Optimization of a Completely Mixed Anaerobic Biofilm Reactor (CMABR), Based on Brewery Wastewater Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of Wastewater
2.2. Reactor Filling Method
2.3. Reactor Setup and Operation
2.4. Condition Optimization
2.5. Analytical Measurements
2.6. DeoxyriboNucleic Acid (DNA) Extraction, Polymerase Chain Reaction (PCR) Amplification, and High-Throughput Sequencing
3. Results and Discussion
3.1. Box–Behnken Design and Statistical Analysis
− 4.83X22 − 0.66X32
3.2. Completely Mixed Anaerobic Biofilm Reactor (CAMBR) Performance under Optimal Conditions
3.3. Microbial Community Study of the CMABR
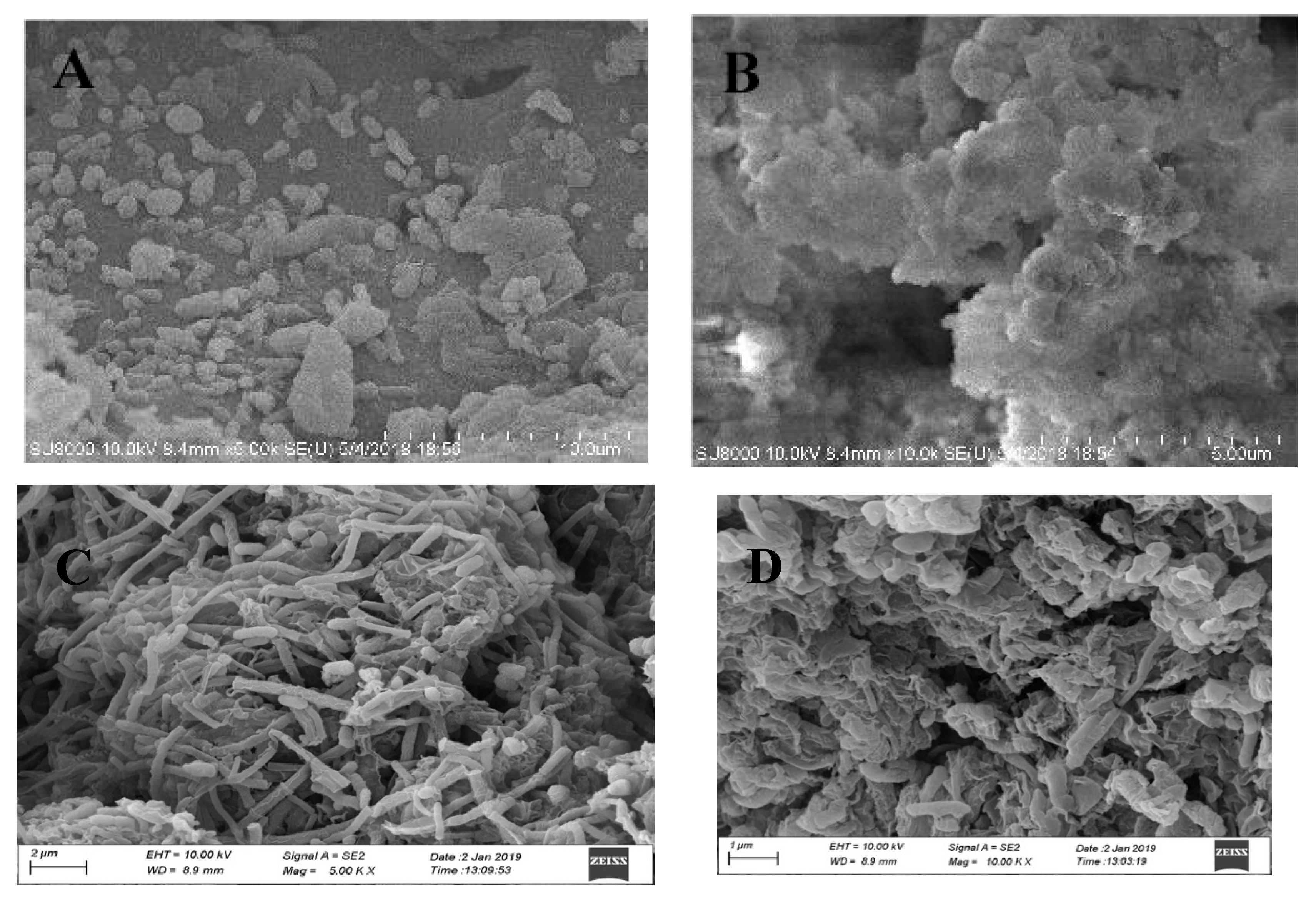
3.3.1. Macrostructure of Reactor Biophase
3.3.2. Microstructure of the CMABR Microbial Phase
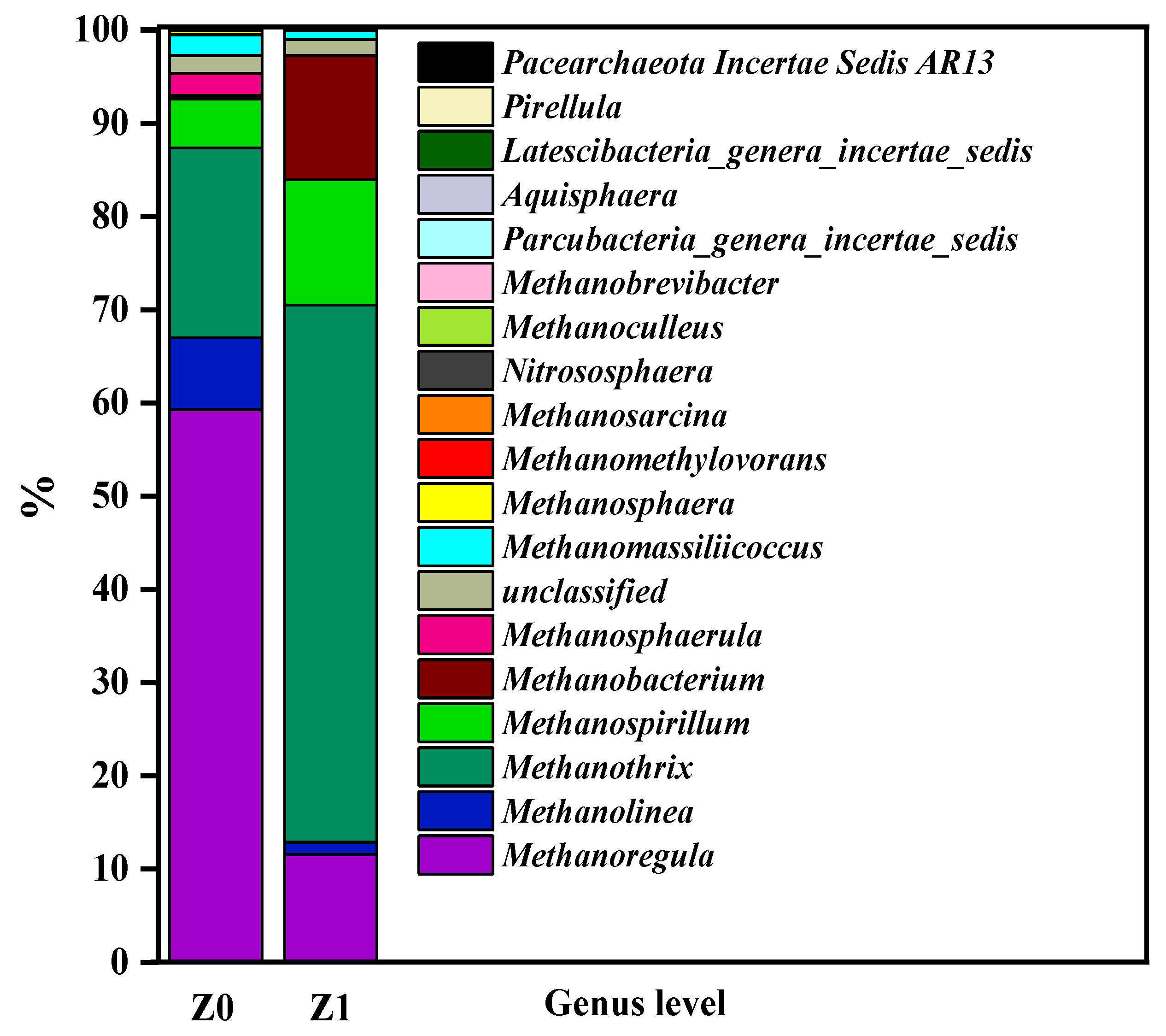
3.3.3. Sequence Analysis of the Microbial Community Structure
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Han, D.M.; Matthew, J.C.; Cao, G.L. Deep challenges for China’s war on water pollution. Environ. Pollut. 2016, 218, 1222–1233. [Google Scholar] [CrossRef] [Green Version]
- Mark, F. A new wastewater treatment technology for developing countries. Filtr. Sep. 2014, 51, 14–17. [Google Scholar]
- Shi, J.X.; Han, Y.X.; Xu, C.Y. Enhanced anaerobic degradation of selected nitrogen heterocyclic compounds with the assistance of carboxymethyl cellulose. Sci. Total Environ. 2019, 689, 781–788. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef]
