Colony-Level Effects of Amygdalin on Honeybees and Their Microbes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Compound Feeding
2.2. RNA Isolation and Sequencing
2.3. Identification of Microbes
2.4. Gene Expression Analysis
3. Results
3.1. Amygdalin Consumption May Change Certain Microbe and Viral Titers
3.2. Honey Bee Transcriptome Was Not Heavily Altered by Amygdalin Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tamer, C.E.; Suna, S.; Özcan-Sinir, G. Toxicological Aspects of Ingredients Used in Nonalcoholic Beverages. In Non-Alcoholic Beverages; Woodhead Publishing: Cambridge, UK, 2019; pp. 441–481. [Google Scholar]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.L.; Adler, L.S.; Leonard, A.S.; Andicoechea, J.; Regan, K.H.; Anthony, W.E.; Manson, J.S.; Irwin, R.E. Secondary metabolites in floral nectar reduce parasite infections in bumblebees. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Laure, M.L.N.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed Wing Virus Implicated in Overwintering Honeybee Colony Losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, J.D.; Schwarz, R.S. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011, 19, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.S.; Huang, Q.; Evans, J.D. Hologenome theory and the honey bee pathosphere. Curr. Opin. Insect Sci. 2015, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A. Genomics of the honey bee microbiome. Curr. Opin. Insect Sci. 2015, 10, 22–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, P.; Bartlett, K.D.; Moran, N.A. The Bacterium Frischella perrara Causes Scab Formation in the Gut of its Honeybee Host. mBio 2015, 6, e00193-15. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, P.C. For antagonists and mutualists: The paradox of insect toxic secondary metabolites in nectar and pollen. Phytochem. Rev. 2019, 19, 603–614. [Google Scholar] [CrossRef] [Green Version]
- Palmer-Young, E.C.; Sadd, B.M.; Stevenson, P.C.; Irwin, R.E.; Adler, L.S. Bumble bee parasite strains vary in resistance to phytochemicals. Sci. Rep. 2016, 6, 37087. [Google Scholar] [CrossRef]
