1. Introduction
As the leading cause of infectious disease death worldwide, the global burden of tuberculosis (TB) disease today stands at 10 million cases per annum, with 12% of individuals dying from TB disease. The causative bacterial agent of TB,
Mycobacterium tuberculosis (
M.tb), is also the most common opportunistic infection and leading cause of death in HIV-infected persons in developing nations [
1]. The inability to rapidly and accurately diagnose TB disease and evaluate resistance with high sensitivity and specificity at the point-of-care (POC) continues to be a confounder in disease transmission, patient mortality and morbidity, and time to treatment initiation in the developing world; this is a major barrier to effective control of TB.
In 2012, the World Health Organization endorsed the nucleic acid amplification-based GeneXpert MTB/RIF assay as a frontline diagnostic and drug resistance test for TB in endemic settings [
2]. Unfortunately, reliance on a special instrument possesses significant limitations that hinder its capacity to serve as a comprehensive screening tool. Most crucially, for example, in South Africa, where Xpert is widely used, Xpert devices are commonly located in centralized tertiary healthcare facilities in densely populated areas [
3]. This leaves individuals in rural, sparsely populated areas without immediate access to Xpert testing, requiring patient specimens be shipped from remote clinics to testing centers, imposing a lengthy turnaround time (days to weeks) for what should be a two-hour diagnostic test. This exacerbates pre-treatment loss to follow-up rates, which can be up to 38% for patients diagnosed with TB in both rural and urban pats of the developing world [
4]. In addition, Xpert cartridge stock-outs and supply chain mismanagement, interruptions in electricity, unpredictable transport of patient specimens from off-site locations, and high start-up costs have impeded implementation of Xpert [
5]. Knowing these barriers, researchers have shown that case detection has not significantly improved with the rollout of Xpert, and that the number of patients lost to follow-up did not decrease following TB diagnosis with Xpert [
5,
6,
7]. Thus, empiric treatment without molecular diagnosis remains commonplace [
4].
In the absence of any reliable biomarker-based POC diagnostic test for TB, the gold standards for accurate TB diagnosis continue to rely on microscopic pathogen detection paired with diagnostic confirmation using bacterial culture methods [
1]. Sputum smear staining and microscopy has the advantages of having high specificity (≈100%) while being inexpensive and easy to perform. Unfortunately, these advantages do not overcome poor diagnostic sensitivity (≈50%), subjectivity, and laboriousness. Manual inspection of stained sputum smear slides requires visual inspection of a minimum 100 fields of view using high-power magnification, which can take up to 3 h, depending on the degree of infection and the microscopist experience [
8]. Understaffed and busy clinics that continue to rely on smear inspection are especially prone to false negatives resulting from improper examination [
9].
Spatial concentration of TB bacteria has the potential to reduce inspection time of patient samples, which could be further improved when paired with TB-specific volumetric concentration strategies. Using what is known about the hydrodynamics of an evaporating drop, spatial concentration is theoretically achievable without the need for specialized equipment. Our group and others have extensively studied the flow and deposition patterns of particles in drying drops and have shown that these microfluidic properties can achieve spatial concentration at the edge or center of the drop [
10,
11,
12,
13].
Our group has previously published two biosensor designs that make use of the radial flows found in evaporating sessile droplets [
11,
13]. The first design used the primary radial flow to deposit polystyrene particles at the edge of an evaporating drop, creating what is colloquially known as a “coffee ring” [
13]. Our second published design used secondary radial flows, known as a Marangoni flows, to concentrate particles in the center of a drying drop. This design used an antibody recognition system to induce particle aggregation in the presence of biomarker, followed by deposition in the center of the evaporating drop [
11]. A larger deposition pattern in the center of the drop corresponded to a greater biomarker quantity, and this could be quantitatively measured. The assay also achieved a limit-of-detection (LOD) of picomolar biomarker concentrations. The system included glycerol to remove interference of salt crystallization and enhance the Marangoni flows. It also used a polydimethylsiloxane (PDMS) substrate instead of glass, which has the required material properties for production of Marangoni flows radially inward along the substrate. Unfortunately, both the substrate and solvent material properties that made this assay work are not compatible with acid-fast staining protocols used at the POC. In addition, both assays use 1 μL drops, a volume too small for suspension of a large-volume sample after volumetric concentration.