- Yang, J.; Wang, D.H.; Luo, Z.F. Influence of reflux ratio on the anaerobic digestion of pig manure in leach beds coupled with continuous stirred tank reactors. Waste Manag. 2019, 97, 115–122. [Google Scholar] [CrossRef]
- Doğan, K.; Emre, O.K.; Bestamin, Ö. A review on anaerobic biofilm reactors for the treatment of dairy industry wastewater. Process Biochem. 2015, 50, 262–271. [Google Scholar]
- Cheng, H.; Lin, H.; Huo, H.; Dong, Y.; Xue, Q.; Cao, L. Continuous removal of ore floatation reagents by an anaerobic–aerobic biological filter. Bioresour. Technol. 2012, 114, 255–261. [Google Scholar] [CrossRef]
- Li, H.; Han, K.Z.; Li, Z.P. Performance, granule conductivity and microbial community analysis of upflow anaerobic sludge blanket (UASB) reactors from mesophilic to thermophilic operation. Biochem. Eng. 2018, 133, 59–65. [Google Scholar] [CrossRef]
- Sen, S.; Demirer, G.N. Anaerobic treatment of real textile wastewater with a fluidized bed reactor. Water Res. 2003, 37, 1868–1878. [Google Scholar] [CrossRef]
- Noor, S.; Soliman, H.M. Experimental assessment of a new device for gas–liquid separation. Chem. Eng. Res. Des. 2019, 149, 45–51. [Google Scholar] [CrossRef]
