Mapping HPV 16 Sub-Lineages in Anal Cancer and Implications for Disease Outcomes
Abstract
:1. Introduction
2. Material and Methods
2.1. Sample Collection
2.1.1. Anal Cancer Cohort, Collection and Annotation
2.1.2. Residual Rectal Swabs from Asymptomatic Men
2.2. Governance
2.3. PCR Target-Enrichment for Deep Sequencing of HPV 16
2.4. Library Preparation
2.5. Quality Control and Quality Analysis
2.6. Bioinformatic Analysis
2.7. Assessment of Variants According to Clinic-Demographic Characteristics and Survival Analysis
3. Results
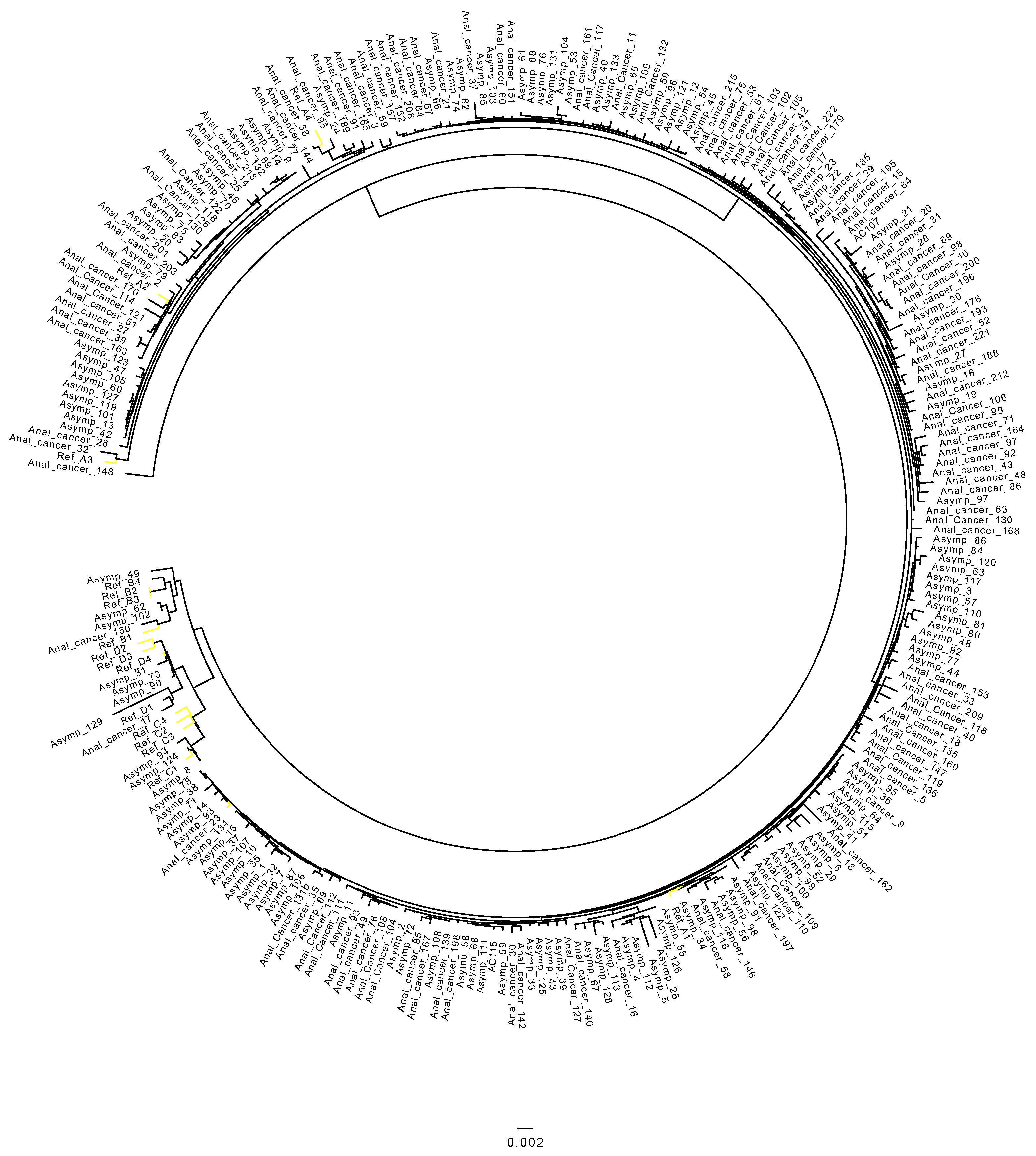
3.1. Distribution of HPV 16 Sub-Lineages in Anal Cancers
3.2. HPV 16 Sub-Lineages in the Control Cohort
3.3. Differences in Prevalence of HPV 16 Sub-Lineages between Anal Cancer and Control Cohort
3.4. Association of HPV 16 Sub-Lineages with Demographic and Clinical Variables
3.5. HPV 16 Sub-Lineages and Overall Survival
3.6. Integration
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- De Sanjosé, S.; Serrano, B.; Tous, S.; Alejo, M.; Lloveras, B.; Quirós, B.; Clavero, O.; Vidal, A.; Ferrándiz-Pulido, C.; Pavón, M.; et al. Burden of Human Papillomavirus (HPV)-Related Cancers Attributable to HPVs 6/11/16/18/31/33/45/52 and 58. JNCI Cancer Spectr. 