Here, we describe a different design for spatial concentration of
M.tb bacilli for use in microscopic sputum smear inspection using Marangoni flow (
Figure 1). In this report, we detail the design and testing of potential materials before comparing our method to direct sputum smear.
Figure 2 demonstrates combined magnetic enrichment and spatial concentration using nanoparticles (NPs) and Marangoni flows, achieving a simple workflow for use in low-resource settings.
3. Results
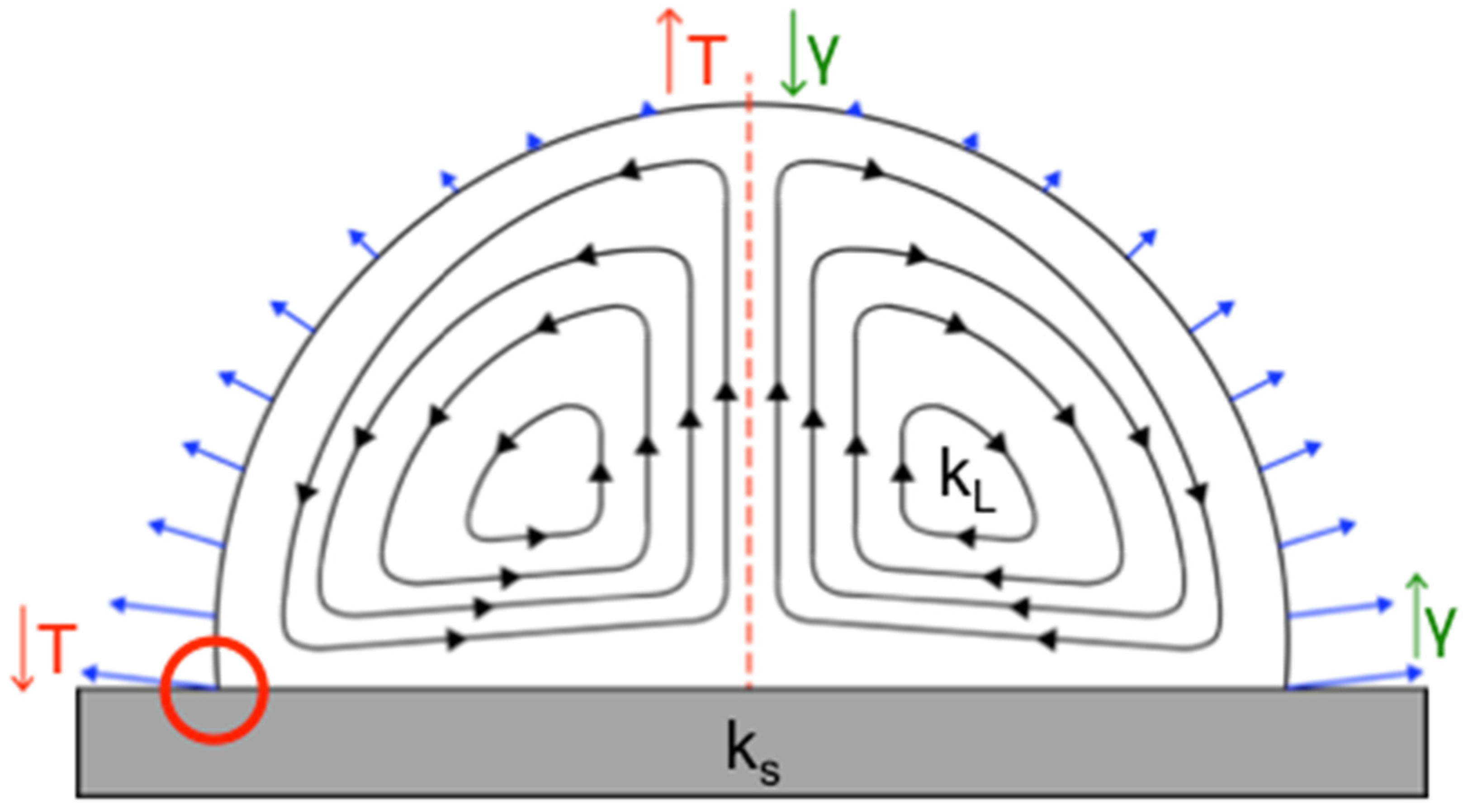
There are two types of radial flows observed in an evaporating sessile droplet [
29,
30,
31,
32,
33,
34]. The first, which are found in droplets containing pure water, are capillary flows responsible for the “coffee ring effect,” carrying suspended colloids to the edge of the droplet radially outward along the substrate, resulting in a nonuniform deposition pattern across the droplet area upon complete drying, with particles at the edge of the droplet upon drying [
29,
30,
31]. A secondary radial flow forms when the nonuniform evaporation rate (indicated by blue arrows in
Figure 1) of the sessile drop, thermal conductivities of the substrate and droplet solvent, and surface tension of the drop all interact synergistically, resulting in a temperature gradient across the droplet volume. This creates predictable, specific flow patterns, known as Marangoni flows (
Figure 1) [
10,
19]. Marangoni flows can move in the same direction or opposite direction of the fluid stream in the coffee ring effect and can result in particles depositing at the edge or center of the drop, or across the entire liquid–solid interface. On the basis of our prior work, we know that when Marangoni flows move inward along a hydrophobic substrate, a clearly defined central deposition of 1 μm particles occurs, forming what is essentially a reverse coffee ring. In addition, Marangoni flows can also be influenced by a nonuniform surface tension gradient [
31].
While there are a variety of biological applications using Marangoni flows in evaporating sessile droplets [
11,
35,
36,
37,
38], unfortunately previous work does not use materials compatible with many microbiological staining protocols. Knowing this, we sought to use established knowledge to intelligently design a new system compatible with cellular biomarkers. This system can then be paired with a volumetric concentration strategy to further improve the results of the method.