- Ding, J.; Wang, X.; Zhou, X.F. CFD optimization of continuous stirred-tank (CSTR) reactor for biohydrogen production. Bioresour. Technol. 2010, 101, 7005–7013. [Google Scholar] [CrossRef]
- Biraj, S.T.; Oie, G.D.; Bhola, T. Flow measurements around guide vanes of Francis turbine: A PIV approach. Renew. Energy 2018, 126, 177–188. [Google Scholar]
- Su, J.F.; Ma, M.; Wei, L. Algicidal and denitrification characterization of Acinetobacter sp. J25 against Microcystis aeruginosa and microbial community in eutrophic landscape water. Mar. Pollut. Bull. 2016, 107, 233–239. [Google Scholar] [CrossRef]
- Marin, P.; Alkalay, D.; Guerrero, L. Design and startup of an anaerobic fluidized bed reactor. Water Sci. Technol. 1999, 40, 63–70. [Google Scholar] [CrossRef]
- Su, J.F.; Luo, X.X.; Wei, L. Performance and microbial communities of Mn(II)-based autotrophic denitrification in a Moving Bed Biofilm Reactor (MBBR). Bioresour. Technol. 2016, 211, 743–750. [Google Scholar] [CrossRef]
- Ceylan, H.; Kubilay, S.; Aktas, N. An approach for prediction of optimum reaction conditions for laccase-catalyzed bio-transformation of 1-naphthol by response surface methodology (RSM). Bioresour. Technol. 2008, 99, 2025–2031. [Google Scholar] [CrossRef]
- Sevgi, P.; Perviz, S. Application of response surface methodology with a Box–Behnken design for struvite precipitation. Adv. Powder Technol. 2019, 30, 2396–2407. [Google Scholar]
