Comparison of the Intestinal Microbiota of Patients with Urticaria and Healthy Controls: The Role of Blastocystis
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. PCR
2.3. 16s rRNA Sequencing
- Amplification of the 1500 bp 16S rRNA region using tailed universal primers (27F and 1492R) with LongAmp Taq 2× Master Mix (NEB M0287) (Oxford Nanopore Technologies, Oxford, UK).
- Purification of the 1500 bp amplicon and quantification of DNA concentration.
- Barcoding of each sample using the PCR Barcoding Expansion Pack 1–96 (EXPPBC096) protocol (Oxford Nanopore Technologies, Oxford, UK).
- DNA end repair and adapter preparation using the LSK109 1D Ligation Kit (Oxford Nanopore Technologies, Oxford, UK).
- Adapter ligation with the NEBNext Quick T4 DNA Ligase (SQK-LSK109, Oxford Nanopore Technologies, Oxford, UK).
- Purification of the ligated products.
- Flow cell priming and sample loading.
- Sequencing initiation using MinKNOW V.5.2.3. software, Oxford Nanopore Technologies, Oxford, UK).
2.4. Bioinformatic Analysis
2.5. Statistical Data
3. Results
3.1. DNA Isolation Results
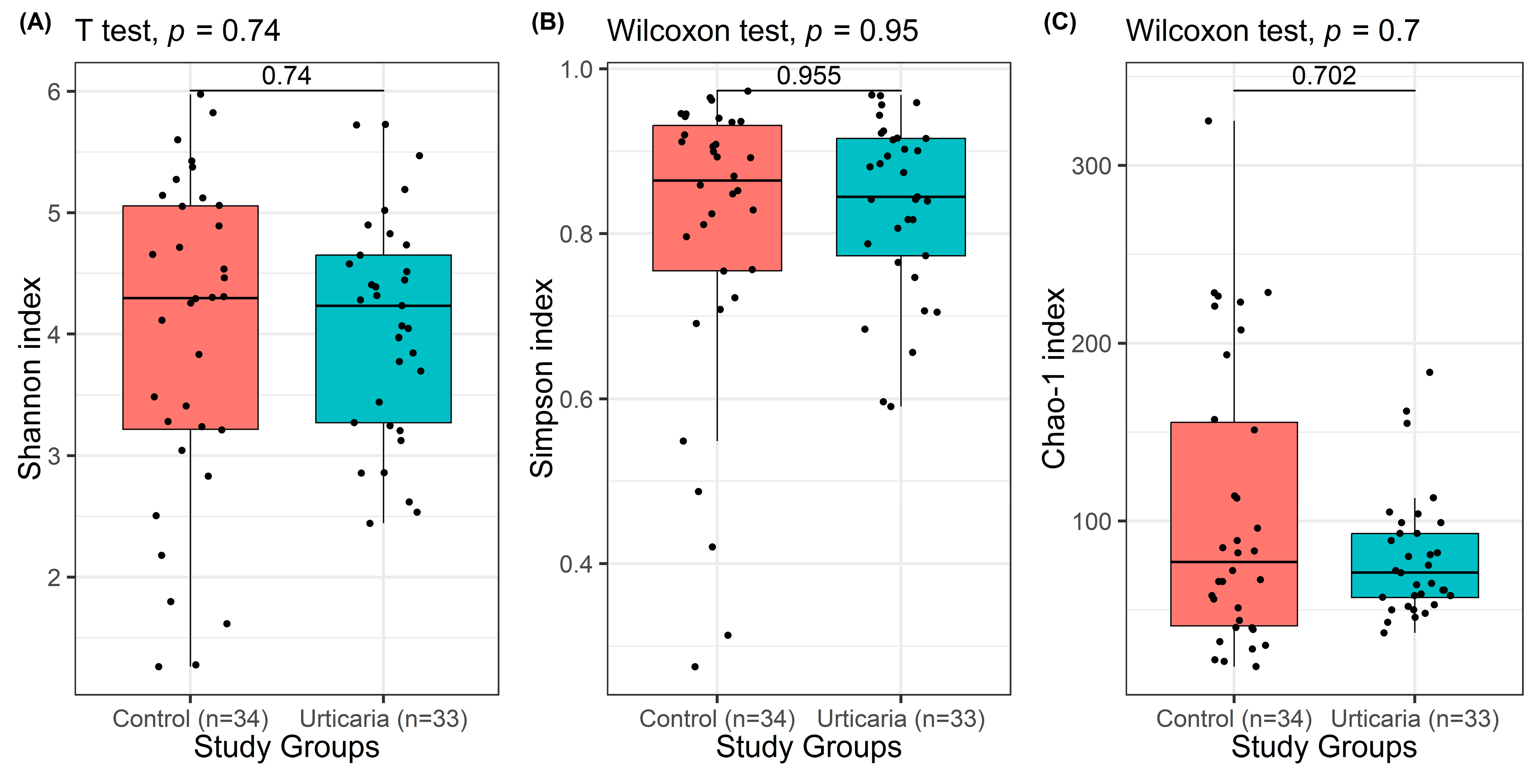
3.2. Comparison of Study Groups According to Shannon, Simpson, and Chao-1 Indices
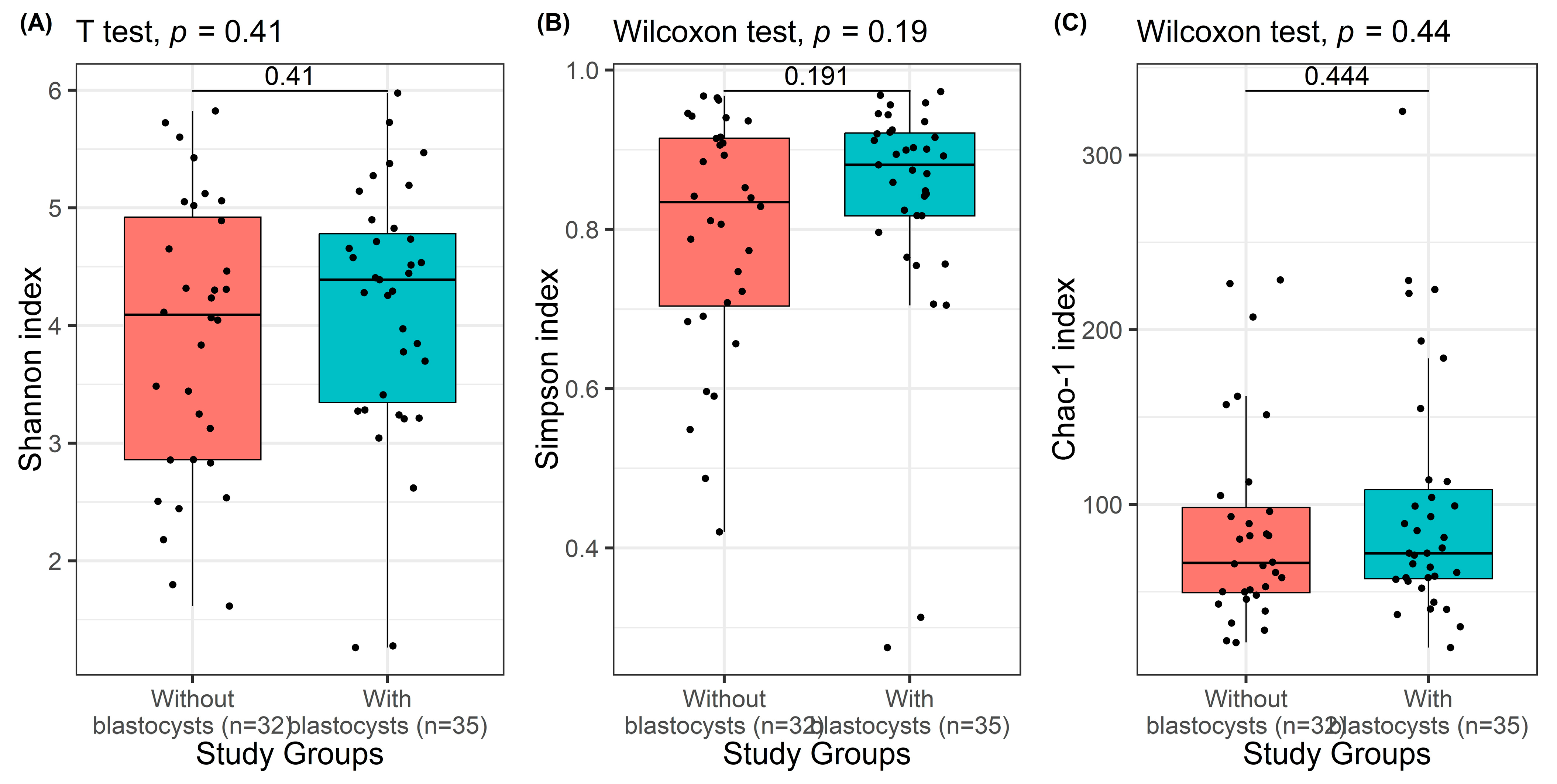