- Costa, C.; Lodesani, M.; Maistrello, L. Effect of thymol and resveratrol administered with candy or syrup on the development of Nosema ceranaeand on the longevity of honeybees (Apis mellifera L.) in laboratory conditions. Apidologie 2009, 41, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Bernklau, E.; Bjostad, L.B.; Hogeboom, A.; Carlisle, A.; Arathi, H.S. Dietary Phytochemicals, Honey Bee Longevity and Pathogen Tolerance. Insects 2019, 10, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer-Young, E.C.; Sadd, B.M.; Irwin, R.E.; Adler, L.S. Synergistic effects of floral phytochemicals against a bumble bee parasite. Ecol. Evol. 2017, 7, 1836–1849. [Google Scholar] [CrossRef] [PubMed]
- Spivak, M.; Goblirsch, M.; Simone-Finstrom, M. Social-medication in bees: The line between individual and social regulation. Curr. Opin. Insect Sci. 2019, 33, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Simone-Finstrom, M.D.; Spivak, M. Increased Resin Collection after Parasite Challenge: A Case of Self-Medication in Honey Bees? PLoS ONE 2012, 7, e34601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simone-Finstrom, M.; Evans, J.D.; Spivak, M. Resin collection and social immunity in honey bees. Evolution 2009, 63, 3016–3022. [Google Scholar] [CrossRef] [PubMed]
- Simone-Finstrom, M.; Borba, R.S.; Wilson, M.; Spivak, M. Propolis Counteracts Some Threats to Honey Bee Health. Insects 2017, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.A.; Baker, D.D.; Palmer, M.J.; Stabler, S.; Mustard, J.A.; Power, E.F.; Borland, A.M.; Stevenson, P.C. Caffeine in floral nectar enhances a pollinator’s memory of reward. Science 2013, 339, 1202–1204. [Google Scholar] [CrossRef] [Green Version]
- Adler, L.S. The ecological significance of toxic nectar. Oikos 2000, 91, 409–420. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Z.; Wen, P.; Qu, Y.; Tan, K.; Nieh, J.C. The reluctant visitor: A terpenoid in toxic nectar can reduce olfactory learning and memory in Asian honey bees. J. Exp. Biol. 2018, 221. [Google Scholar] [CrossRef] [Green Version]
- Detzel, A.; Wink, M. Attraction, deterrence or intoxication of bees (Apis mellifera) by plant allelochemicals. Chemoecology 1993, 4, 8–18. [Google Scholar] [CrossRef]
- Charpentier, G.; Vidau, C.; Ferdy, J.-B.; Tabart, J.; Vetillard, A. Lethal and sub-lethal effects of thymol on honeybee (Apis mellifera) larvae reared in vitro. Pest Manag. Sci. 2013, 70, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Simone-Finstrom, M.; Spivak, M. Propolis and bee health: The natural history and significance of resin use by honey bees. Apidologie 2010, 41, 295–311. [Google Scholar] [CrossRef] [Green Version]
