Occurrence and Distribution of Antibiotic Resistance Genes in Municipal Wastewater Treatment Plants with D-Type Filters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Pretreatment
2.2. DNA Extraction and Library Construction
2.3. Metagenomic Sequencing and Taxonomy Analysis
2.4. Data Analysis Method
2.5. Network Analysis
3. Results and Discussion
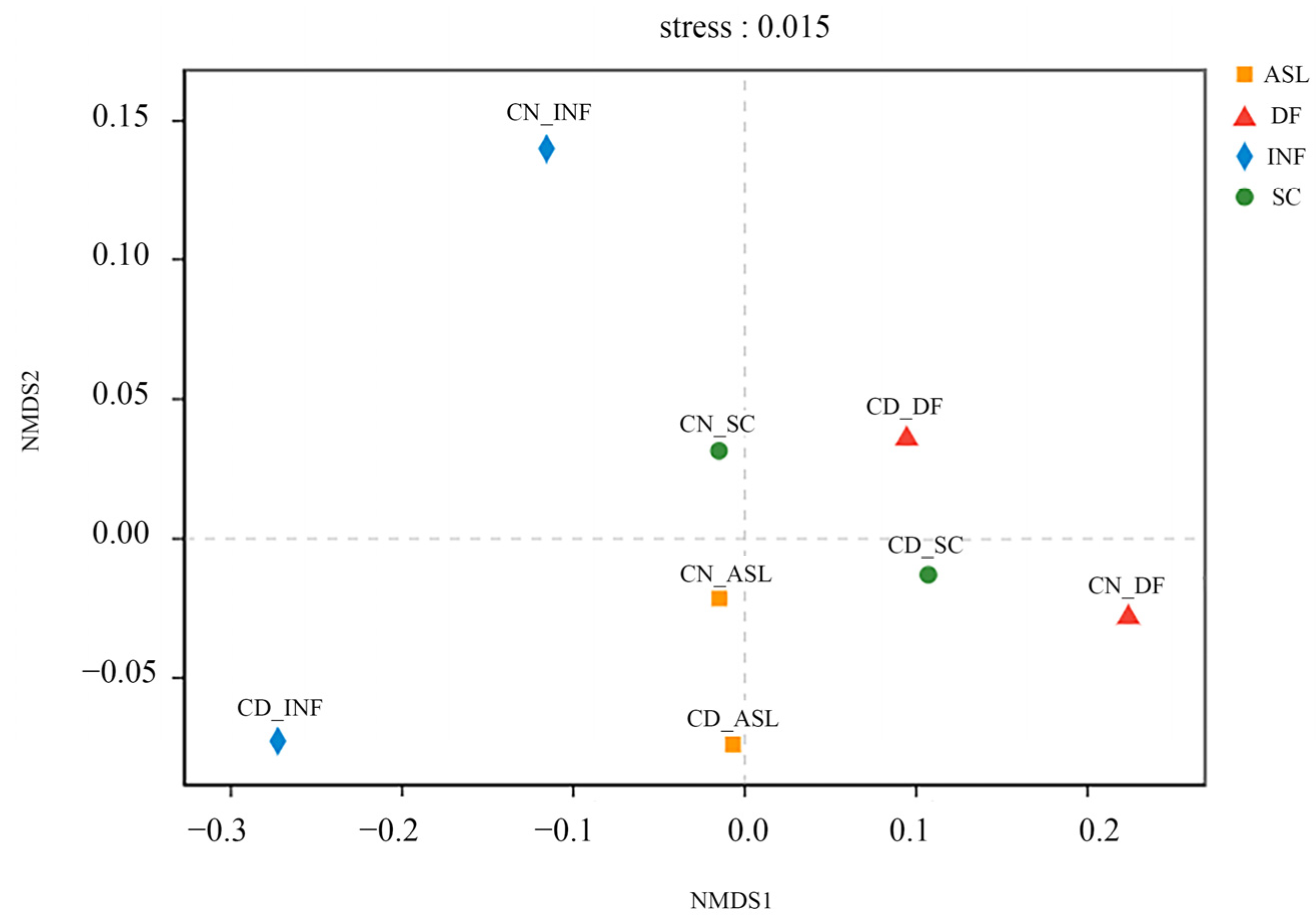
3.1. Diversity and Structure of Bacterial Communities in Wastewater and Sludge
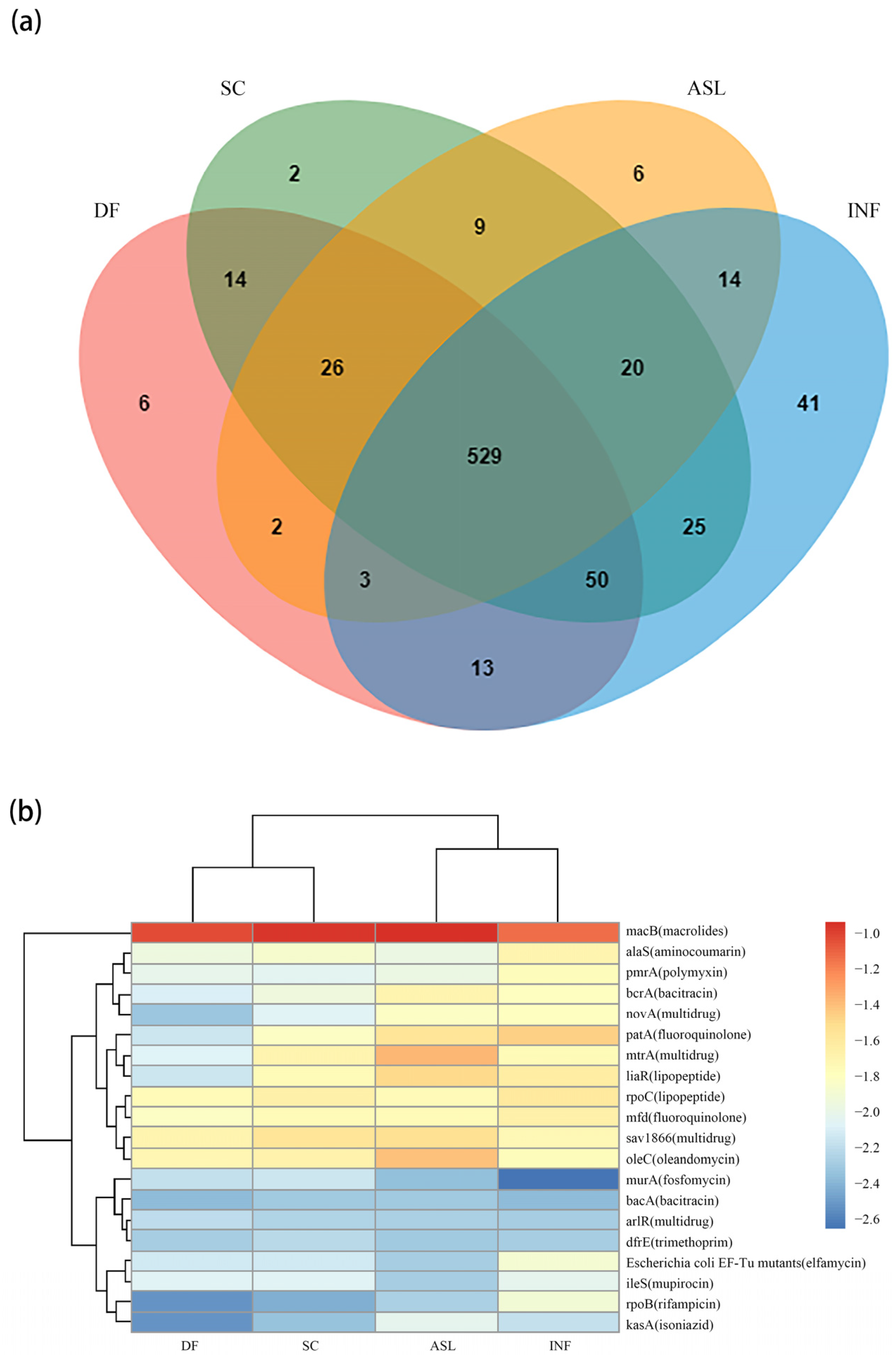
3.2. ARG Distribution along the Treatment Processes
3.3. ARG Removal Specifically in D-Type Filters
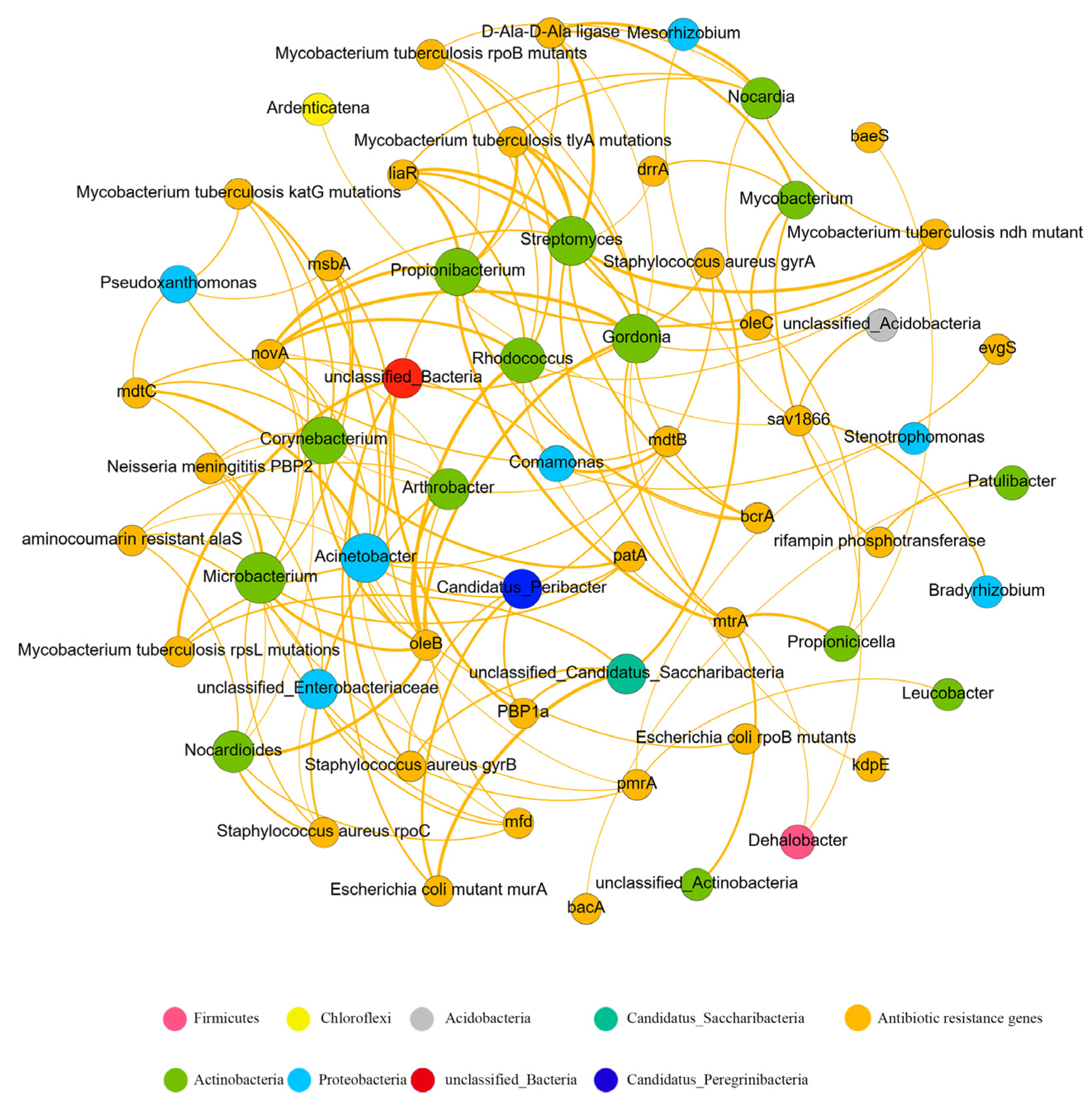
3.4. Relationship and Co-Occurrence Patterns between ARG and Bacterial Taxa
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arulkumaran, N.; Routledge, M.; Schlebusch, S.; Lipman, J.; Conway Morris, A. Antimicrobial-associated harm in critical care: A narrative review. Intensive Care Med. 2020, 46, 225–235. [Google Scholar] [CrossRef]
- DuPont, H.L.; Steffen, R. Use of antimicrobial agents for treatment and prevention of travellers’ diarrhoea in the face of enhanced risk of transient fecal carriage of multi-drug resistant enterobacteriaceae: Setting the stage for consensus recommendations. J. Travel. Med. 2017, 24, S57–S62. [Google Scholar] [CrossRef]
- Peak, N.; Knapp, C.W.; Yang, R.K.; Hanfelt, M.M.; Smith, M.S.; Aga, D.S.; Graham, D.W. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 2007, 9, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Yuan, K.; Chen, X.; Yang, Y.; Zhang, T.; Wang, Y.; Luan, T.; Zou, S.; Li, X. Metagenomic Analysis Revealing Antibiotic Resistance Genes (ARGs) and Their Genetic Compartments in the Tibetan Environment. Environ. Sci. Technol. 2016, 50, 6670–6679. [Google Scholar] [CrossRef]