3.3. Comparison of Study Groups According to the Presence of Blastocystis spp.
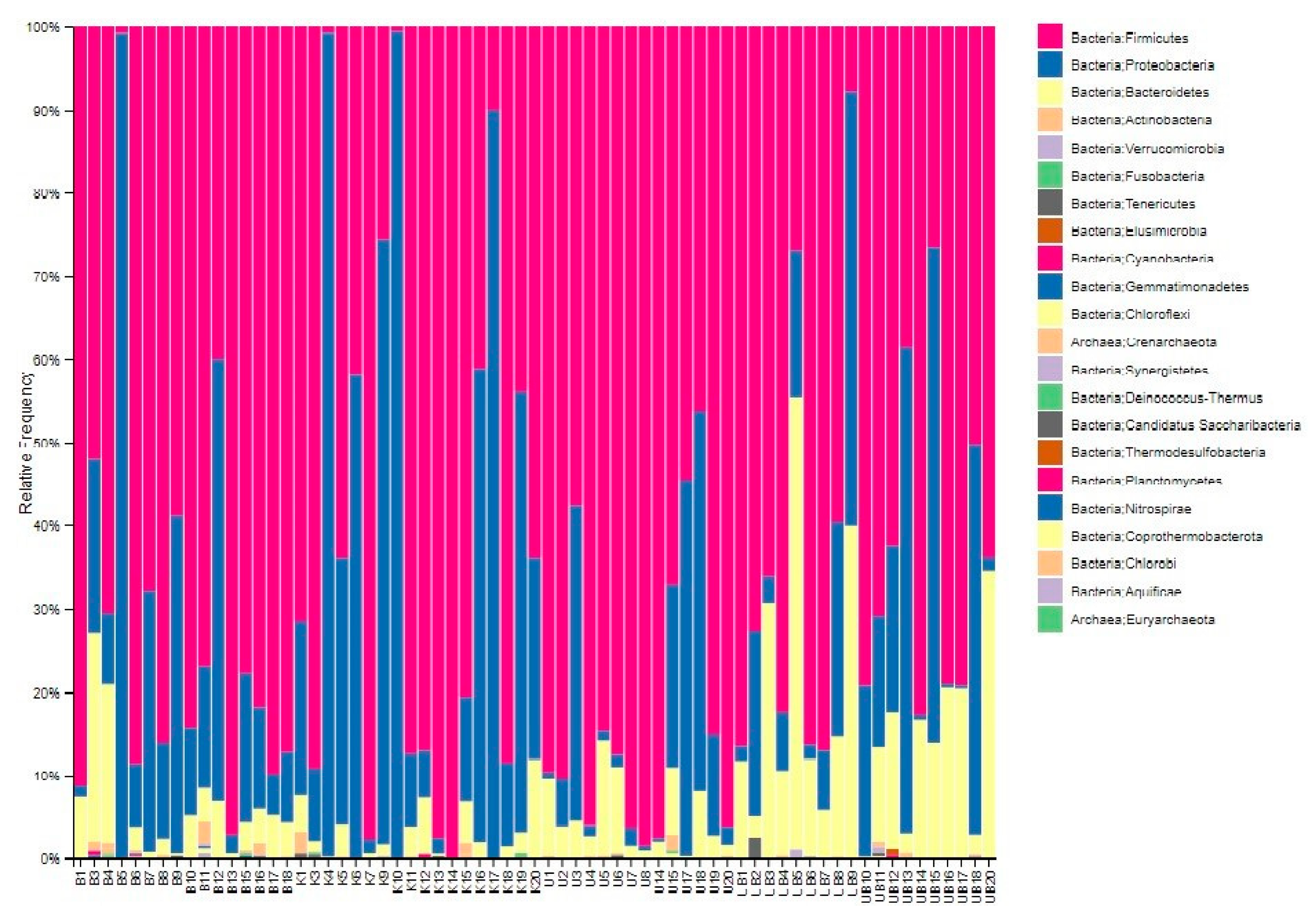
3.4. Displaying Microbial Community Diversity and Quantity Graphs of Samples at Phylum Level in Percentage (Level 2 and Level 6)
3.5. Dendrogram Based on the Diversity of Samples
3.6. Microbiota Diversity Between Control and Urticaria Patients
3.7. Microbiota Differences Between Blastocystis-Positive and -Negative Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kocatürk Göncü, E.; Aktan, Ş.; Atakan, N.; Bülbül Başkan, E.; Erdem, T.; Koca, R.; Şavk, E.; Taşkapan, O.; Utaş, S. Türkiye ürtiker tanı ve tedavi kılavuzu-2016. Turkderm-Arch. Turk. Dermatol. Venerol. 2016, 50, 82–98. [Google Scholar] [CrossRef]
- Korkmaz Adiyaman, G.; Al Doğruman, F.; Mumcuoğlu, I. Dışkı örneklerinde Blastocystis spp. varlığının mikroskobik, kültür ve moleküler yöntemlerle araştırılması. Mikrobiyoloji Bülteni 2015, 49, 85–97. [Google Scholar] [CrossRef]
- Jiménez, P.A.; Jaimes, J.E.; Ramírez, J.D. A summary of Blastocystis subtypes in North and South America. Parasites Vectors 2019, 12, 376. [Google Scholar] [CrossRef]