- Liu, H.J.; Jiang, W.; Wan, D.J. Study of a combined heterotrophic and sulfur autotrophic denitrification technology for removal of nitrate in water. J. Hazard. Mater. 2009, 169, 23–28. [Google Scholar] [CrossRef]
- Cheng, G.D.; Wang, F.; Li, C. Experimental Study on Modification of Polyurethane Foam Plastics Carriers. China Water Wastewater 2012, 28, 86–89. [Google Scholar]
- Hao, T.W.; Wei, L.; Lu, H. Characterization of sulfate-reducing granular sludge in the SANI® process. Water Res. 2013, 47, 7042–7052. [Google Scholar] [CrossRef]
- Rajneesh, S.; Puspendu, B.; Rajesh, R.D. Optimization of organics removal and understanding the impact of HRT on vermifiltration of brewery wastewater. Sci. Total Environ. 2019, 651, 1283–1293. [Google Scholar]
- Ravichandrana, P.; Balajib, K. Effect of HRT on performance of hybrid upflow anaerobic sludge blanket (HUASB) reactor using bio balls in treatment of pulp and paper mill bagasse wash water. Mater. Today Proc. 2020, 22, 627–632. [Google Scholar] [CrossRef]
- Mahin, M.; Parviz, M.; Nasim, K. Efficient hydrogen gas production from molasses in hybrid anaerobic-activated sludge-rotating biological contactor. Int. J. Hydrog. Energy 2019, 44, 2592–2602. [Google Scholar]
- Shao, X.W.; Peng, D.C.; Teng, Z.H. Treatment of brewery wastewater using anaerobic sequencing batch reactor (ASBR). Bioresour. Technol. 2008, 99, 3182–3186. [Google Scholar] [CrossRef]