We first focused on identifying a new substrate material. Identifying the substrate required that multiple design requirements be taken into consideration. For acid-fast staining, materials should be optically clear, flame-resistant (will not burn with prolonged flame exposure), nonporous, inexpensive, and readily available. It was also important that we preferentially concentrate the NPs and bacteria into the center of the drying droplet, since this would create a small, well-defined area for inspection under a microscope.
It is known that both the droplet contact angle and the ratio of the substrate, k
s, and droplet solvent, k
L, thermal conductivities are important in determining the circulation direction of Marangoni flows (
Figure 1) [
10] in evaporating droplets. Established theory by Ristenpart et al. states that for the Marangoni flows to move inward along the liquid–solid substrate interface and outward along a drop’s liquid–air interface (referenced from here on as the direction of fluid flow along the substrate) can be achieved by keeping the ratio of the thermal conductivities
less than 1.45. When the ratio is greater than two, the flow field reverses and instead flows outward along the substrate surface. When a ratio between 1.45 and 2 is achieved, the Marangoni flow direction is dependent on the droplet contact angle [
10,
32]. The direction of the fluid flow [
10] and forces applied to the colloids ultimately impact where colloids deposit, and for this reason it was critically important to maintain fluid flow inward along the substrate. Although the contact angle is irrelevant for Marangoni flow direction when
is less than 1.45 or greater than 2, it can also impact the final particle deposition pattern [
10], and therefore we preserved both the contact angle and Marangoni flow direction in our new design. Logically, glass slides used for sputum smear microscopy, and PDMS, the substrate known to yield the desired Marangoni flow patterns [
11], were chosen as candidate substrates. Beyond these, surface coating materials containing PTFE or polysiloxanes were selected for initial testing because of their low thermal conductivity (
Table 1) and high heat resistance. ITO-coated glass, which meets all the design criteria, with the exception that its high thermal conductivity would produce outward-oriented flow patterns in aqueous droplets, was selected for comparison to the candidate substrates with lower conductivity.
We started substrate characterization by measuring the contact angle of water on the substrates. This provided information about how a substrates’ hydrophobicity/hydrophilicity will impact surface tension of the droplet.
