Synthetic Protein Circuits and Devices Based on Reversible Protein-Protein Interactions: An Overview
Abstract
:1. Introduction
2. Protein Modules for Protein-Protein Interaction (PPI)-Based Synthetic Circuit Design
3. Peptides as Powerful Synthetic Modules for Protein Circuit Design
4. Peptides as Building Blocks for Targeted Proteolysis
5. Combinatorial Libraries as Platforms for Peptides Isolation
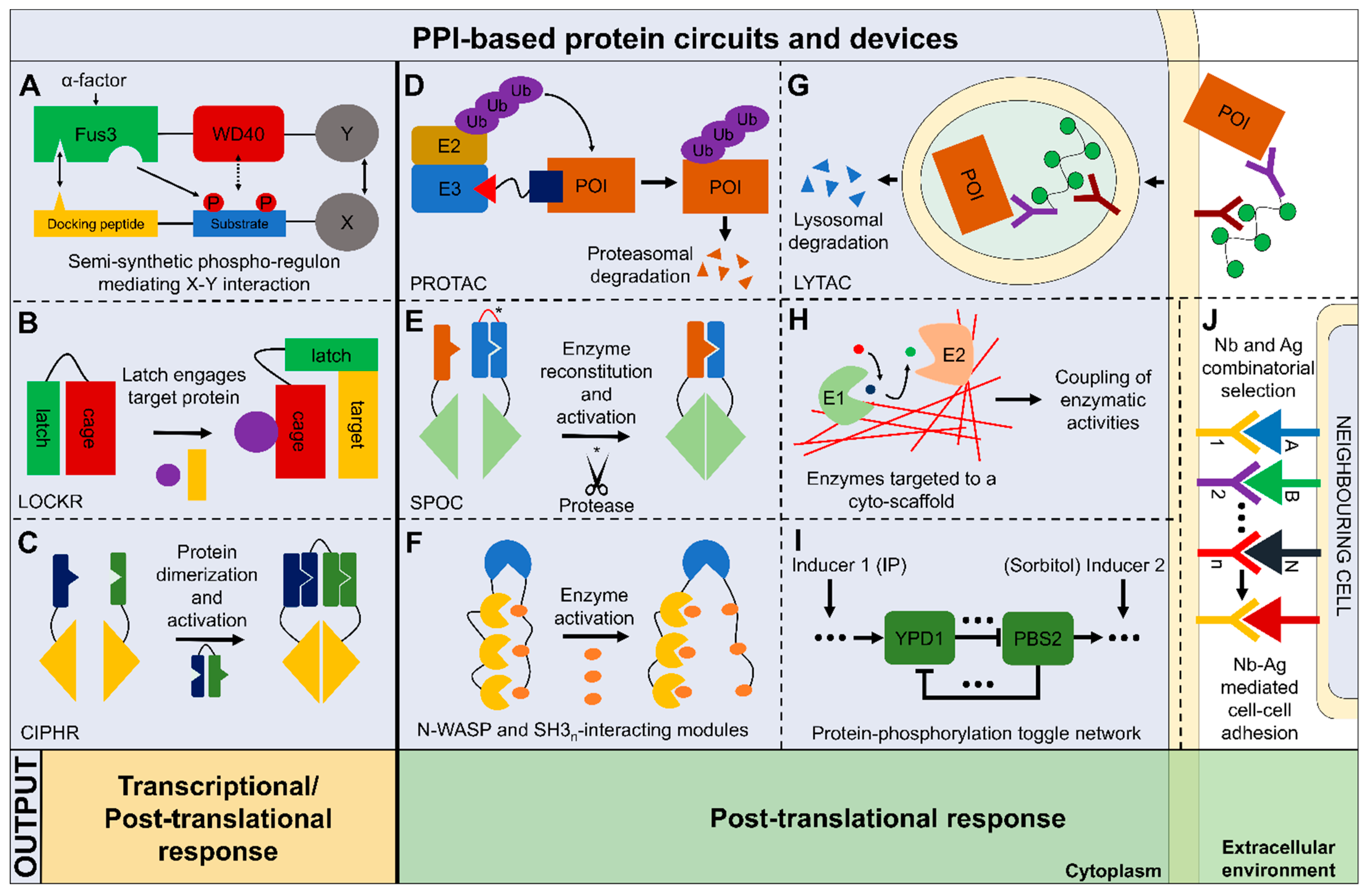
6. Perspectives on the Application of PPI-Based Synthetic Protein Circuits
Funding
Acknowledgments
Conflicts of Interest
References
- Kholodenko, B.N. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006, 7, 165–176. [Google Scholar] [CrossRef]
- Nair, A.; Chauhan, P.; Saha, B.; Kubatzky, K.F. Molecular Sciences Conceptual Evolution of Cell Signaling. Int. J. Mol. Sci. 2019, 20, 3292. [Google Scholar] [CrossRef] [Green Version]
- Kaelin, W.G.; Ratcliffe, P.J. Oxygen Sensing by Metazoans: The Central Role of the HIF Hydroxylase Pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 342, 742–744. [Google Scholar] [CrossRef] [Green Version]
- Ricci-Tam, C.; Ben-Zion, I.; Wang, J.; Palme, J.; Li, A.; Savir, Y.; Springer, M. Decoupling transcription factor expression and activity enables dimmer switch gene regulation. Science 2021, 372, 292–295. [Google Scholar] [CrossRef]
- Mishra, D.; Bepler, T.; Teague, B.; Berger, B.; Broach, J.; Weiss, R. An engineered protein-phosphorylation toggle network with implications for endogenous network discovery. Science 2021, 373, eaav0780. [Google Scholar] [CrossRef]
- Zhang, Q.; Bhattacharya, S.; Pi, J.; Clewell, R.A.; Carmichael, P.L.; Andersen, M.E. Adaptive posttranslational control in cellular stress response pathways and its relationship to toxicity testing and safety assessment. Toxicol. Sci. 2015, 147, 302–316. [Google Scholar] [CrossRef] [Green Version]
- Khalil, A.S.; Collins, J.J. Synthetic biology: Applications come of age. Nat. Rev. Genet. 2010, 11, 367–379. [Google Scholar] [CrossRef]
- Hicks, M.; Bachmann, T.T.; Wang, B. Synthetic Biology Enables Programmable Cell-Based Biosensors. ChemPhysChem 2020, 21, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Slomovic, S.; Pardee, K.; Collins, J.J. Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl. Acad. Sci. USA 2015, 112, 14429–14435. [Google Scholar] [CrossRef] [Green Version]
- David, F.; Davis, A.M.; Gossing, M.; Hayes, M.A.; Romero, E.; Scott, L.H.; Wigglesworth, M.J. A Perspective on Synthetic Biology in Drug Discovery and Development—Current Impact and Future Opportunities. SLAS Discov. 2021, 26, 581–603. [Google Scholar] [CrossRef]