- Yu, K.F.; Li, P.; Chen, Y.H.; Zhang, B.; Huang, Y.S.; Huang, F.Y.; He, Y.L. Antibiotic resistome associated with microbial communities in an integrated wastewater reclamation system. Water Res. 2020, 173, 115541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Guan, Y.T.; Zhao, R.X.; Feng, J.; Huang, J.; Ma, L.P.; Li, B. Metagenomic and network analyses decipher profiles and co-occurrence patterns of antibiotic resistome and bacterial taxa in the reclaimed wastewater distribution system. J. Hazard. Mater. 2020, 400, 123170. [Google Scholar] [CrossRef]
- Gandolfi, I.; Bertolini, V.; Ambrosini, R.; Bestetti, G.; Franzetti, A. Unravelling the bacterial diversity in the atmosphere. Appl. Microbiol. Biotechnol. 2013, 97, 4727–4736. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.H.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z.G. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef]
- Kim, S.; Yun, Z.; Ha, U.H.; Lee, S.; Park, H.; Kwon, E.E.; Cho, Y.; Choung, S.; Oh, J.; Medriano, C.A.; et al. Transfer of antibiotic resistance plasmids in pure and activated sludge cultures in the presence of environmentally representative micro-contaminant concentrations. Sci. Total Environ. 2014, 468–469, 813–820. [Google Scholar] [CrossRef]
- Warnes, S.L.; Highmore, C.J.; Keevil, C.W. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: Implications for public health. mBio 2012, 3, e00489-12. [Google Scholar] [CrossRef] [Green Version]