- Duan, J.; Liu, C.; Bai, X.; Zhao, X.; Jiang, T. Global trends and hotspots of gastrointestinal microbiome and toxicity based on bibliometrics. Front. Microbiol. 2023, 14, 1231372. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V.; Ditu, L.M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 2018, 9, 1830. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X.; Zhang, C.; Liu, Z.; Li, C.; Ren, Z. Gut microbiota and bone diseases: A growing partnership. Front. Microbiol. 2022, 13, 877776. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Luk, B.; Chang-Graham, A.L.; Hall, A.; Herrmann, B.; Ruan, W.; Endres, B.T.; Shi, Z.; Garey, K.W.; Hyser, J.M.; et al. Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. mBio 2019, 10, e01087-19. [Google Scholar] [CrossRef]
- Goto, Y. Epithelial cells as a transmitter of signals from commensal bacteria and host immune cells. Front. Immunol. 2019, 10, 2057. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Kunst, C.; Schmid, S.; Michalski, M.; Tümen, D.; Buttenschön, J.; Müller, M.; Gülow, K. The influence of gut microbiota on oxidative stress and the immune system. Biomedicines 2023, 11, 1388. [Google Scholar] [CrossRef]
- Chu, J.; Feng, S.; Guo, C.; Xue, B.; He, K.; Li, L. Immunological mechanisms of inflammatory diseases caused by gut microbiota dysbiosis: A review. Biomed. Pharmacother. 2023, 164, 114985. [Google Scholar] [CrossRef]