- Baloch, M.I.; Akunna, J.C.; Collier, P.J. The performance of a phase separated granular bed bioreactor treating brewery wastewater. Bioresour. Technol. 2007, 98, 1849–1855. [Google Scholar] [CrossRef]
- Parawira, W.I.; Kudita, M.G.; Nyandoroh, R.; Zvauya, A. A study of industrial anaerobic treatment of opaque beer brewery wastewater in a tropical climate using a full-scale UASB reactor seeded with activated sludge. Process Biochem. 2005, 40, 593–599. [Google Scholar] [CrossRef]
- Miqueleto, A.P.; Rodrigues, J.A.; Ratusznei, S.M. Treatment of easily degradable wastewater in a stirred anaerobic sequencing batch biofilm reactor. Water Res. 2005, 39, 2376–2384. [Google Scholar] [CrossRef]
- Cubas, S.A.; Foresti, E.; Rodrigues, J.A.D. Influence of the liquid-phase mass transfer on the performance of a stirred anaerobic sequencing batch reactor containing immobilized biomass. Biochem. Eng. J. 2004, 17, 100–105. [Google Scholar] [CrossRef]
- Shi, J.X.; Xu, C.Y.; Han, Y.X. Enhanced anaerobic degradation of nitrogen heterocyclic compounds with methanol, sodium citrate, chlorella, spirulina, and carboxymethylcellulose as co-metabolic substances. J. Hazard. Mater. 2020, 384, 121496. [Google Scholar] [CrossRef]
- Wang, C.; Jin, Y.G.; Ji, C. An additive effect of elevated atmospheric CO2 and rising temperature on methane emissions related to methanogenic community in rice paddies. Agric. Ecosyst. Environ. 2018, 257, 165–174. [Google Scholar] [CrossRef]