- Tang, T.C.; An, B.; Huang, Y.; Vasikaran, S.; Wang, Y.; Jiang, X.; Lu, T.K.; Zhong, C. Materials design by synthetic biology. Nat. Rev. Mater. 2021, 6, 332–350. [Google Scholar] [CrossRef]
- Hanczyc, M.M. Engineering life: A review of synthetic biology. Artif. Life 2020, 26, 260–273. [Google Scholar] [CrossRef]
- Del Vecchio, D.; Dy, A.J.; Qian, Y. Control theory meets synthetic biology. J. R. Soc. Interface 2016, 13, 20160380. [Google Scholar] [CrossRef]
- Li, Z.; Yang, Q. Systems and synthetic biology approaches in understanding biological oscillators. Quant. Biol. 2018, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef]
- Verbič, A.; Praznik, A.; Jerala, R. A guide to the design of synthetic gene networks in mammalian cells. FEBS J. 2021, 288, 5265–5288. [Google Scholar] [CrossRef]
- Chen, Z.; Elowitz, M.B. Programmable protein circuit design. Cell 2021, 184, 2284–2301. [Google Scholar] [CrossRef]
- Fields, S.; Song, O.K. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Kortemme, T.; Baker, D. Computational design of protein-protein interactions. Curr. Opin. Chem. Biol. 2004, 8, 91–97. [Google Scholar] [CrossRef]
- Mandell, D.J.; Kortemme, T. Computer-aided design of functional protein interactions. Nat. Chem. Biol. 2009, 5, 797–807. [Google Scholar] [CrossRef]
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. Protacs: Great opportunities for academia and industry. Signal Transduct. Target. Ther. 2019, 4, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Grünberg, R.; Serrano, L. Strategies for protein synthetic biology. Nucleic Acids Res. 2010, 38, 2663–2675. [Google Scholar] [CrossRef] [Green Version]
- Dove, S.L.; Hochschild, A. Conversion of the ω subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998, 12, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Atsumi, S.; Little, J.W. Regulatory circuit design and evolution using phage λ. Genes Dev. 2004, 18, 2086–2094. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Zeng, W.; Zong, Y.; Chen, Z.; Miao, C.; Wang, B.; Lou, C. Engineering the Ultrasensitive Transcription Factors by Fusing a Modular Oligomerization Domain. ACS Synth. Biol. 2018, 7, 1188–1194. [Google Scholar] [CrossRef] [Green Version]
- Younger, A.K.D.; Dalvie, N.C.; Rottinghaus, A.G.; Leonard, J.N. Engineering Modular Biosensors to Confer Metabolite-Responsive Regulation of Transcription. ACS Synth. Biol. 2017, 6, 311–325. [Google Scholar] [CrossRef]
- Inobe, T.; Nukina, N. Rapamycin-induced oligomer formation system of FRB-FKBP fusion proteins. J. Biosci. Bioeng. 2016, 122, 40–46. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, X.F.; Brown, E.J.; Schreiber, S.L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 1995, 92, 4947–4951. [Google Scholar] [CrossRef] [Green Version]