- Reilly, J.R.; Artz, D.R.; Biddinger, D.; Bobiwash, K.; Boyle, N.K.; Brittain, C.; Brokaw, J.; Campbell, J.W.; Daniels, J.; Elle, E.; et al. Crop production in the USA is frequently limited by a lack of pollinators. Proc. R. Soc. B Biol. Sci. 2020, 287, 20200922. [Google Scholar] [CrossRef] [PubMed]
- DeGrandi-Hoffman, G.; Thorp, R.; Loper, G.; Eisikowitch, D. Identification and Distribution of Cross-Pollinating Honey-Bees on Almonds. J. Appl. Ecol. 1992, 29, 238. [Google Scholar] [CrossRef]
- Berenbaum, M.R.; Johnson, R.M. Xenobiotic detoxification pathways in honey bees. Curr. Opin. Insect Sci. 2015, 10, 51–58. [Google Scholar] [CrossRef] [Green Version]
- London-Shafir, I.; Shafir, S.; Eisikowitch, D. Amygdalin in almond nectar and pollen—Facts and possible roles. Plant Syst. Evol. 2003, 238, 87–95. [Google Scholar] [CrossRef]
- Kevan, P.G.; Ebert, T. Can almond nectar & pollen poison honey bees? Am. Bee J. 2005, 145, 507–509. [Google Scholar]
- Lecocq, A.; Green, A.A.; Pinheiro De Castro, É.C.; Olsen, C.E.; Jensen, A.B.; Zagrobelny, M. Honeybees Tolerate Cyanogenic Glucosides from Clover Nectar and Flowers. Insects 2018, 9, 31. [Google Scholar] [CrossRef] [Green Version]
- Hurst, V.; Stevenson, P.C.; Wright, G.A. Toxins induce ‘malaise’ behaviour in the honeybee (Apis mellifera). J. Comp. Physiol. A 2014, 200, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, P.C.; Nicolson, S.W.; Wright, G.A. Plant secondary metabolites in nectar: Impacts on pollinators and ecological functions. Funct. Ecol. 2016, 31, 65–75. [Google Scholar] [CrossRef]
- Tauber, J.P.; Collins, W.R.; Schwarz, R.S.; Chen, Y.; Grubbs, K.; Huang, Q.; Lopez, D.; Peterson, R.; Evans, J.D. Natural Product Medicines for Honey Bees: Perspective and Protocols. Insects 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Program, M.B. Beekeeping calendar for the Northeast; Cornell University: Ithaca, NY, USA, 2020. [Google Scholar]
- Evans, J.D.; Schwarz, R.S.; Chen, Y.; Budge, G.; Cornman, R.S.; De La Rua, P.; De Miranda, J.R.; Foret, S.; Foster, L.; Gauthier, L.; et al. Standard methods for molecular research in Apis mellifera. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef] [Green Version]