- Maciel-Fiuza, M.F.; Muller, G.C.; Campos, D.M.S.; do Socorro Silva Costa, P.; Peruzzo, J.; Bonamigo, R.R.; Veit, T.; Viana, F.S.L. Role of gut microbiota in infectious and inflammatory diseases. Front. Microbiol. 2023, 14, 1098386. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, R.; Leghissa, P.; Greco, V. Cutaneous lesions in Blastocystis hominis infection. Acta Derm.-Venereol. 2004, 84, 322–323. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.X.; Png, C.W.; Tay, C.Y.B.; Teo, J.D.W.; Jiao, H.; Lehming, N.; Tan, K.S.T.; Zhang, Y. Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infect. Immun. 2014, 82, 4789–4801. [Google Scholar] [CrossRef] [PubMed]
- Ragavan, N.D.; Kumar, S.; Chye, T.T.; Mahadeva, S.; Shiaw-Hooi, H. Blastocystis sp. in irritable bowel syndrome (IBS)-detection in stool aspirates during colonoscopy. PLoS ONE 2015, 10, e0121173. [Google Scholar] [CrossRef]
- Uranga, J.A.; Martínez, V.; Abalo, R. Mast cell regulation and irritable bowel syndrome: Effects of food components with potential nutraceutical use. Molecules 2020, 25, 4314. [Google Scholar] [CrossRef]