- Rhodius, V.A.; Segall-Shapiro, T.H.; Sharon, B.D.; Ghodasara, A.; Orlova, E.; Tabakh, H.; Burkhardt, D.H.; Clancy, K.; Peterson, T.C.; Gross, C.A.; et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 2013, 9, 1–13. [Google Scholar] [CrossRef]
- Smith, A.J.; Thomas, F.; Shoemark, D.; Woolfson, D.N.; Savery, N.J. Guiding Biomolecular Interactions in Cells Using de Novo Protein-Protein Interfaces. ACS Synth. Biol. 2019, 8, 1284–1293. [Google Scholar] [CrossRef]
- Grover, A.; Pande, A.; Choudhary, K.; Gupta, K.; Sundar, D. Re-programming DNA-binding specificity in zinc finger proteins for targeting unique address in a genome. Syst. Synth. Biol. 2010, 4, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Giesecke, A.V.; Fang, R.; Joung, J.K. Synthetic protein-protein interaction domains created by shuffling Cys 2His2 zinc-fingers. Mol. Syst. Biol. 2006, 2, 2006.0011. [Google Scholar] [CrossRef] [Green Version]
- Pu, J.; Zinkus-Boltz, J.; Dickinson, B.C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 2017, 13, 432–438. [Google Scholar] [CrossRef] [Green Version]
- Groß, A.; Hashimoto, C.; Sticht, H.; Eichler, J. Synthetic peptides as protein mimics. Front. Bioeng. Biotechnol. 2016, 3, 211. [Google Scholar] [CrossRef] [Green Version]
- Gordley, R.M.; Williams, R.E.; Bashor, C.J.; Toettcher, J.E.; Yan, S.; Lim, W.A. Engineering dynamical control of cell fate switching using synthetic phospho-regulons. Proc. Natl. Acad. Sci. USA 2016, 113, 13528–13533. [Google Scholar] [CrossRef] [Green Version]
- Wei, P.; Wong, W.W.; Park, J.S.; Corcoran, E.E.; Peisajovich, S.G.; Onuffer, J.J.; Weiss, A.; Lim, W.A. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature 2012, 488, 384–388. [Google Scholar] [CrossRef] [Green Version]
- Groves, B.; Khakhar, A.; Nadel, C.M.; Gardner, R.G.; Seelig, G. Rewiring MAP kinases in Saccharomyces cerevisiae to regulate novel targets through ubiquitination. ELife 2016, 5, 1–22. [Google Scholar] [CrossRef]
- Dueber, J.E.; Mirsky, E.A.; Lim, W.A. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat. Biotechnol. 2007, 25, 660–662. [Google Scholar] [CrossRef]
- Dai, Y.; Abbasi, K.; Bandyopadhyay, S.; Liu, C.C. Dynamic Control of Peptide Strand Displacement Reaction Using Functional Biomolecular Domain for Biosensing. ACS Sens. 2019, 4, 1980–1985. [Google Scholar] [CrossRef]
- Stein, V.; Alexandrov, K. Protease-based synthetic sensing and signal amplification. Proc. Natl. Acad. Sci. USA 2014, 111, 15934–15939. [Google Scholar] [CrossRef] [Green Version]
- Stein, V.; Nabi, M.; Alexandrov, K. Ultrasensitive Scaffold-Dependent Protease Sensors with Large Dynamic Range. ACS Synth. Biol. 2017, 6, 1337–1342. [Google Scholar] [CrossRef]