- Hiroyuki, I.; Sakai, S.; Sekiguchi, Y.J. Methanolinea tarda gen. nov., sp. nov., a methaneproducing archaeon isolated from a methanogenic digester sludge. Int. J. Syst. Evol. Microbiol. 2008, 58, 294–301. [Google Scholar]
- Su, C.Y.; Deng, Q.J.; Lu, Y.X. Effect of circulation and micro-aeration on sludge characteristics and microbial community in an ABR for treating traditional Chinese medicine wastewater. Environ. Technol. 2020, 41, 3284–3296. [Google Scholar] [CrossRef]
- Lauwers, A.M.; Heinen, W.; Gorris, L.G. Early stages in biofilm development in methanogenic fluidized-bed reactors. Appl. Microbiol. Biotechnol. 1990, 33, 352–358. [Google Scholar] [CrossRef]
- Rw, R.; De, A.; Ra, N. Light and electron microscopic examinations of methane-producing biofilms from anaerobic fixed-bed reactors. Appl. Environ. Microbiol. 1984, 48, 127–136. [Google Scholar]
- Gorris, L.G.; Van, D.J.M.; Van, D.D.C. Biofilm development in laboratory methanogenic fluidized bed reactors. Biotechnol. Bioeng. 1989, 33, 687–693. [Google Scholar] [CrossRef]

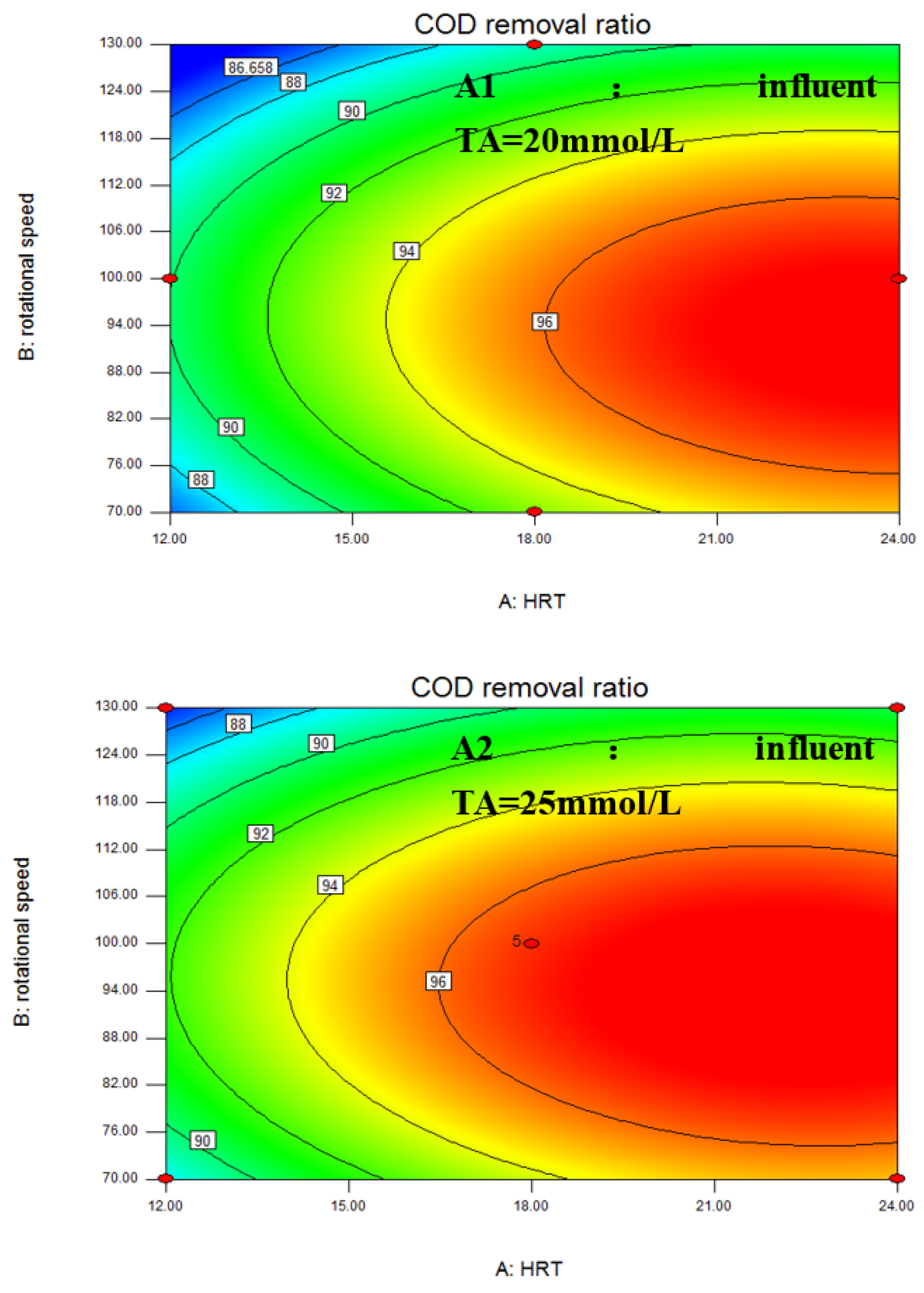
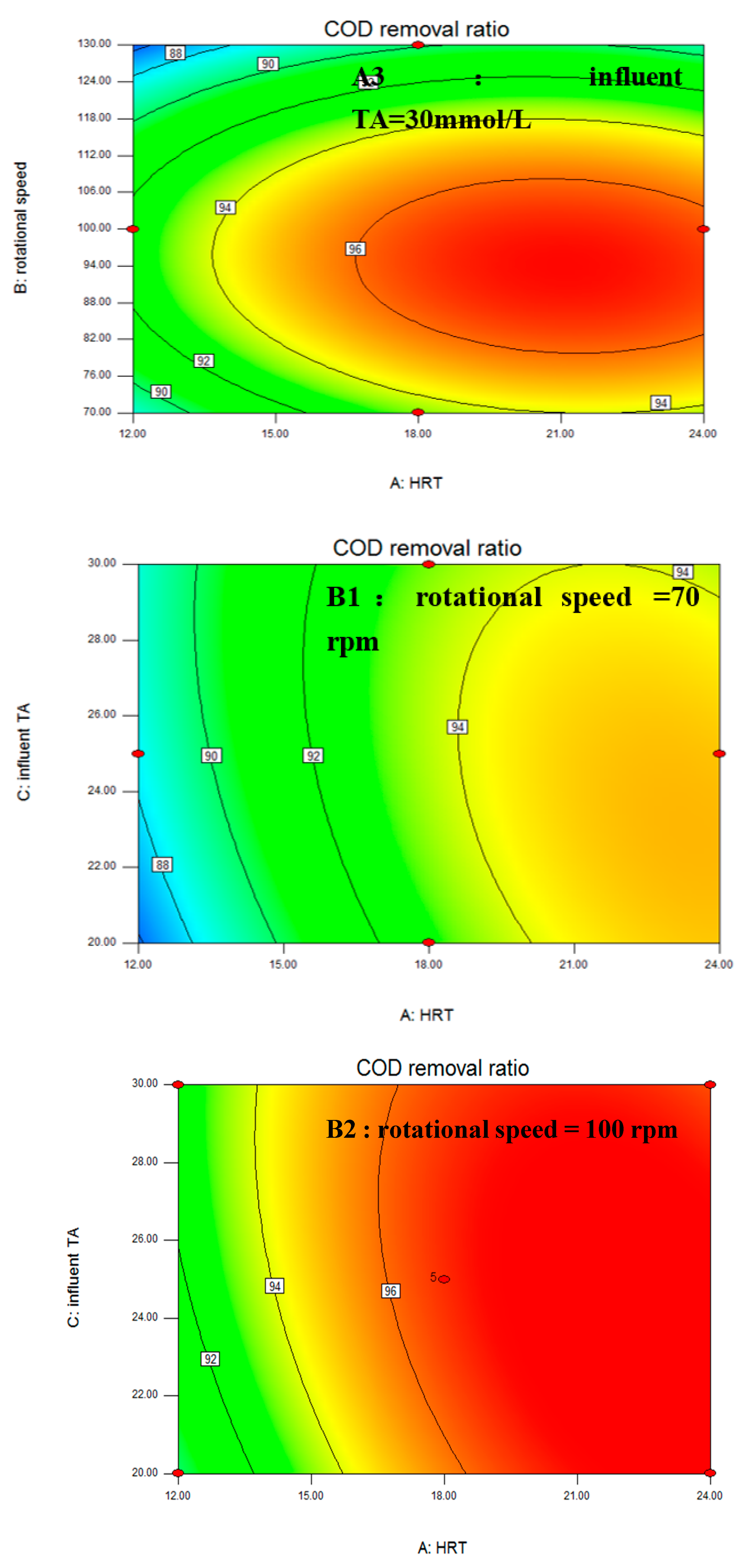
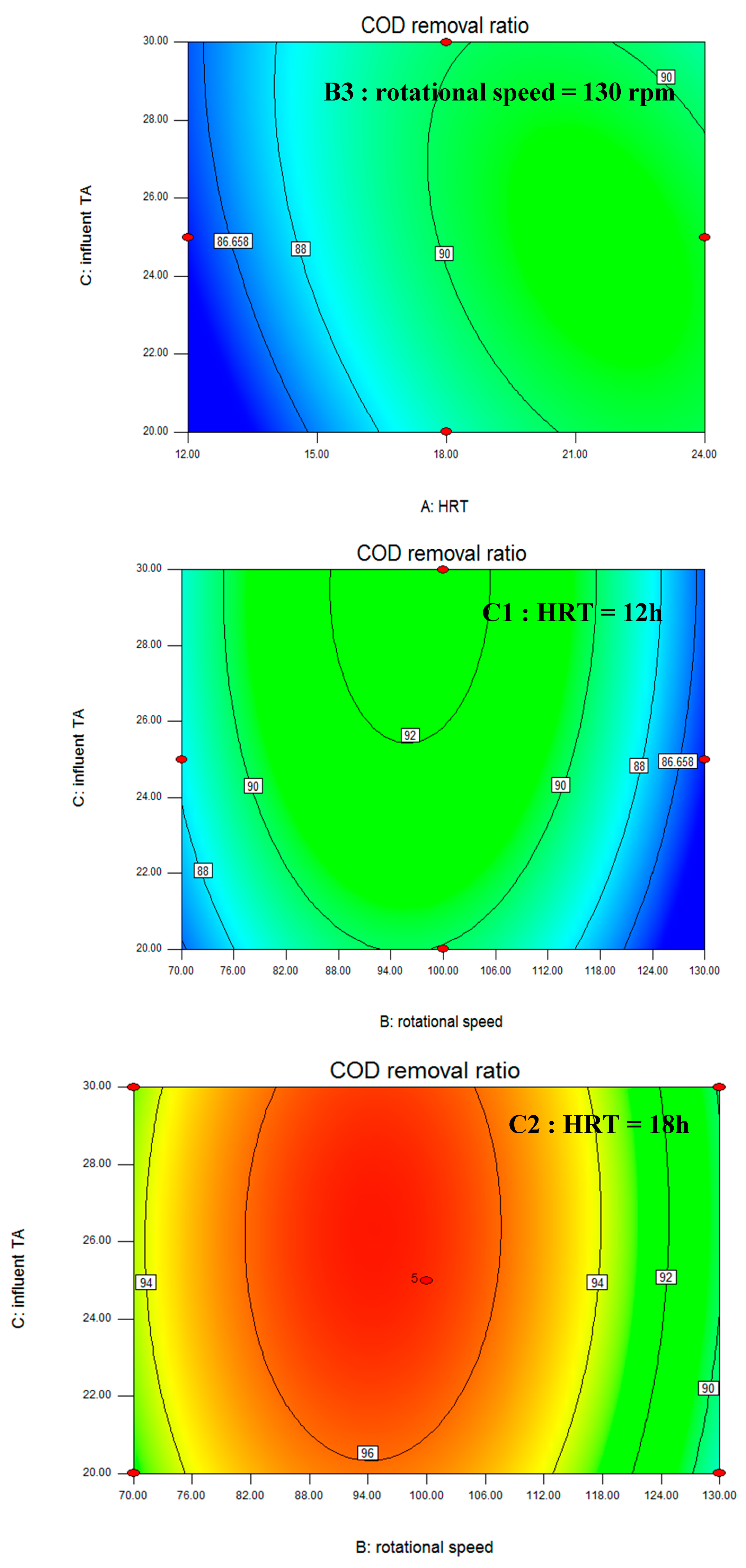
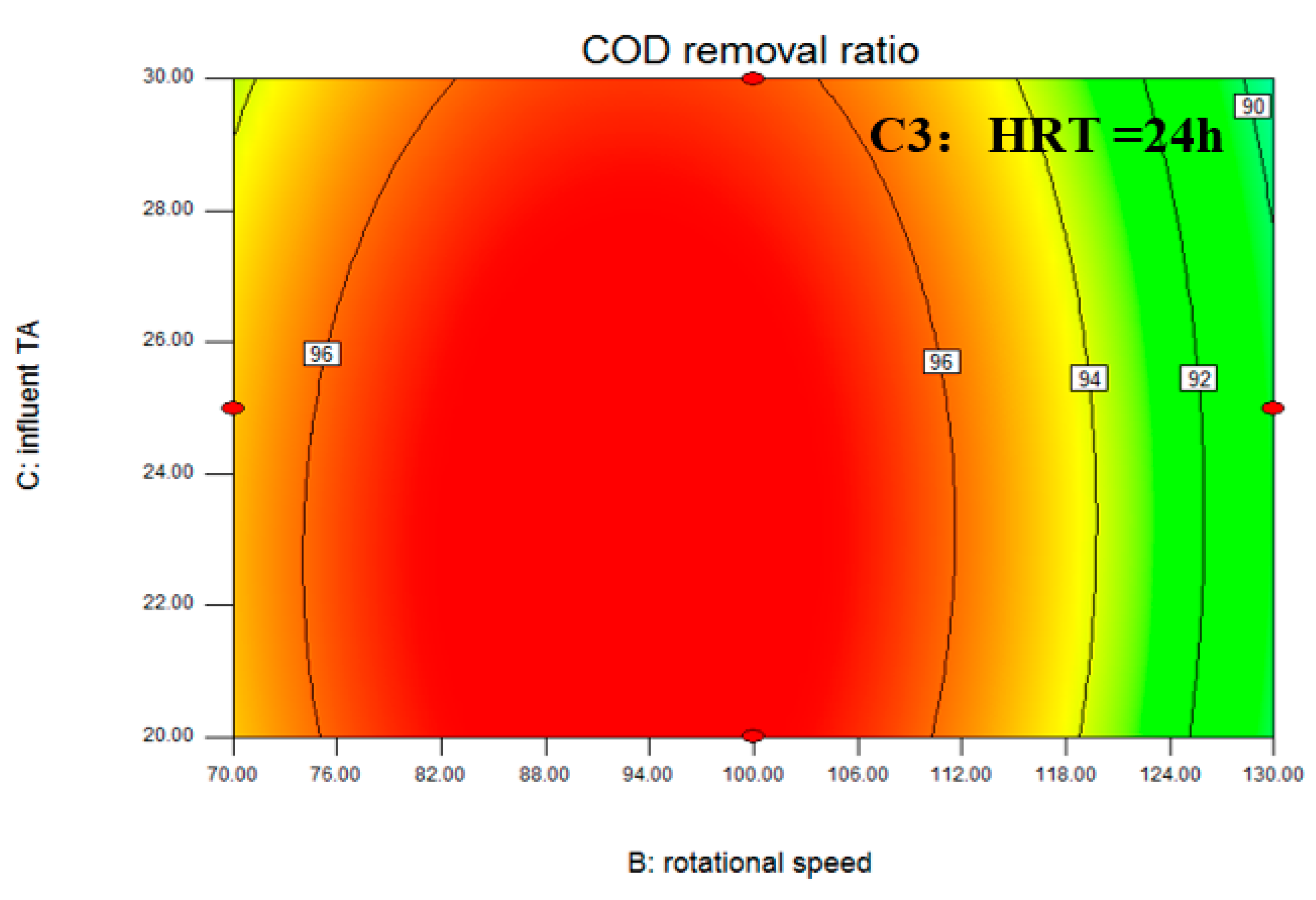

| Stage | Days | HRT (h) | Rotational Speed (rpm) | Influent TA (mmol/L) | COD Removal Ratio (%) |