- Ju, F.; Beck, K.; Yin, X.; Maccagnan, A.; McArdell, C.S.; Singer, H.P.; Johnson, D.R.; Zhang, T.; Burgmann, H. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J. 2019, 13, 346–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsberg, K.J.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Knight, R.; Fierer, N.; Dantas, G. Bacterial phylogeny structures soil resistomes across habitats. Nature 2014, 509, 612–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, J. Application of a Fiber Filter in Chengdu Shahe Sewage Treatment Plant. Environ. Pollut. Control 2006, 28, 388–390. [Google Scholar]
- Wang, Z.Z.; Sun, Y.X.; Wu, G.X.; Wu, Q.Y.; Hu, H.Y.; Wu, Y.H.; Guo, F.; Guo, Y.M. Analysis of performance and cost of a micro-flocculation-D type filter process applied in wastewater tertiary treatment process. Chin. J. Environ. Eng. 2014, 8, 3132–3136. [Google Scholar]
- Tong, J.; Tang, A.P.; Wang, H.Y.; Liu, X.X.; Huang, Z.; Wang, Z.H.; Wang, Z.Y.; Zhang, J.Y.; Wei, Y.S.; Su, Y.Y.; et al. Microbial community evolution and fate of antibiotic resistance genes along six different full-scale municipal wastewater treatment processes. Bioresour. Technol. 2019, 272, 489–500. [Google Scholar] [CrossRef]
- Hayward, J.L.; Huang, Y.N.; Yost, C.K.; Hansen, L.T.; Lake, C.; Tong, A.; Jamieson, R.C. Lateral flow sand filters are effective for removal of antibiotic resistance genes from domestic wastewater. Water Res. 2019, 162, 482–491. [Google Scholar] [CrossRef]
- Yuan, L.; Ma, Q.; Zhao, J. Meta-omics and its application in biological wastewater treatment system. Acta Sci. Circumstantiae 2020, 40, 2690–2699. [Google Scholar]
- Yang, Y.; Li, B.; Zou, S.C.; Fang, H.H.P.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef]