- Holt, B.A.; Kwong, G.A. Protease circuits for processing biological information. Nat. Commun. 2020, 11, 5201. [Google Scholar] [CrossRef]
- Fletcher, J.M.; Boyle, A.L.; Bruning, M.; Bartlett, S.J.; Vincent, T.L.; Zaccai, N.R.; Armstrong, C.T.; Bromley, E.H.C.; Booth, P.J.; Brady, R.L.; et al. A basis set of de novo coiled-Coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 2012, 1, 240–250. [Google Scholar] [CrossRef]
- Thompson, K.E.; Bashor, C.J.; Lim, W.A.; Keating, A.E. SYNZIP Protein Interaction Toolbox: In Vitro and in Vivo Specifications of Heterospecific Coiled-Coil Interaction Domains. ACS Synth. Biol. 2012, 1, 118–129. [Google Scholar] [CrossRef]
- Fink, T.; Lonzarić, J.; Praznik, A.; Plaper, T.; Merljak, E.; Leben, K.; Jerala, N.; Lebar, T.; Strmšek, Ž.; Lapenta, F.; et al. Design of fast proteolysis-based signaling and logic circuits in mammalian cells. Nat. Chem. Biol. 2019, 15, 115–122. [Google Scholar] [CrossRef]
- Chen, Z.; Kibler, R.D.; Hunt, A.; Busch, F.; Pearl, J.; Jia, M.; VanAernum, Z.L.; Wicky, B.I.M.; Dods, G.; Liao, H.; et al. De novo design of protein logic gates. Science 2020, 368, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Langan, R.A.; Boyken, S.E.; Ng, A.H.; Samson, J.A.; Dods, G.; Westbrook, A.M.; Nguyen, T.H.; Lajoie, M.J.; Chen, Z.; Berger, S.; et al. De novo design of bioactive protein switches. Nature 2019, 572, 205–210. [Google Scholar] [CrossRef]
- Kirkpatrick, R.L.; Lewis, K.; Langan, R.A.; Lajoie, M.J.; Boyken, S.E.; Eakman, M.; Baker, D.; Zalatan, J.G. Conditional Recruitment to a DNA-Bound CRISPR-Cas Complex Using a Colocalization-Dependent Protein Switch. ACS Synth. Biol. 2020, 9, 2316–2323. [Google Scholar] [CrossRef]
- Edgell, C.L.; Smith, A.J.; Beesley, J.L.; Savery, N.J.; Woolfson, D.N. De novo designed protein-interaction modules for in-cell applications. ACS Synth. Biol. 2020, 9, 427–436. [Google Scholar] [CrossRef]
- Lee, M.J.; Mantell, J.; Hodgson, L.; Alibhai, D.; Fletcher, J.M.; Brown, I.R.; Frank, S.; Xue, W.F.; Verkade, P.; Woolfson, D.N.; et al. Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm. Nat. Chem. Biol. 2018, 14, 142–147. [Google Scholar] [CrossRef]
- Alabi, S.B.; Crews, C.M. Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 2021, 296, 100647. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, K.M.; Kim, K.B.; Verma, R.; Ransick, A.; Stein, B.; Crews, C.M.; Deshaies, R.J. Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol. Cell. Proteom. MCP 2003, 2, 1350–1358. [Google Scholar] [CrossRef] [Green Version]