- Aydemir, S.; Arvas, Y.E.; Aydemir, M.E.; Barlık, F.; Gürbüz, E.; Yazgan, Y.; Ekici, A. Antiprotozoal Effects of Pediococcus acidilactici-Derived Postbiotic on Blastocystis Subtypes ST1/ST3. Pathogens 2025, 14, 664. [Google Scholar] [CrossRef]
- Nabizadeh, E.; Jazani, N.H.; Bagheri, M.; Shahabi, S. Association of altered gut microbiota composition with chronic urticaria. Ann. Allergy Asthma Immunol. 2017, 119, 48–53. [Google Scholar] [CrossRef]
- Fasano, A. All disease begins in the (leaky) gut: Role of zonulin-mediated gut permeability in the pathogenesis of some chronic inflammatory diseases. F1000Research 2020, 9, 69. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. eBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Furet, J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.A.; Ragab, S.H.; Abd ElBaky, A.; Shoeib, A.R.; Alhosary, Y.; Fekry, D. Frequency of Firmicutes and Bacteroidetes in gut microbiota in obese and normal weight Egyptian children and adults. Arch. Med. Sci. 2011, 7, 501–507. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- Muñoz-Yáñez, C.; Méndez-Hernández, A.; González-Galarza, F.F.; Prieto-Hinojosa, A.I.; Guangorena-Gómez, J.O. Diet Quality Modulates Gut Microbiota Structure in Blastocystis-Colonised Individuals from Two Distinct Cohorts with Contrasting Sociodemographic Profiles. Microorganisms 2025, 13, 1949. [Google Scholar] [CrossRef]