- Tozkar, C.Ö.; Kence, M.; Kence, A.; Huang, Q.; Evans, J.D. Metatranscriptomic analyses of honey bee colonies. Front. Genet. 2015, 6, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, A.; Bunikis, I.; Pettersson, O.V.; Mosbech, M.-B.; Childers, A.K.; Evans, J.D.; Mikheyev, A.S.; Robertson, H.M.; Robinson, G.E.; Webster, M.T. A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome-length scaffolds. BMC Genom. 2019, 20, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauber, J.P.; Nguyen, V.; Lopez, D.; Evans, J.D. Effects of a Resident Yeast from the Honeybee Gut on Immunity, Microbiota, and Nosema Disease. Insects 2019, 10, 296. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Evans, J.D. Targeting the honey bee gut parasite Nosema ceranae with siRNA positively affects gut bacteria. BMC Microbiol. 2020, 20, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 2019, 36, 1303–1304. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K. BLAST plus: Architecture and applications. BMC Bioinformatics. BioMed Cent. 2009, 10, 1. [Google Scholar]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 002832. [Google Scholar] [CrossRef] [Green Version]
- Elsik, C.G.; Tayal, A.; Diesh, C.M.; Unni, D.R.; Emery, M.L.; Nguyen, H.N.; Hagen, D.E. Hymenoptera Genome Database: Integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 2016, 44, D793–D800. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Anders, S.; Kim, V.; Huber, W. RNA-Seq workflow: Gene-level exploratory analysis and differential expression. F1000Research 2015, 4, 1070. [Google Scholar] [CrossRef]
- Li, J.L.; Cornman, R.S.; Evans, J.D.; Pettis, J.S.; Zhao, Y.; Murphy, C.; Peng, W.J.; Wu, J.; Hamilton, M.; Boncristiani, H.F.; et al. Systemic Spread and Propagation of a Plant-Pathogenic Virus in European Honeybees, Apis mellifera. mBio 2014, 5, e00898–e00913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornman, R. Available Genetic Data Do Not Support Adaptation of Tobacco Ringspot Virus to an Arthropod Host. mBio 2017, 8, e01875–e01916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, A.M.; Lopez, D.L.; Iturralde Martinez, J.F.; Galbraith, D.A.; Rose, R.; van Engelsdorp, D.; Rosa, C.; Evans, J.D.; Grozinger, C.M. Distribution of recently identified bee-infecting viruses in managed honey bee (Apis mellifera) populations in the USA. Apidologie 2020, 171, 736–745. [Google Scholar] [CrossRef]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.-L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doublet, V.; Poeschl, Y.; Gogol-Doering, A.; Alaux, C.; Annoscia, D.; Aurori, C.; Barribeau, S.M.; Bedoya-Reina, O.C.; Brown, M.J.F.; Bull, J.C.; et al. Unity in defence: Honeybee workers exhibit conserved molecular responses to diverse pathogens. BMC Genom. 2017, 18, 207. [Google Scholar] [CrossRef] [PubMed]