- Yoo, K.; Yoo, H.; Lee, J.; Choi, E.J.; Park, J. Exploring the antibiotic resistome in activated sludge and anaerobic digestion sludge in an urban wastewater treatment plant via metagenomic analysis. J. Microbiol. 2020, 58, 123–130. [Google Scholar] [CrossRef]
- Strange, J.E.S.; Leekitcharoenphon, P.; Moller, F.D.; Aarestrup, F.M. Metagenomics analysis of bacteriophages and antimicrobial resistance from global urban sewage. Sci. Rep. 2021, 11, 1600. [Google Scholar] [CrossRef] [PubMed]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.M.; Zhao, D.Y.; Huang, R.; Cao, X.Y.; Zeng, J.; Yu, Z.B.; Hooker, K.V.; Hambright, K.D.; Wu, Q.L. Contrasting Network Features between Free-Living and Particle-Attached Bacterial Communities in Taihu Lake. Microb. Ecol. 2018, 76, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.F.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.Y.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Li, R.Q.; Li, Y.R.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [Green Version]
- Lawson, C.E.; Wu, S.; Bhattacharjee, A.S.; Hamilton, J.J.; McMahon, K.D.; Goel, R.; Noguera, D.R. Metabolic network analysis reveals microbial community interactions in anammox granules. Nat. Commun. 2017, 8, 15416. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.Y.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J.G. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, Y.H.; Oono, Y. Relational patterns of gene expression via non-metric multidimensional scaling analysis. Bioinformatics 2005, 21, 730–740. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Yang, Y.; Ma, L.P.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Schmitz, B.W.; Caton, K.; Zhang, B.; Zabaleta, J.; Garai, J.; Taylor, C.M.; Romanchishina, T.; Gerba, C.P.; Pepper, I.L.; et al. Assessing the spatial and temporal variability of bacterial communities in two Bardenpho wastewater treatment systems via Illumina MiSeq sequencing. Sci. Total Environ. 