Figure 4 measured contact angle of 2 μL water droplets on the candidate substrates. The contact angles measured for PDMS (109.6° ± 1.9°), DuPont
® PTFE (104.6° ± 6.1°), Water Repellent Rain-X
® (101° ± 1.8°), Sigmacote
® (86.3° ± 1.2°), plain glass (23.5° ± 4.7°), and Probe On
TM glass (53.1° ± 5.6°) are consistent with literature values [
39,
40,
41]. The positive charge of the Probe On
TM slides creates a more hydrophobic surface, with polar water molecules being partially repelled by the surface, creating a larger contact angle when compared to a standard glass microscope slide.
When measuring the contact angle for the WD-40
® PTFE coating, the dried film readily rehydrated upon drop deposition, and vortices of PTFE coating mixing into the droplets were observed through the side of the droplet on the goniometer during measurement (data not shown). This would explain the smaller contact angle (40.8° ± 2.6°) compared to the DuPont
® PTFE coating and reported literature values [
41].
The Anti-Fog Rain-X® performed as intended in everyday usage, such as on windshields or glasses, although this was the opposite of the other polysiloxane coatings; by creating a superhydrophilic surface, any water droplets that form on the glass surface are flattened to reduce visual interference. Anti-Fog Rain-X® had the smallest reported contact angle (6.9° ± 3.5°), although within 10 s of droplet deposition, the contact angle was reduced to 0°.
Our goal was to create a highly repeatable, well-defined, small central deposition area that was also easy to find under a microscope, and therefore it was important that we observed central deposition for both nano- and micron-scale particles, colloid sizes expected to be in our final droplet, before moving forward to biological experiments using rod-shaped bacilli. After measuring the contact angle of the selected substrates, we allowed particle-containing drops to evaporate and assessed the deposition pattern of PS particles and magnetic NPs. As shown
Figure 5, the materials with contact angles less than 85° failed to concentrate particles in the center of the evaporating droplet. The contact angle is dependent on cohesive and adhesive intermolecular forces within the liquid and between the liquid–solid interfaces. These forces are also at play between the liquid and suspended colloids. These can influence particle deposition directly on the basis of the particles’ chemical composition, not only indirectly by changes in the contact angle and surface tension. Furthermore, surface forces and inertial body forces acting on the particles control the particle trajectory within the flow velocity field and their deposition location. The balance of these forces change with the size, density, shape, and surface composition of the particles, as well as the velocity of the fluid flow, which result in different particle deposition patterns upon drying.
Three substrate surfaces in
Figure 5 showed deposition in the center of the droplet for both 1 μm and 200 nm colloidal solutions: Water Repellent Rain-X
®, PDMS, and Sigmacote
®. Water Repellent Rain-X
® was ultimately selected as the final substrate coating for the design for a few reasons. First, Water Repellent Rain-X
® covalently modifies the surface of glass without leaving a residue behind, which is a beneficial design for downstream assay packaging; the slides could be prepared by the manufacturer and would be stable for long-term storage. Second, unlike the Rain-X
®, Sigmacote
® left a residue on the slide that could be disturbed during handling. This would make pre-coated slides more difficult to package in a deliverable assay. It also increases the chance that a sample is lost during the staining process after deposition and drying and makes it more difficult for the sample to physically heat fix to the glass. Moreover, simply, Water Repellent Rain-X
® is less expensive and very easy to acquire.
ITO-coated glass demonstrates how the reverse flow pattern changes the applied forces and impact on colloidal deposition. With a high thermal conductivity, theory states that the Marangoni flows are oriented outward along the substrate [
10], not inward as desired. This is regardless of its measured contact angle (98.5° ± 1.9°), which is similar to that of PDMS and other candidate surfaces that have highly efficient central deposition. As shown in
Figure 5, PS particles (a similar size to individual bacteria) settle at the edges of the drop, and NPs are deposited across the total drop interface, although with some concentration in the middle of the drop.