- Barroso-Arévalo, S.; Vicente-Rubiano, M.; Puerta, F.; Molero, F.; Sánchez-Vizcaíno, J.M. Immune related genes as markers for monitoring health status of honey bee colonies. BMC Veter. Res. 2019, 15, 72. [Google Scholar] [CrossRef] [Green Version]
- Erban, T.; Sopko, B.; Kadlikova, K.; Talacko, P.; Harant, K. Varroa destructor parasitism has a greater effect on proteome changes than the deformed wing virus and activates TGF-beta signaling pathways. Sci. Rep. 2019, 9, 9400. [Google Scholar] [CrossRef]
- Chan, Q.W.; Melathopoulos, A.P.; Pernal, S.F.; Foster, L.J. The innate immune and systemic response in honey bees to a bacterial pathogen, Paenibacillus larvae. BMC Genom. 2009, 10, 387. [Google Scholar] [CrossRef] [Green Version]
- Corona, M.; Branchiccela, B.; Madella, S.; Chen, Y.P.; Evans, J.D. Decoupling the effects of nutrition, age and behavioral caste on honey bee physiology and immunity. BioRxiv 2019, 667931. [Google Scholar] [CrossRef]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey bees as models for gut microbiota research. Lab Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef]
- Fridman, S.; Izhaki, I.; Gerchman, Y.; Halpern, M. Bacterial communities in floral nectar. Environ. Microbiol. Rep. 2011, 4, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Ricigliano, V.A.; Fitz, W.; Copeland, D.C.; Mott, B.M.; Maes, P.; Floyd, A.S.; Dockstader, A.; Anderson, K.E. The impact of pollen consumption on honey bee (Apis mellifera) digestive physiology and carbohydrate metabolism. Arch. Insect Biochem. Physiol. 2017, 96, e21406. [Google Scholar] [CrossRef] [PubMed]
- Pontoh, J.; Low, N.H. Purification and characterization of beta-glucosidase from honey bees (Apis mellifera). Insect Biochem. Mol. Biol. 2002, 32, 679–690. [Google Scholar] [CrossRef]
- Daughenbaugh, K.F.; Martin, M.; Brutscher, L.M.; Cavigli, I.; Garcia, E.; Lavin, M.; Flenniken, M.L. Honey Bee Infecting Lake Sinai Viruses. Viruses 2015, 7, 3285–3309. [Google Scholar] [CrossRef] [Green Version]