- Yason, J.A.; Liang, Y.R.; Png, C.W.; Zhang, Y.; Tan, K.S.W. Interactions between a pathogenic Blastocystis subtype and gut microbiota: In vitro and in vivo studies. Microbiome 2019, 7, 30. [Google Scholar] [CrossRef]
- Yañez, C.M.; Hernández, A.M.; Sandoval, A.M.; Dominguez, M.A.M.; Muniz, S.A.Z.; Gomes, J.A.G. Prevalence of Blastocystis and its association with Firmicutes/Bacteroidetes ratio in clinically healthy and metabolically ill subjects. BMC Microbiol. 2021, 21, 339. [Google Scholar] [CrossRef] [PubMed]
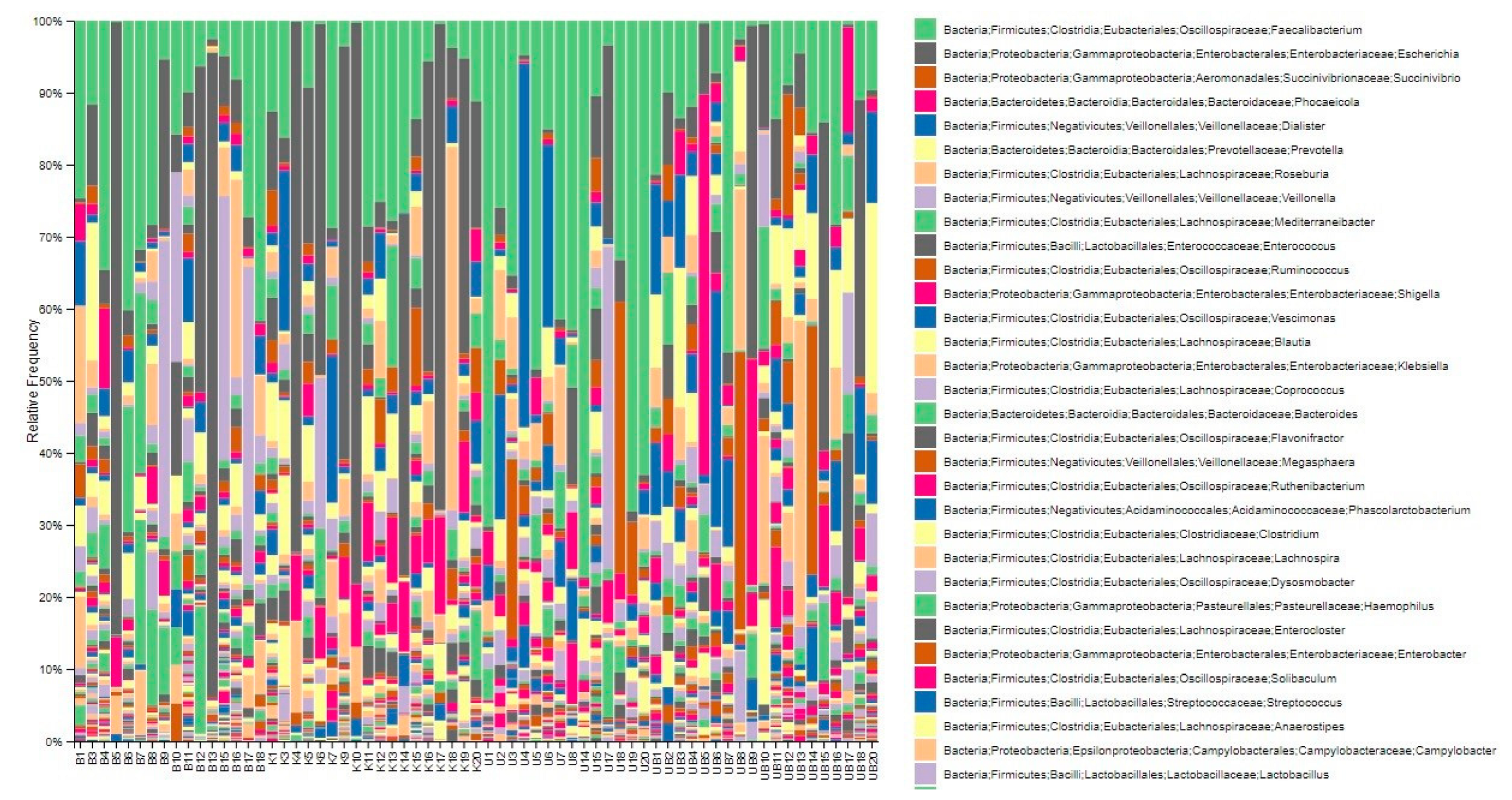
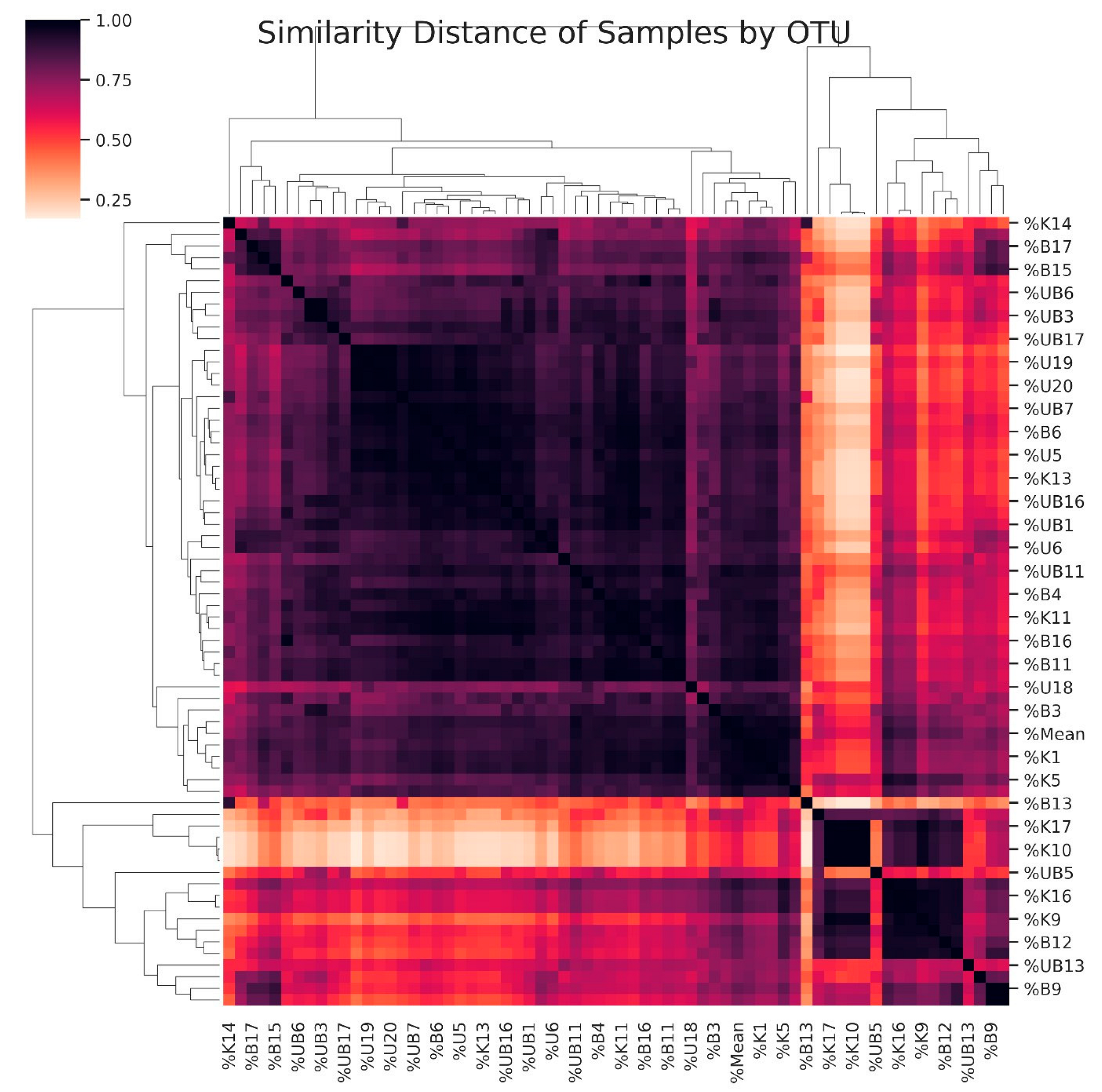
| Study Groups | |||
|---|---|---|---|
| Healthy Control (n = 34) | Urticaria (n = 33) | p-Value | |
| Shannon entropy index | 0.740 1 | ||
| Mean ± SD (range) | 3.98 ± 1.33 (1.26–5.98) | 4.07 ± 0.91 (2.44–5.73) | |
| Median (IQR) | 4.30 (3.22–5.06) | 4.23 (3.27–4.65) | |
| Simpson index | 0.955 2 | ||
| Mean ± SD (range) | 0.80 ± 0.19 | 0.83 ± 0.11 | |
| Median (IQR) | 0.86 (0.75–0.93) | 0.84 (0.77–0.92) | |
| Chao-1 index | 0.702 2 | ||
| Mean ± SD (range) | 105.09 ± 80.53 | 79.34 ± 34.71 | |
| Median (IQR) | 77 (41–155.63) | 71 (57–93) | |
| Study Groups | |||
|---|---|---|---|