With a microscopy-compatible substrate determined, testing was then performed to establish a compatible solvent system. Trantum et al. had demonstrated that glycerol was important, although not necessary, for formation of Marangoni flows. In addition, glycerol was added to prevent salt crystallization that interfered with imaging [
11]. Unfortunately, glycerol is difficult to remove or evaporate due to its viscosity and high boiling point, making it incompatible with microscopy staining. Because of the acid-fast staining process, salt crystals are rinsed away, eliminating this problem. As shown in
Figure S2 and as observed by Trantum et al., removal of glycerol from the droplet still formed centrally located Marangoni deposition in 2 μL droplets of 1 μm PS particles; 200 nm NPs; and
M. smeg, a nonpathogenic mycobacterial cousin of
M.tb. Again, theory states that these flows should be directed radially inward along the substrate.
After identifying a new substrate material and new solvent system that were both compatible with microscopic staining techniques and met the criteria established by Ristenpart et al., we further confirmed that the final deposition pattern was due to Marangoni flows oriented inward along the substrate using OCT, as we have demonstrated previously [
11,
12].
Figure 6A shows composite, time-lapse images through the diameter of a 1 μL evaporating droplet containing 10
6 polystyrene particles/μL.
Video S1 uses the same image series to dynamically show that the flow fields are oriented as expected in the inward direction along the substrate, the same direction shown by the black arrows in
Figure 1. Reflection off the particles is indicated by the white dots moving within the droplet. During the first minute of evaporation, the microfluidic flows produced by the thermal gradient were initially unstable, possibly due to Bénard–Marangoni instability, though this is just speculative [
42]. As time progresses, the chaotic flows stabilized into radially symmetric flows, as seen at 3 min. Due to the small size of the droplet, the accumulation of particles in the center of the droplet was not visible via OCT, but it was clearly observed upon total drop evaporation.
Microscopy was also used to visualize the microfluidic flows. While microscopy cannot image the entirety of droplet flows vertically, the images and videos gathered help to support the data shown in
Figure 6A. In
Figure 6B, a singular image with a 4 s exposure time was taken near the solvent–substrate interface of a 5 μL droplet containing 10
4 particles/μL, demonstrating movement radially inward.
Video S2 captured a series of images of the same droplet, which dynamically shows the flow field orientation. Images for
Video S2 were taken at 1 fps for 40 s.
With promising results in scale experiments and flow confirmation using OCT and microscopy, we performed trials with larger volume droplets containing NPs to confirm that the Marangoni flows occurred independently of droplet size, using the final deposition pattern to assess performance (
Figure S3). For droplets 1–100 μL in diameter, all drops exhibited central deposition of the suspended NPs. A 50 μL drop was chosen because it created a small enough central inspection area for efficient inspection without completely obstructing view, whereas smaller droplets resulted in central areas that were too dense to observe bacterial clusters among the NPs. Droplets larger than 50 μL had an extremely long drying time, making it not feasible to perform, given the drying time for a 50 uL droplet already takes a minimum of 5 h. A time series showing the evaporation of a 50 μL drop, demonstrating the formation of the concentrated area during evaporation can be found in
Figure S4.
While it seemed the system was ready for use, a critical flaw in the substrate design, which was previously alluded to, became clear and required further design refinement. Because the surface of the microscope slide was covalently modified to become hydrophobic, even with heat fixation, the samples of NPs with and without bacteria were almost completely lost during the staining process; this required testing of a variety of fixation techniques to solve the problem. Specialty microscope slides provided inspiration. Specifically, companies commercially make positively charged slides that aid in sample adherence (such as the Probe-OnTM slides used earlier in this study), and at-home recipes for coating slides in poly-L-lysine (PLL) are readily available online. The PLL polymer helps enhance electrostatic interactions between the microscope slide surface and applied sample. We found that coating the entire Rain-X®-coated slide in PLL impacted the deposition pattern, leading to uniform deposition across the liquid–solid interface of the drop, likely caused by the positive charge of PLL ionically attracting the bacteria and NPs to the surface with enough force to overcome forces exerted by the radial Marangoni flows.