2018, 2, pky045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerendiain, D.; Grigorescu, R.; Kirk, A.; Stevenson, A.; Holden, M.T.G.; Pan, J.; Kavanagh, K.; Graham, S.V.; Cuschieri, K. HPV status and HPV16 viral load in anal cancer and its association with clinical outcome. Cancer Med. 2022, 11, 4193–4203. [Google Scholar] [CrossRef] [PubMed]
- Cuschieri, K.; Brewster, D.; Williams, A.R.W.; Millan, D.; Murray, G.; Nicoll, S.; Imrie, J.; Hardie, A.; Graham, C.; Cubie, H.A. Distribution of HPV types associated with cervical cancers in Scotland and implications for the impact of HPV vaccines. Br. J. Cancer 2010, 102, 930–932. [Google Scholar] [CrossRef] [Green Version]
- Islami, F.; Ferlay, J.; Lortet-Tieulent, J.; Bray, F.; Jemal, A. International trends in anal cancer incidence rates. Int. J. Epidemiol. 2017, 46, 924–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, D.; Coupland, V.; Moller, H. An analysis of temporal and generational trends in the incidence of anal and other HPV-related cancers in Southeast England. Br. J. Cancer 2009, 100, 527–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anal Cancer Incidence Statistics|Cancer Research, UK. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/anal-cancer/incidence#heading-Two (accessed on 11 September 2021).
- Anal Cancer—Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/anus.html (accessed on 11 September 2021).