| Blastocystis-Negative (n = 32) | Blastocystis-Positive (n = 35) | p-Value | |
| Shannon entropy index | 0.408 1 | ||
| Mean ± SD (range) | 3.91 ± 1.18 (1.62–5.83) | 4.14 ± 1.09 (1.26–5.98) | |
| Median (IQR) | 4.09 (2.86–4.92) | 4.39 (3.35–4.78) | |
| Simpson index | 0.191 2 | ||
| Mean ± SD (range) | 0.80 ± 0.15 (0.42–0.97) | 0.84 ± 0.15 (0.27–0.68) | |
| Median (IQR) | 0.83 (0.70–0.91) | 0.88 (0.82–0.92) | |
| Chao-1 index | 0.444 2 | ||
| Mean ± SD (range) | 86.10 ± 56.90 (21–228.5) | 98.18 ± 68.78 (18–325.1) | |
| Median (IQR) | 66.5 (49.5–98.25) | 72 (57.5–108.5) | |
| Bacteria | Statistic | p-Value |
|---|---|---|
| Firmicutes | 536.5 | 0.763 |
| Proteobacteria | 355 | 0.015 |
| Bacteroidetes | 290 | 0.008 |
| Faecalibacterium | 446 | 0.151 |
| Escherichia | 311.5 | 0.005 |
| Succinivibrionaceae | 92.5 | 0.099 |
| Phocaelcola | 236 | 0.043 |
| Diallister | 317.5 | 0.557 |
| Provetella | 173.5 | 0.047 |
| Roseburia | 374.5 | 0.973 |
| Veilonella | 106.5 | 0.901 |
| FtoBratio | 321 | 0.026 |
| Bacteria | Statistic | p Value |
|---|---|---|
| Firmicutes | 452.5 | 0.179 |
| Proteobacteria | 517 | 0.748 |
| Bacteroidetes | 263 | 0.003 |
| Faecalibacterium | 435.5 | 0.120 |
| Escherichia | 495.5 | 0.684 |
| Succinivibrionaceae | 115 | 0.348 |