We then investigated if functionalizing the Rain-X
®-coated slides in the center of the droplet deposition area with PLL, not the entire slide, could create a “landing zone” for the sample, helping secure it for staining while not significantly changing the deposition pattern. With this, arrays of increasing size PLL spots were placed onto Rain-X
®-coated slides, followed by NP deposition on top of the array, ensuring the center of the drop was placed over the PLL spot, as indicated by graph paper taped the back of the microscope slide (
Figure 3). The grid was removed for staining and imaging, and after analysis of the droplets, we determined that depositing a 10 μL PLL spot in the center of the 50 μL drop deposition area was the best option because it resulted in improved uniformity of the central deposition area (
Figure S5). The drawback to using such a large PLL spot is that a small amount of sample does deposit outside the most centrally concentrated area, across other regions of the liquid–solid interface of the drop. However, it was decided that this trade-off is worthwhile, since a larger PLL spot size allows for error as the operator manually deposits the final sample on the microscope slide. The implication is that the most concentrated center created by the Marangoni flows will still land on the PLL area, ensuring the majority of the sample is not lost. Although there are alternative methods to functionally achieve the same thing, such as derivatizing the surface of the microscope slides with antibodies that target both the bacteria or NPs, the protocols to perform this are much more complex, require specialized reagents, and could require specialized storage conditions that in our opinion negate any potential selectivity it would provide.
With the Marangoni flow system finally designed, combined volumetric concentration and spatial enrichment of
M. bovis BCG was performed. The bacteria were nonspecifically captured on streptavidin-coated NPs and isolated on a stationary magnet. After washing the particles, they were either applied over the standard 2 cm
2 area, creating a volumetric concentration-only control, or were suspended in 50 μL and applied as a drop to the PLL-Rain-X
®-functionalized slide. These were both visually and semi-quantitatively compared to a direct, unconcentrated smear (
Figure 7). All samples, regardless of processing method, were stained using the gold standard Ziehl–Neelsen staining protocol. Initially, we had designed the method using anti-
M.tb immunospecific NPs but found that for both naked NPs and isotype control NPs, just as many bacteria were nonspecifically captured by the NPs during volumetric concentration. Those with experience working with mycobacteria know that they are extremely “sticky,” and it can be difficult to prevent nonspecific binding of bacteria to surfaces. It is likely that creating an immunospecific particle will further improve the results shown here.
With the current limit-of-detection for sputum smear microscopy being 10
4 bacilli/mL [
43], as expected, it was generally difficult to find any bacteria on the direct smears for all samples except 10
6 bacilli/mL. When samples were volumetrically concentrated, higher concentration samples benefitted significantly more than lower concentration samples. Even with volumetric concentration, only 2% of the smear area was inspected, and there were few or no bacteria seen by microscopy in the 10
3 bacilli/mL and 10
4 bacilli/mL samples. When volumetric concentration and spatial enrichment were combined, the 10
3 bacilli/mL and 10
4 bacilli/mL samples had visible, stained mycobacterial clusters in the central deposition area of the droplet. No bacteria were seen in any of the samples containing 0 bacteria. On a plain glass slide, central deposition of sample NPs did not occur (
Figure 5 and
Figure S6).
The benefit of combining spatial and volumetric concentration was further seen when samples were semi-quantitatively analyzed using the same examination methods used for diagnostic sputum grading during sputum smear microscopy (
Table 2) [
28]. Grading of the smears was performed conservatively, given the difficulties of achieving a uniformly single-bacillus suspension with accurate quantification of bacteria in the sample. For the samples with higher concentrations of bacteria, volumetric concentration improved the grading by one grading, but this was not seen in lower concentration samples, particularly the 10
3 bacilli/mL. Specifically, volumetric concentration on its own failed to improve detection of these samples. With the addition of spatial concentration, these samples all had detectable amounts of bacteria, even with some visible sample loss during staining. Even in areas where bulk sample was lost, stained bacteria were observed, showing that the addition of the PLL spot helped prevent sample loss not just macroscopically but microscopically (data not shown).
4. Discussion
Our Marangoni flow-enabled spatial concentration system, combined with volumetric magnetic NP concentration, resulted in a design that outperformed the current technique and was able to visualize bacteria in samples that contained 10× fewer bacteria than the current limit-of-detection for direct sputum smear microscopy (
Table 2). This study is the first of its kind to use the microfluidic principles of Marangoni flows to improve microscopic visualization of whole pathogens using standard microscopic staining techniques.