2019, 657, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.G.; Li, B.X.; Li, N.; Sardar, M.F.; Song, T.T.; Zhu, C.X.; Lv, X.W.; Li, H.N. Effects of UV disinfection on phenotypes and genotypes of antibiotic-resistant bacteria in secondary effluent from a municipal wastewater treatment plant. Water Res. 2019, 157, 546–554. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, T. Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ. Sci. Technol. 2013, 47, 5433–5441. [Google Scholar] [CrossRef]
- Kummerer, K. Resistance in the environment. J. Antimicrob. Chemother. 2004, 54, 311–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paton, R.; Miles, R.S.; Hood, J.; Amyes, S.G.B.; Miles, R.S.; Amyes, S.G.B. ARI 1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 1993, 2, 81–87. [Google Scholar] [CrossRef]
- Lyytikäinen, O.; Köljalg, S.; Härmä, M.; Vuopio-Varkila, J. Outbreak caused by two multi-resistant Acinetobacter baumannii clones in a burns unit: Emergence of resistance to imipenem. J. Hosp. Infection 1995, 31, 41–54. [Google Scholar] [CrossRef]
- Ye, L.; Zhang, T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2013, 97, 2681–2690. [Google Scholar] [CrossRef] [Green Version]
- Ju, F.; Xia, Y.; Guo, F.; Wang, Z.P.; Zhang, T. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ. Microbiol. 2014, 16, 2421–2432. [Google Scholar] [CrossRef]
- Ju, F.; Zhang, T. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISME J. 2015, 9, 683–695. [Google Scholar] [CrossRef]