- Palmer-Young, E.C.; Tozkar, C.Ö.; Schwarz, R.S.; Chen, Y.; E Irwin, R.; Adler, L.S.; Evans, J.D. Nectar and Pollen Phytochemicals Stimulate Honey Bee (Hymenoptera: Apidae) Immunity to Viral Infection. J. Econ. Entomol. 2017, 110, 1959–1972. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103 (Suppl. 1), S48–S61. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Dead or Alive: Deformed Wing Virus and Varroa destructor Reduce the Life Span of Winter Honeybees. Appl. Environ. Microbiol. 2011, 78, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Miller, W.A.; Carrillo-Tripp, J.; Bonning, B.C.; Dolezal, A.G.; Toth, A.L. Conclusive Evidence of Replication of a Plant Virus in Honeybees Is Lacking. mBio 2014, 5, e00985–e01014. [Google Scholar] [CrossRef] [Green Version]
- Li, J.L.; Cornman, R.S.; Evans, J.D.; Pettis, J.S.; Zhao, Y.; Murphy, C.; Peng, W.J.; Wu, J.; Hamilton, M.; Boncristiani, H.F.; et al. Reply to “Conclusive Evidence of Replication of a Plant Virus in Honeybees Is Lacking”. mBio 2014, 5, e01250–e01314. [Google Scholar] [CrossRef] [Green Version]
- D’Alvise, P.; Seeburger, V.; Gihring, K.; Kieboom, M.; Hasselmann, M. Seasonal dynamics and co-occurrence patterns of honey bee pathogens revealed by high-throughput RT-qPCR analysis. Ecol. Evol. 2019, 9, 10241–10252. [Google Scholar] [CrossRef] [Green Version]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020, 14, 801–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vejnovic, B.; Stevanovic, J.; Schwarz, R.S.; Aleksic, N.; Mirilovic, M.; Jovanovic, N.M.; Stanimirovic, Z. Quantitative PCR assessment of Lotmaria passim in Apis mellifera colonies co-infected naturally with Nosema ceranae. J. Invertebr. Pathol. 2018, 151, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Glenny, W.; Cavigli, I.; Daughenbaugh, K.F.; Radford, R.; Kegley, S.E.; Flenniken, M.L. Honey bee (Apis mellifera) colony health and pathogen composition in migratory beekeeping operations involved in California almond pollination. PLoS ONE 2017, 12, e0182814. [Google Scholar] [CrossRef] [PubMed]
- Meixner, M.D.; Francis, R.M.; Gajda, A.; Kryger, P.; Andonov, S.; Uzunov, A.; Topolska, G.; Costa, C.; Amiri, E.; Berg, S.; et al. Occurrence of parasites and pathogens in honey bee colonies used in a European genotype-environment interactions experiment. J. Apic. Res. 2014, 53, 215–229. [Google Scholar] [CrossRef]