We designed the method with the goal of introducing an improvement to the current workflow, rather than establishing an entirely new diagnostic method. Unlike our previous design, particles that do not aggregate were designed to still deposit in the center of the droplet. We chose this design because it creates a clearly defined, small area for inspection for the presence or absence of
M.tb bacilli under the microscope. A significant majority of the time and labor that goes into sputum smear microscopic diagnosis of TB is inspection of the smear itself, which is applied over a large 2 cm
2 area. Concentrating the sample into a smaller known area increases the chances of observing TB bacilli, especially when paired with volumetric concentration. The current smear inspection looks at 100 fields of view, which is equivalent to 2% of the standard smear area. Use of Marangoni concentration deposits the majority of the sample into the most central deposition area (as indicated in
Figure 7), increasing the inspected sample area to ≈90%. For a 1 mL sample, volumetric concentration alone into a 20 μL smear concentrates the sample 50×. Spatially, while the sample is primarily concentrated into the center of the droplet, the total area occupied by the sample was 5.3× smaller than the standard smear area, while the most central deposition area was 45× smaller. Combined volumetric and spatial concentration into a 50 μL droplet yielded a sample that was enriched 105-fold. If the method were improved to concentrate 100% of the sample in the most central deposition area of the droplet, perhaps through improvements in the fixation protocol, total enrichment could be up to ≈900×.
To overcome the limitations of bright field microscopy-based inspection of sputum smears, researchers have made a substantial effort to develop fluorescence microscopy (FM) for use with volumetric sample concentration-based methods for TB detection [
44,
45,
46,
47,
48]. These approaches, centrifugation and magnetic bead separation, unfortunately use time-intensive and nonspecific isolation procedures, increasing labor requirements to the already lengthy inspection protocols. As a consequence, these concentration-based FM smear methods show only modest improvements in clinical sensitivity [
49], but reduced specificity [
49,
50]. In addition, FM inadvertently creates a second barrier for implementation because of its reliance on FM technology that is frequently unavailable in low-resource settings [
9]. Other magnetic bead separation strategies use larger particles that obscure visualization of the bacteria, requiring that FM be used for visualization. While these methods have failures that prevent implementation, our design works to overcome the barriers presented. Our method demonstrates the potential for what is achievable with further design of an antigen-specific NP. For the purposes of this study, the “stickiness” of mycobacteria allowed for the use of nonspecific isolation on a simple streptavidin particle, but in practice, other contaminants, such as non-tuberculous bacteria and human cells found in the airways, would also stick to the particles, creating a sample that is impossible to accurately and effectively visualize without providing any specificity. The use of NPs smaller than the diffraction limit of light enabled improved visualization of the sample using bright field microscopy and gold standard acid-fast staining techniques. Although not demonstrated, fluorescent-staining techniques such as auramine O [
27] could theoretically be used with our concentration method, but it is not a requirement for implementation such as with prior methods. The newly developed DMN-tre stain would provide the added benefit of not subjecting the sample to repeated harsh chemicals as occurs with acid-fast staining techniques; this would help resolve the issue of sample loss, but fluorescence microscopy would be required for visualization [
51].
There are two core phenomena that are important elements of our method. First, during volumetric concentration, the capture of bacteria on NPs form aggregates that are clearly visualized upon staining. Second, thermocapillary Marangoni flows oriented inward along the substrate are important for ensuring central deposition of captured bacteria and NPs. The formation of bacterial aggregates aid in the final visualization of the bacteria in the center of the drop, which are seen as large, three-dimensional pink or red networks of bacteria amongst the NPs (
Figure 7). Regardless of the presence or absence of bacteria, the NPs settle in the center of the droplet during evaporation, creating a clearly defined area for inspection.