- De Villiers, E.-M.; Fauquet, C.; Broker, T.R.; Bernard, H.-U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Burk, R.D.; Harari, A.; Chen, Z. Human papillomavirus genome variants. Virology 2013, 445, 232–243. [Google Scholar] [CrossRef] [Green Version]
- Cornet, I.; Gheit, T.; Iannacone, M.R.; Vignat, J.; Sylla, B.S.; Del Mistro, A.; Franceschi, S.; Tommasino, M.; Clifford, G.M.; on behalf of the IARC HPV Variant Study Group. HPV16 genetic variation and the development of cervical cancer worldwide. Br. J. Cancer 2012, 108, 240–244. [Google Scholar] [CrossRef] [Green Version]
- Clifford, G.M.; Tenet, V.; Georges, D.; Alemany, L.; Pavón, M.A.; Chen, Z.; Yeager, M.; Cullen, M.; Boland, J.F.; Bass, S.; et al. Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: Whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res. 2019, 7, 67–74. [Google Scholar] [CrossRef]
- Volpini, L.P.B.; Boldrini, N.A.T.; de Freitas, L.B.; Miranda, A.E.; Spano, L.C. The high prevalence of HPV and HPV16 European variants in cervical and anal samples of HIV-seropositive women with normal Pap test results. PLoS ONE 2017, 12, e0176422. [Google Scholar] [CrossRef]
- Ferreira, M.T.; Gonçalves, M.G.; López, R.V.M.; Sichero, L. Genetic variants of HPV-16 and their geographical and anatomical distribution in men: A systematic review with meta-analysis. Virology 2021, 558, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Cullen, M.; Boland, J.F.; Chen, Z.; Wentzensen, N.; Zhang, X.; Yu, K.; Yang, Q.; Mitchell, J.; et al. HPV16 Sublineage Associations with Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J. Natl. Cancer Inst. 2016, 108, djw100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Joint Committee on Cancer. AJCC Cancer Staging Manual, 8th ed.; Springer: Berlin/Heidelberg, Germany, 2017; p. 275. Available online: www.cancerstaging.orgajcc@facs.org (accessed on 11 September 2021).
- Glynne-Jones, R.; Nilsson, P.; Aschele, C.; Goh, V.; Peiffert, D.; Cervantes, A.; Arnold, D. Anal cancer: ESMO-ESSO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, iii10–iii20. [Google Scholar] [CrossRef] [PubMed]
- Cameron, R.L.; Cuschieri, K.; Pollock, K.G.J. Baseline HPV prevalence in rectal swabs from men attending a sexual health clinic in Scotland: Assessing the potential impact of a selective HPV vaccination programme for men who have sex with men. Sex. Transm. Infect. 2019, 96, 55–57. [Google Scholar] [CrossRef] [PubMed]
- Cullen, M.; Boland, J.F.; Schiffman, M.; Zhang, X.; Wentzensen, N.; Yang, Q.; Chen, Z.; Yu, K.; Mitchell, J.; Roberson, D.; et al. Deep sequencing of HPV16 genomes: A new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection. Papillomavirus Res. 2015, 1, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo-Mühr, L.S.; Lagheden, C.; Hultin, E.; Eklund, C.; Adami, H.-O.; Dillner, J.; Sundström, K. Human papillomavirus type 16 genomic variation in women with subsequent in situ or invasive cervical cancer: Prospective population-based study. Br. J. Cancer 2018, 119, 1163–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babraham Bioinformatics—FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 April 2022).
- PaVE. Available online: https://pave.niaid.nih.gov/explore/reference_genomes/human_genomes (accessed on 31 October 2022).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team, R. Rstudio: Integrated Development for r. Rstudio, pbc, Boston, MA. 2020. Available online: http://www.Rstudio.Com (accessed on 3 March 2022).