|---|---|---|---|---|---|
| 1 | 1–6 | 18.00 | 70.00 | 20.00 | 92.53 |
| 2 | 7–12 | 12.00 | 100.00 | 20.00 | 90.19 |
| 3 | 13–18 | 24.00 | 100.00 | 20.00 | 97.25 |
| 4 | 19–24 | 18.00 | 130.00 | 20.00 | 89.10 |
| 5 | 25–30 | 12.00 | 70.00 | 25.00 | 88.29 |
| 6 | 31–36 | 24.00 | 70.00 | 25.00 | 95.33 |
| 7 | 37–42 | 18.00 | 100.00 | 25.00 | 96.33 |
| 8 | 43–48 | 18.00 | 100.00 | 25.00 | 96.99 |
| 9 | 49–54 | 18.00 | 100.00 | 25.00 | 97.05 |
| 10 | 55–60 | 18.00 | 100.00 | 25.00 | 96.63 |
| 11 | 61–66 | 18.00 | 100.00 | 25.00 | 96.57 |
| 12 | 67–72 | 12.00 | 130.00 | 25.00 | 85.29 |
| 13 | 73–78 | 24.00 | 130.00 | 25.00 | 90.42 |
| 14 | 79–84 | 18.00 | 70.00 | 30.00 | 93.15 |
| 15 | 85–90 | 12.00 | 100.00 | 30.00 | 92.50 |
| 16 | 91–96 | 24.00 | 100.00 | 30.00 | 96.09 |
| 17 | 97–102 | 18.00 | 130.00 | 30.00 | 90.12 |
| Source | Sum of Squares | Mean Square | F Value | p-Value Prob > F |
|---|---|---|---|---|
| Model | 221.13 | 24.57 | 181.82 | <0.0001 *** |
| A-HRT | 65.09 | 65.09 | 481.70 | <0.0001 *** |
| B-rotational speed | 25.81 | 25.81 | 191.01 | <0.0001 *** |
| C-TA | 0.97 | 0.97 | 7.20 | 0.0314 * |
| AB | 0.91 | 0.91 | 6.75 | 0.0355 * |
| AC | 3.01 | 3.01 | 22.28 | 0.0022 ** |
| BC | 0.040 | 0.040 | 0.30 | 0.6033 |
| A2 | 17.69 | 17.69 | 130.88 | <0.0001 *** |
| B2 | 98.31 | 98.31 | 727.48 | <0.0001 *** |