- Schneekloth, J.S.; Fonseca, F.N.; Koldobskiy, M.; Mandal, A.; Deshaies, R.; Sakamoto, K.; Crews, C.M. Chemical Genetic Control of Protein Levels: Selective in Vivo Targeted Degradation. J. Am. Chem. Soc. 2004, 126, 3748–3754. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Liu, T.; Jiao, Q.; Ji, J.; Tao, M.; Liu, Y.; You, Q.; Jiang, Z. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 2018, 146, 251–259. [Google Scholar] [CrossRef]
- Banik, S.M.; Pedram, K.; Wisnovsky, S.; Ahn, G.; Riley, N.M.; Bertozzi, C.R. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 2020, 584, 291–297. [Google Scholar] [CrossRef]
- Neklesa, T.K.; Tae, H.S.; Schneekloth, A.R.; Stulberg, M.J.; Corson, T.W.; Sundberg, T.B.; Raina, K.; Holley, S.A.; Crews, C.M. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat. Chem. Biol. 2011, 7, 538–543. [Google Scholar] [CrossRef] [Green Version]
- Röth, S.; Fulcher, L.J.; Sapkota, G.P. Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci. 2019, 76, 2761–2777. [Google Scholar] [CrossRef] [Green Version]
- Gao, N.; Chu, T.T.; Li, Q.Q.; Lim, Y.J.; Qiu, T.; Ma, M.R.; Hu, Z.W.; Yang, X.F.; Chen, Y.X.; Zhao, Y.F.; et al. Hydrophobic tagging-mediated degradation of Alzheimer’s disease related Tau. RSC Adv. 2017, 7, 40362–40366. [Google Scholar] [CrossRef] [Green Version]
- Naseri, G.; Koffas, M.A.G. Application of combinatorial optimization strategies in synthetic biology. Nat. Commun. 2020, 11, 2446. [Google Scholar] [CrossRef] [PubMed]
- Glass, D.S.; Riedel-Kruse, I.H. A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns. Cell 2018, 174, 649–658.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gräwe, A.; Ranglack, J.; Weyrich, A.; Stein, V. IFLinkC: An iterative functional linker cloning strategy for the combinatorial assembly and recombination of linker peptides with functional domains. Nucleic Acids Res. 2020, 48, E24. [Google Scholar] [CrossRef]
- Huang, P.S.; Boyken, S.E.; Baker, D. The coming of age of de novo protein design. Nature 2016, 537, 320–327. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosa, S.; Bertaso, C.; Pesaresi, P.; Masiero, S.; Tagliani, A. Synthetic Protein Circuits and Devices Based on Reversible Protein-Protein Interactions: An Overview. Life 2021, 11, 1171. https://doi.org/10.3390/life11111171
Rosa S, Bertaso C, Pesaresi P, Masiero S, Tagliani A. Synthetic Protein Circuits and Devices Based on Reversible Protein-Protein Interactions: An Overview. Life. 2021; 11(11):1171. https://doi.org/10.3390/life11111171
Chicago/Turabian StyleRosa, Stefano, Chiara Bertaso, Paolo Pesaresi, Simona Masiero, and Andrea Tagliani. 2021. "Synthetic Protein Circuits and Devices Based on Reversible Protein-Protein Interactions: An Overview" Life 11, no. 11: 1171. https://doi.org/10.3390/life11111171