- Gonçalves, M.G.; Ferreira, M.T.; López, R.V.M.; Ferreira, S.; Sirak, B.; Baggio, M.L.; Lazcano-Ponce, E.; Nyitray, A.G.; Giuliano, A.R.; Villa, L.L.; et al. Prevalence and persistence of HPV-16 molecular variants in the anal canal of men: The HIM study. J. Clin. Virol. 2022, 149, 105128. [Google Scholar] [CrossRef] [PubMed]
- Nicolás-Párraga, S.; Gandini, C.; Pimenoff, V.N.; Alemany, L.; Sanjosé, S.; Bosch, F.X.; Bravo, I.G.; the RIS HPV TT and HPV VVAP Study Groups. HPV16 variants distribution in invasive cancers of the cervix, vulva, vagina, penis, and anus. Cancer Med. 2016, 5, 2909–2919. [Google Scholar] [CrossRef] [PubMed]
- Kuhs, K.L.; Faden, D.; Chen, L.; Smith, D.; Pinheiro, M.; Wood, C.; Davis, S.; Yeager, M.; Boland, J.; Cullen, M.; et al. Genetic variation within the human papillomavirus type 16 genome is associated with oropharyngeal cancer prognosis. Ann. Oncol. 2022, 33, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Godi, A.; Kemp, T.J.; Pinto, L.A.; Beddows, S. Sensitivity of Human Papillomavirus (HPV) Lineage and Sublineage Variant Pseudoviruses to Neutralization by Nonavalent Vaccine Antibodies. J. Infect. Dis. 2019, 220, 1940–1945. [Google Scholar] [CrossRef] [PubMed]

| Variable | Level | n = 119 | % |
|---|---|---|---|
| Sex | Female | 90 | 75.6 |
| Male | 29 | 24.4 | |
| Age | <50 | 15 | 12.6 |
| 50–59 | 32 | 26.9 | |
| 60–69 | 39 | 32.8 | |
| 70 and over | 33 | 27.7 | |
| Stage | I | 18 | 15.1 |
| II | 48 | 40.3 | |
| III | 36 | 30.2 | |
| IV | 16 | 13.4 | |
| Unknown | 1 | 0.8 | |
| Response to treatment | Yes | 95 | 79.8 |
| No | 17 | 14.3 | |
| Unknown | 7 | 5.9 | |
| Vital status | Alive | 87 | 73.1 |
| Deceased | 30 | 25.2 | |
| Unknown | 2 | 1.7 |
| Anal Cancer | Asymptomatic Group | |||
|---|---|---|---|---|
| Sub-Lineage | N | % (N = 119) | N | % (N = 134) |
| HPV 16 A1 | 91 | 76.5% | 102 | 76.1% |
| HPV 16 A2 | 20 | 16.8% | 23 | 17.2% |
| HPV 16 A3 | 1 | 0.8% | 0 | 0% |
| HPV 16 A4 | 5 | 4.2% | 0 | 0% |
| HPV 16 B1 | 2 | 1.7% | 2 | 1.5% |
| HPV 16 B2 | 0 | 0.0% | 1 | 0.7% |
| HPV 16 B3 | 0 | 0.0% | 0 | 0% |
| HPV 16 B4 | 0 | 0.0% | 0 | 0% |
| HPV 16 C1 | 0 | 0.0% | 2 | 1.5% |
| HPV 16 C2 | 0 | 0.0% | 0 | 0% |
| HPV 16 C3 | 0 | 0.0% | 0 | 0% |
| HPV 16 C4 | 0 | 0.0% | 0 | 0% |
| HPV 16 D1 | 1 | 0.8% | 4 | 3.0% |
| HPV 16 D2 | 0 | 0.0% | 0 | 0% |
| HPV 16 D3 | 0 | 0.0% | 0 | 0% |
| HPV 16 D4 | 0 | 0.0% | 0 | 0% |
| Total | 119 | 134 | ||
| Variable | Level | Unadjusted OR (95% Cis) | p Value | Adjusted OR (95% Cis) | p Value |
|---|---|---|---|---|---|
| Sex | Male | 1 | 1 | ||
| Female | 1.12 (0.39–2.92) | 0.827 | 1.09 (0.37–3.00) | 0.87 | |
| Age | <50 | 1 | 1 | ||
| 50–59 | 1.20 (0.27–4.80) | 0.8 | 1.11 (0.24–4.56) | 0.89 | |
| 60–69 | 1.50 (0.34–5.93) | 0.57 | 1.82 (0.40–7.67) | 0.416 | |