- Runckel, C.; Flenniken, M.L.; Engel, J.C.; Ruby, J.G.; Ganem, D.; Andino, R.; DeRisi, J.L. Temporal Analysis of the Honey Bee Microbiome Reveals Four Novel Viruses and Seasonal Prevalence of Known Viruses, Nosema, and Crithidia. PLoS ONE 2011, 6, e20656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornman, R.S. Relative abundance and molecular evolution of Lake Sinai Virus (Sinaivirus) clades. PeerJ 2019, 7, e6305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, B.R.; Atallah, J.; Plachetzki, D.C. The importance of tissue specificity for RNA-seq: Highlighting the errors of composite structure extractions. BMC Genom. 2013, 14, 586. [Google Scholar] [CrossRef] [Green Version]
- Williams, G.R.; Alaux, C.; Costa, C.; Doublet, V.; Eisenhardt, D.; Kuhn, R.; Murray, T.E.; Neumann, P.; Oliver, R.; Tanner, G.; et al. Standard methods for maintaining adult Apis mellifera in cages under in vitro laboratory conditions. J. Apic. Res. 2013, 52, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Maia, M.F.; Moore, S.J. Plant-based insect repellents: A review of their efficacy, development and testing. Malar. J. 2011, 10, S11–S14. [Google Scholar] [CrossRef] [Green Version]
- Barlow, S.E.; Wright, G.A.; Ma, C.; Barberis, M.; Farrell, I.W.; Marr, E.C.; Brankin, A.; Pavlik, B.M.; Stevenson, P.C. Distasteful Nectar Deters Floral Robbery. Curr. Biol. 2017, 27, 2552–2558.e3. [Google Scholar] [CrossRef]
- Irwin, R.E.; Bronstein, J.L.; Manson, J.S.; Richardson, L. Nectar Robbing: Ecological and Evolutionary Perspectives. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 271–292. [Google Scholar] [CrossRef]
- Singaravelan, N.; Nee’Man, G.; Inbar, M.; Izhaki, I. Feeding Responses of Free-flying Honeybees to Secondary Compounds Mimicking Floral Nectars. J. Chem. Ecol. 2005, 31, 2791–2804. [Google Scholar] [CrossRef] [PubMed]
- Tiedeken, E.J.; Stout, J.C.; Stevenson, P.C.; Wright, G.A. Bumblebees are not deterred by ecologically relevant concentrations of nectar toxins. J. Exp. Biol. 2014, 217, 1620–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, H.; Stevenson, P.C. Do linden trees kill bees? Reviewing the causes of bee deaths on silver linden (Tilia tomentosa). Biol. Lett. 2017, 13, 20170484. [Google Scholar] [CrossRef] [Green Version]
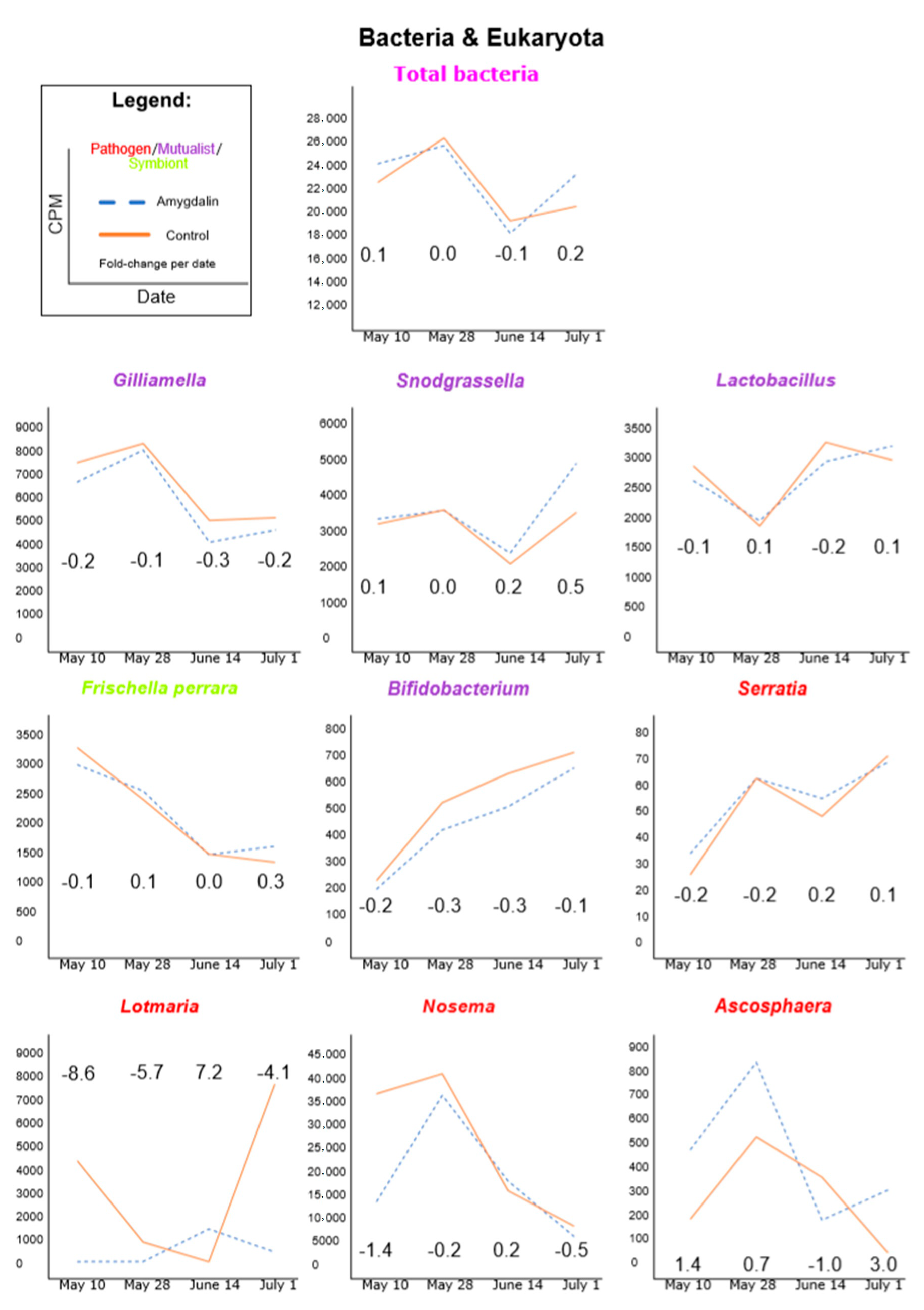
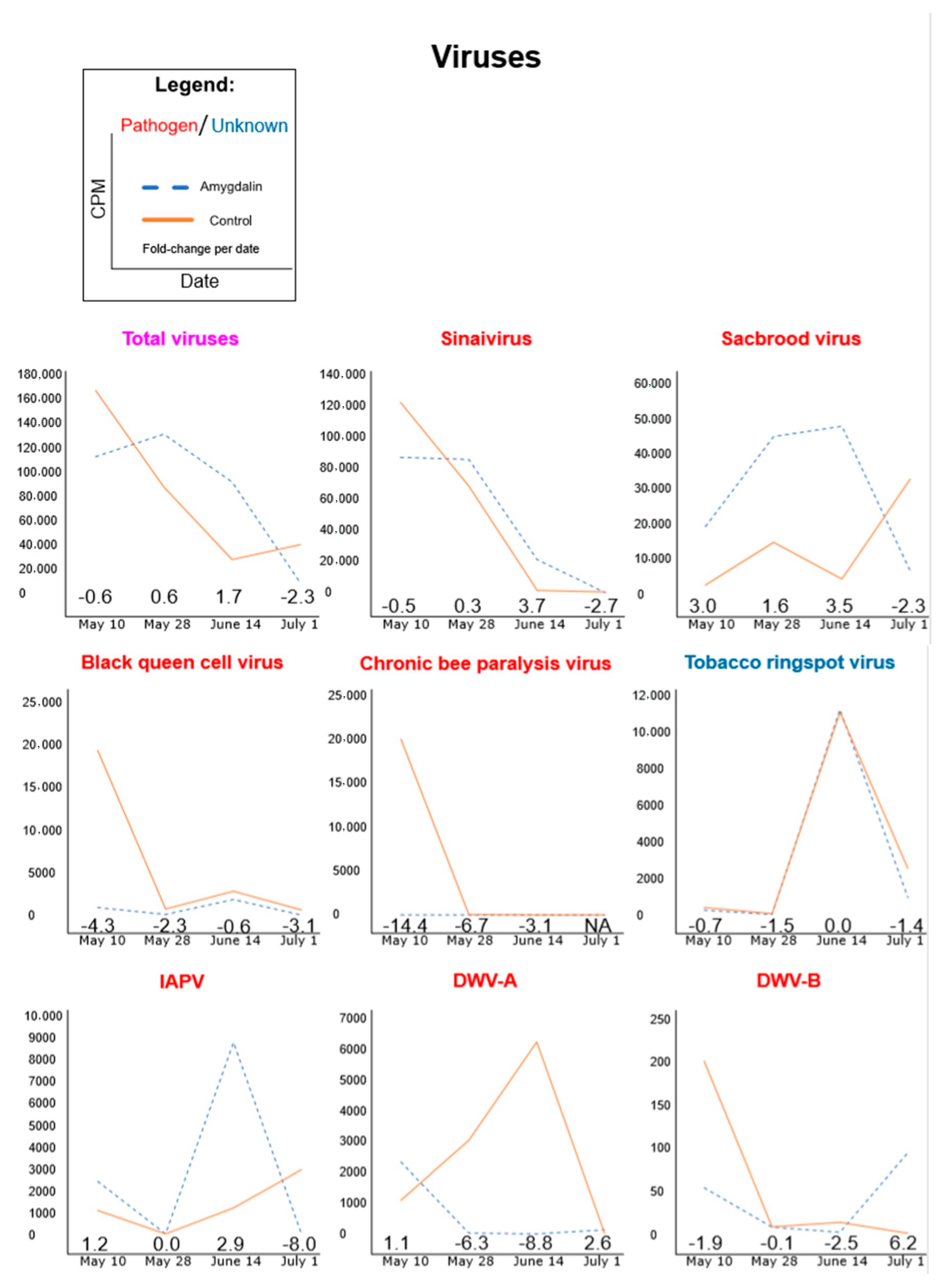
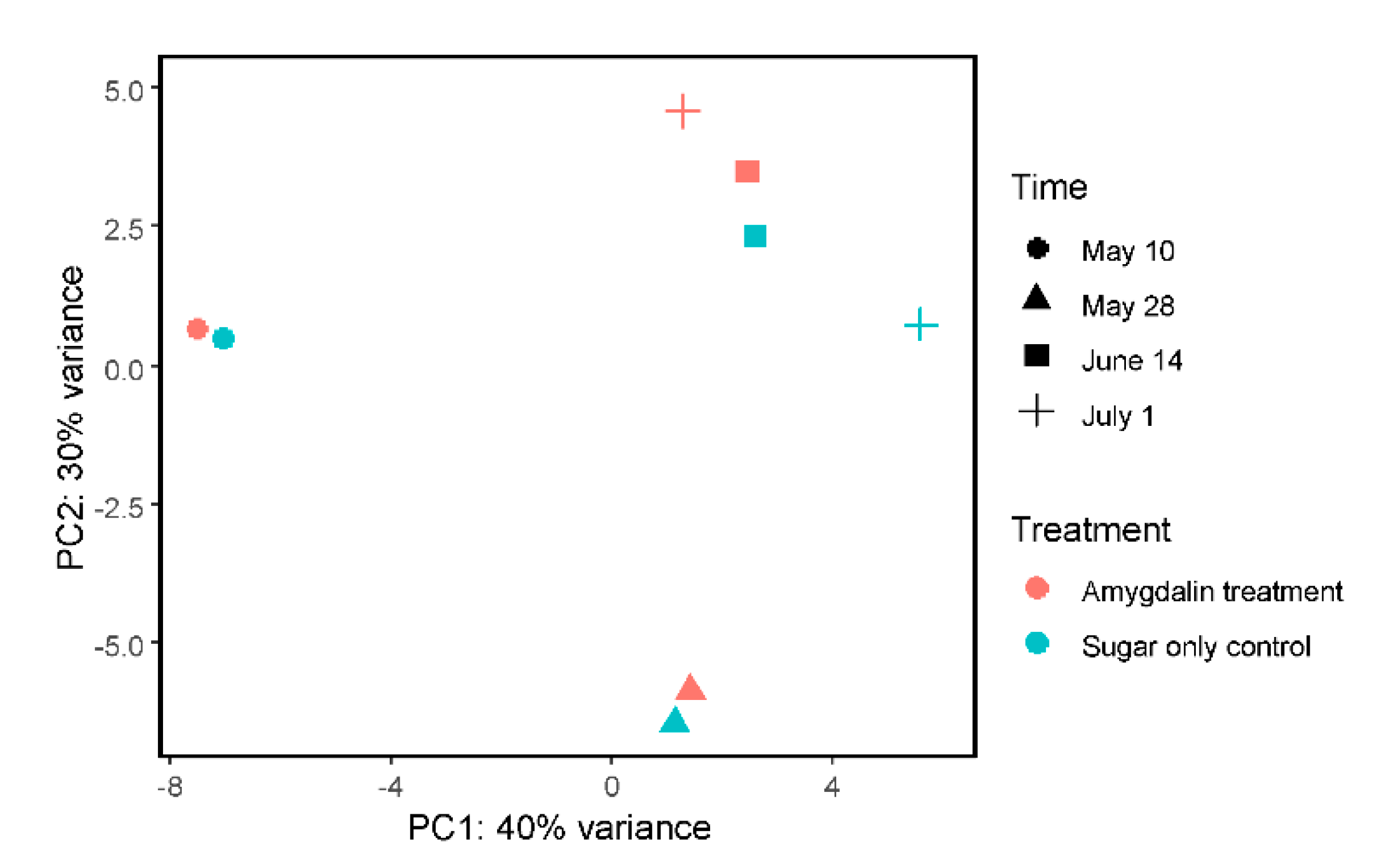
| 10 May 2013 | 28 May 2013 | 14 June 2013 | 1 July 2013 | |||||
|---|---|---|---|---|---|---|---|---|
| Alignment/count details | Amygdalin | Control | Amygdalin | Control | Amygdalin | Control | Amygdalin | Control |
