Microorganisms 3, no. 3: 391-406. https://doi.org/10.3390/microorganisms3030391