| 70 and over | 1.37 (0.30–5.67) | 0.666 | 1.63 (0.34–7.41) | 0.529 | |
| Response to treatment | No | 1 | 1 | ||
| Yes | 1.02 (0.26–3.26) | 0.968 | 1.18 (0.34–7.41) | 0.528 | |
| Stage | I | 1 | 1 | ||
| II | 1.36 (0.37–4.62) | 0.625 | 1.28 (0.33–4.51) | 0.706 | |
| III | 1.60 (0.40–6.11) | 0.486 | 1.56 (0.38–6.06) | 0.522 | |
| IV | 1.80 (0.36–10.40) | 0.478 | 3.03 (0.42–29.47) | 0.289 | |
| Vital Status | Alive | 1 | 1 | ||
| Deceased | 1.01 (0.39–2.85) | 0.983 | 0.92 (0.25–3.81) | 0.907 |
| Variable | Level | Unadjusted HR (95% Cis) | p Value | Adjusted HR (95% Cis) | p Value |
|---|---|---|---|---|---|
| HPV 16 sub-lineage | A1 (n = 88) | 1 | 1 | ||
| Non-A1 (n = 27) | 0.87 (0.37–2) | 0.751 | 0.83 (0.28–2.46) | 0.743 | |
| Sex | Male | 1 | 1 | ||
| Female | 1.2 (0.48–2.9) | 0.71 | 0.88 (0.32–2.39) | 0.795 | |
| Age | <50 | 1 | 1 | ||
| 50–59 | 1.10 (0.33–3.70) | 0.877 | 0.83 (0.21–3.26) | 0.788 | |
| 60–69 | 0.85 (0.26–2.8) | 0.795 | 2.67 (0.607–11.72) | 0.194 | |
| 70 and over | 1.54 (0.48–5.0) | 0.466 | 5.56 (1.082–28.58) | 0.04 | |
| Stage | I | 1 | 1 | ||
| II | 1.7 (0.37–8.1) | 0.49 | 2.34 (0.47–11.74) | 0.302 | |
| III | 2.4 (0.50–11.6) | 0.274 | 2.26 (0.42–12.27) | 0.344 | |
| IV | 15.7 (3.38–72.8) | <0.001 | 15.95 (2.45–10.3.82) | 0.004 | |
| Response to treatment | No | 1 | 1 | ||
| Yes | 0.11 (0.05–0.25) | <0.001 | 0.12 (0.03–0.39) | <0.001 |
| HPV Genes Integration in the Anal Cancer Samples (n = 13) | N |
|---|---|
| L1 only | 1 |
| E1 only | 1 |
| E1, E2, E4 | 1 |
| E2, E5, part E2 | 1 |
| E2, E4, E5, L2, L1 | 3 |
| E1, E2, E4, E5 and part L2 | 2 |
| E1, E2, E4, E5, L2 and part L1 | 3 |
| E1, E2, E4, E5, L2 and L1 complete | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerendiain, D.; Mühr, L.S.A.; Grigorescu, R.; Holden, M.T.G.; Cuschieri, K. Mapping HPV 16 Sub-Lineages in Anal Cancer and Implications for Disease Outcomes. Diagnostics 2022, 12, 3222. https://doi.org/10.3390/diagnostics12123222
Guerendiain D, Mühr LSA, Grigorescu R, Holden MTG, Cuschieri K. Mapping HPV 16 Sub-Lineages in Anal Cancer and Implications for Disease Outcomes. Diagnostics. 2022; 12(12):3222. https://doi.org/10.3390/diagnostics12123222
Chicago/Turabian StyleGuerendiain, Daniel, Laila Sara Arroyo Mühr, Raluca Grigorescu, Matthew T. G. Holden, and Kate Cuschieri. 2022. "Mapping HPV 16 Sub-Lineages in Anal Cancer and Implications for Disease Outcomes" Diagnostics 12, no. 12: 3222. https://doi.org/10.3390/diagnostics12123222
APA StyleGuerendiain, D., Mühr, L. S. A., Grigorescu, R., Holden, M. T. G., & Cuschieri, K. (2022). Mapping HPV 16 Sub-Lineages in Anal Cancer and Implications for Disease Outcomes. Diagnostics, 12(12), 3222. https://doi.org/10.3390/diagnostics12123222