This study was designed to use known principles of Marangoni flow formation to engineer a new design for microscopic application. Changes from our prior work in both the substrate and solvent were determined. The solutions tested were based on the work of Ristenpart et al. [
10] and our prior work [
11,
12,
13]. Having clearly defined goals and design constraints allowed us to focus materials testing while also easily identifying inexpensive solutions for these improvements. The knowledge that Marangoni flows are not formed in pure water [
52,
53] helped us in modifying the droplet solvent, allowing us to remove what was incompatible with microscopy while maintaining a system that created the desired flow pattern. The addition of the PLL droplet array was a critical design requirement for making our chosen substrate and solvent system useable.
The use of inexpensive, commonly available PLL and Rain-X® for the microscope slide substrate means the cost of the microscope slide is nominally more expensive. Moreover, the cost of the magnetic NPs was only USD 0.56/test at time of publication. Even with the addition of an antigen recognition system to the NPs, this assay could easily be produced at <USD 1.00/test, in addition to the cost of staining reagents already used at the POC. In addition, because Rain-X® covalently modifies the surface of the microscope slide, and PLL-coated slides have a year-long shelf life if stored in a closed container, the slides could ship to decentralized testing sites prepared and ready for use with no required preparation. While PLL spots were applied by hand in this study, automated processing could create highly repeatable and reproducible microscope slides.
This report was designed to be a “proof-of-principle” study, and some limitations remain. As mentioned, it is important that the NPs be immunospecific in the final application, because as prior work demonstrates, nonspecific volumetric concentration of samples does not significantly improve sensitivity and also reduces specificity [
49,
50]. The use of nonspecific isolation techniques in prior studies and the continued issues seen here demonstrate the difficulty in creating an immunospecific particle for specific capture of mycobacteria. The gap that remains by not solving this problem is not lost, since failing to specifically isolate mycobacteria from a sputum sample will make it impossible to visualize bacteria. Specifically, it is extremely common for other biological contaminants to significantly outnumber TB bacteria in sputum samples; isolation of all biological contaminants will result in a dark blue stain that cannot be accurately read by the microscopist. For methods such as this to work, it is imperative that more development focus on methods to rapidly and specifically isolate mycobacteria. Even better, design and use of an antigen recognition element that can discern between tuberculous and nontuberculous infections would create a diagnostic that satisfies another diagnostic need that is currently insufficiently met using culture [
54].
Second, magnetic NPs were chosen specifically because they are smaller than the theoretical diffraction limit of light and are unable to individually obscure bacilli visualization such as with larger magnetic particles. However, together, if spatially concentrated enough, they can obscure imaging. The particle volume used in this study was chosen to yield efficient volumetric concentration while not completely obscuring visualization following spatial enrichment, but in other applications, these parameters would need to be optimized to prevent bulk NP imaging interference.
Third, given this is study intended to demonstrate the improvement of a rapid, POC diagnostic method, it is important that it can be performed in a period no greater than a few hours. In the current iteration, both the time it takes for magnetic separation and the amount of time it takes for the droplet to dry are too long to meet this requirement, and changes to the method design are required. The lower magnetic susceptibility of the NPs, compared to more common ≥1 μm magnetic microparticles used in magnetic bioseparations, significantly extends the time it takes to magnetically separate the sample. When adding the increased viscosity of sputum in a patient sample, the separation using a stationary magnet is not feasible for use at the POC. The use of high-gradient magnetic separation, a technique that has been shown to rapidly and efficiently separate both micro- and NPs from large-volume, high-viscosity samples [
55,
56], has the potential to solve this problem and should be explored as an alternate to stationary magnetic pooling. This technique may also allow for use of even smaller NPs, providing another solution to bulk NP imaging issues. The use of entire sputum samples, which averages 3 mL in volume [
57], can further improve method sensitivity.
To solve the issue of the droplet drying time, the use of multiple smaller droplets instead of a single large droplet can significantly reduce the time to dry, although this could increase the inspection time and/or area. In addition, as demonstrated in microarray development, the use of a greater number of smaller drops rather than fewer larger drops can improve the sensitivity of the method [
58]. Further studies should be performed to determine if this theory applies to our application and what the tradeoff between inspection area and sensitivity is. It also may provide a solution to potential NP visualization issues previously mentioned.