| A. mellifera Hisat2 overall alignment rate | 87.08 | 95.4 | 89.19 | 97.06 | 91.1 | 97.91 | 90.99 | 88.61 |
| Kraken2-HoloBee total viral reads | 4.30 | 23.69 | 7.04 | 23.65 | 6.30 | 10.78 | 0.64 | 2.74 |
| Kraken2-RefSeq total viral reads | 4.66 | 25.20 | 7.29 | 24.36 | 6.62 | 11.93 | 0.68 | 2.80 |
| Kraken2-HoloBee total bacterial reads | 0.90 | 3.14 | 1.32 | 6.71 | 1.23 | 7.4 | 1.85 | 1.40 |
| Kraken2-RefSeq total bacterial reads | 1.08 | 3.48 | 1.49 | 7.31 | 1.47 | 8.14 | 2.15 | 1.61 |
| Kraken2-HoloBee total fungal reads | 0.54 | 5.20 | 1.92 | 10.6 | 1.21 | 6.13 | 0.46 | 0.54 |
| Kraken2-RefSeq total fungal reads | 0.07 | 0.29 | 0.13 | 0.55 | 0.08 | 0.44 | 0.07 | 0.05 |
| May 10 | May 28 | June 14 | July 1 | ||
|---|---|---|---|---|---|
| Antimicrobial peptides | Hymenoptaecin | 1.02 | 0.64 | 2.53 | 0.53 |
| Abaecin | 1.21 | 1.17 | 1.80 | 0.90 | |
| Apidaecin | 1.27 | 1.25 | 1.61 | 0.91 | |
| Defensin-1 | 0.89 | 0.82 | 0.88 | 0.77 | |
| Upstream toll and Imd/JNK pathways | Peptidoglycan-recognition protein 1 | 1.17 | 0.96 | 0.99 | 1.06 |
| Peptidoglycan recognition protein S2 | 1.13 | 1.00 | 1.07 | 0.74 | |
| Beta-1,3-glucan-binding protein 1 (gnbp-1) | 1.19 | 1.21 | 1.22 | 0.83 | |
| Nuclear factor NF-kappa-B p100 subunit (relish) | 1.15 | 0.94 | 1.03 | 0.84 | |
| Hormone (nutrition/behavior) | Vitellogenin | 2.30 | 0.97 | 1.23 | 4.76 |
| Major royal jelly protein 1 | 1.057 | 0.89 | 0.95 | 0.83 | |
| Apisimin | 1.08 | 0.94 | 0.88 | 0.80 | |
| Other immunity | Apidermin 3 | 3.23 | 3.19 | 1.73 | 0.53 |
| Lysozyme | 0.83 | 0.89 | 0.70 | 0.86 | |
| RNAi | Protein argonaute-2 | 1.30 | 1.04 | 1.15 | 1.05 |
| RISC-loading complex subunit TARBP2 | 1.47 | 1.03 | 1.18 | 0.61 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tauber, J.P.; Tozkar, C.Ö.; Schwarz, R.S.; Lopez, D.; Irwin, R.E.; Adler, L.S.; Evans, J.D. Colony-Level Effects of Amygdalin on Honeybees and Their Microbes. Insects 2020, 11, 783. https://doi.org/10.3390/insects11110783
Tauber JP, Tozkar CÖ, Schwarz RS, Lopez D, Irwin RE, Adler LS, Evans JD. Colony-Level Effects of Amygdalin on Honeybees and Their Microbes. Insects. 2020; 11(11):783. https://doi.org/10.3390/insects11110783
Chicago/Turabian StyleTauber, James P., Cansu Ö. Tozkar, Ryan S. Schwarz, Dawn Lopez, Rebecca E. Irwin, Lynn S. Adler, and Jay D. Evans. 2020. "Colony-Level Effects of Amygdalin on Honeybees and Their Microbes" Insects 11, no. 11: 783. https://doi.org/10.3390/insects11110783