- Wu, L.W.; Ning, D.L.; Zhang, B.; Li, Y.; Zhang, P.; Shan, X.Y.; Zhang, Q.T.; Brown, M.R.; Li, Z.X.; Van Nostrand, J.D.Y.; et al. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 2019, 4, 1183–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef] [Green Version]
- Otto, M.; Götz, F. ABC transporters of staphylococci. Res. Microbiol. 2001, 152, 351–356. [Google Scholar] [CrossRef]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421–422, 173–183. [Google Scholar] [CrossRef]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef]
- Zhang, X.X.; Zhang, T.; Fang, H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Leila, H.; Shahriar, S.; Mohammad, D.; Reyhaneh, J. Molecular analysis of PmrA and PmrB genes in colistin-resistant Pseudomonas aeruginosa strains via PCR method. Pak. J. Pharm. Sci. 2019, 32, 1175–1177. [Google Scholar]
- Murray, S.R.; Ernst, R.K.; Bermudes, D.; Miller, S.I.; Low, K.B. PmrA(Con) confers pmrHFIJKL-dependent EGTA and polymyxin resistance on msbB Salmonella by decorating lipid A with phosphoethanolamine. J. Bacteriol. 2007, 189, 5161–5169. [Google Scholar] [CrossRef] [Green Version]
- Gotkowska-Płachta, A.; Filipkowska, Z.; Korzeniewska, E.; Janczukowicz, W.; Dixon, B.; Gołaś, I.; Szwalgin, D. Airborne Microorganisms Emitted from Wastewater Treatment Plant Treating Domestic Wastewater and Meat Processing Industry Wastes. CLEAN-Soil Air Water 2013, 41, 429–436. [Google Scholar] [CrossRef]
- Elena, E.; Aurora, B.; Miguel, E.; Ramón, G. Isolation of Salmonella serotypes in wastewater and effluent: Effect of treatment and potential risk. Int. J. Hyg. Environ. Health 2006, 209, 103–107. [Google Scholar]