| C2 | 1.82 | 1.82 | 13.45 | 0.0080 ** |
| Index | Value |
|---|---|
| Average filler film amount per unit (g/g) | 0.574 ± 0.059 |
| Filler weight with 30% rate (g) | 301.908 |
| Average amount of hanging film (g) | 173.295 ± 17.813 |
| Average sludge concentration equivalent (g/L) | 8.252 ± 0.848 |
| Sample Number | Sample Description | No. of Sequences | Shannon | ACE | Chao1 | Coverage | Simpson |
|---|---|---|---|---|---|---|---|
| Z0 | Raw sludge during anaerobic sludge inoculation | 101,014 | 2.54 | 98,003 | 34,662 | 0.97 | 0.24 |
| Z1 | Biofilm on fillers during stable operation | 70,858 | 1.56 | 221,517 | 75,698 | 0.98 | 0.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, D.; Zhu, K.; Ma, W.; Li, J.; Li, K.; Dai, C. Optimization of a Completely Mixed Anaerobic Biofilm Reactor (CMABR), Based on Brewery Wastewater Treatment. Water 2021, 13, 606. https://doi.org/10.3390/w13050606
Zhong D, Zhu K, Ma W, Li J, Li K, Dai C. Optimization of a Completely Mixed Anaerobic Biofilm Reactor (CMABR), Based on Brewery Wastewater Treatment. Water. 2021; 13(5):606. https://doi.org/10.3390/w13050606
Chicago/Turabian StyleZhong, Dan, Kai Zhu, Wencheng Ma, Jinxin Li, Kefei Li, and Changlei Dai. 2021. "Optimization of a Completely Mixed Anaerobic Biofilm Reactor (CMABR), Based on Brewery Wastewater Treatment" Water 13, no. 5: 606. https://doi.org/10.3390/w13050606