| Phocaelcola | 227.5 | 0.032 |
| Diallister | 335 | 0.823 |
| Provetella | 179.5 | 0.064 |
| Roseburia | 363 | 0.807 |
| Veilonella | 82.5 | 0.143 |
| FtoBratio | 292 | 0.009 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciftci, N.; Macin, S.; Saylam Kurtipek, G.; Arslan, U. Comparison of the Intestinal Microbiota of Patients with Urticaria and Healthy Controls: The Role of Blastocystis. Pathogens 2025, 14, 1140. https://doi.org/10.3390/pathogens14111140
Ciftci N, Macin S, Saylam Kurtipek G, Arslan U. Comparison of the Intestinal Microbiota of Patients with Urticaria and Healthy Controls: The Role of Blastocystis. Pathogens. 2025; 14(11):1140. https://doi.org/10.3390/pathogens14111140
Chicago/Turabian StyleCiftci, Nurullah, Salih Macin, Gülcan Saylam Kurtipek, and Uğur Arslan. 2025. "Comparison of the Intestinal Microbiota of Patients with Urticaria and Healthy Controls: The Role of Blastocystis" Pathogens 14, no. 11: 1140. https://doi.org/10.3390/pathogens14111140
APA StyleCiftci, N., Macin, S., Saylam Kurtipek, G., & Arslan, U. (2025). Comparison of the Intestinal Microbiota of Patients with Urticaria and Healthy Controls: The Role of Blastocystis. Pathogens, 14(11), 1140. https://doi.org/10.3390/pathogens14111140