- Bruning, J.B.; Murillo, A.C.; Chacon, O.; Barletta, R.G.; Sacchettini, J.C. Structure of the Mycobacterium tuberculosis D-alanine:D-alanine ligase, a target of the antituberculosis drug D-cycloserine. Antimicrob. Agents Chemother. 2011, 55, 291–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.Y.; Barletta, R.G. Roles of Mycobacterium smegmatis D-alanine:D-alanine ligase and D-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor D-cycloserine. Antimicrob. Agents Chemother. 2003, 47, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halouska, S.; Fenton, R.J.; Zinniel, D.K.; Marshall, D.D.; Barletta, R.G.; Powers, R. Metabolomics Analysis Identifies d-Alanine-d-Alanine Ligase as the Primary Lethal Target of d-Cycloserine in Mycobacteria. J. Proteome Res. 2014, 13, 1065–1076. [Google Scholar] [CrossRef] [Green Version]

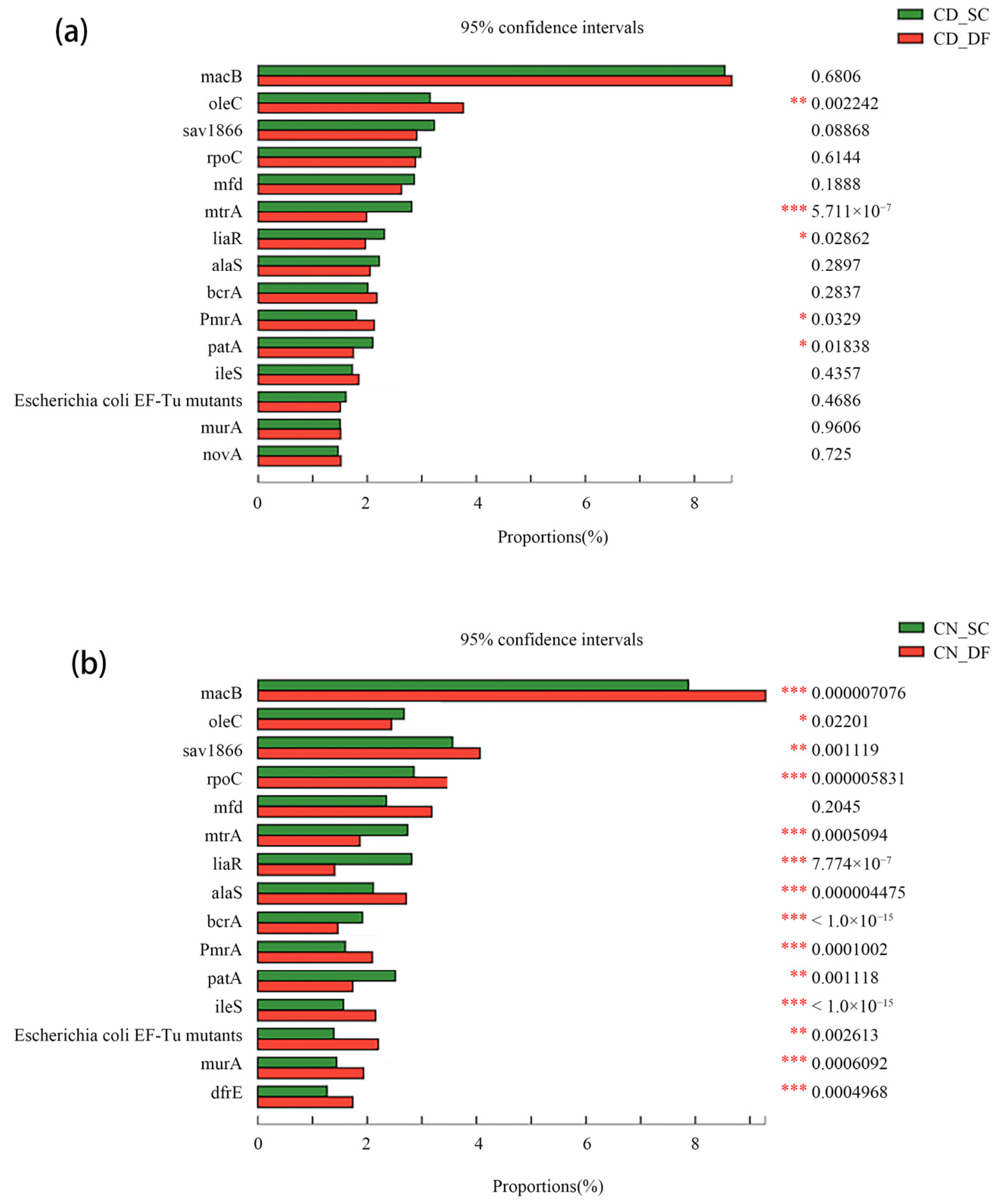

| ARGs Name | SC1 | DF1 | SC2 | DF2 |
|---|---|---|---|---|
| PmrA | 1.8% | 2.1% | 1.6% | 2.1% |
| mtrA | 2.8% | 2.0% | 2.7% | 1.9% |
| liaR | 2.3% | 2.0% | 2.8% | 1.4% |
| patA | 2.1% | 1.7% | 2.5% | 1.7% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Li, B.; Zhao, J.; Tian, Y.; Qiu, Y. Occurrence and Distribution of Antibiotic Resistance Genes in Municipal Wastewater Treatment Plants with D-Type Filters. Water 2021, 13, 3398. https://doi.org/10.3390/w13233398
Wang H, Li B, Zhao J, Tian Y, Qiu Y. Occurrence and Distribution of Antibiotic Resistance Genes in Municipal Wastewater Treatment Plants with D-Type Filters. Water. 2021; 13(23):3398. https://doi.org/10.3390/w13233398
Chicago/Turabian StyleWang, Haoze, Bing Li, Jiaheng Zhao, Yongjing Tian, and Yong Qiu. 2021. "Occurrence and Distribution of Antibiotic Resistance Genes in Municipal Wastewater Treatment Plants with D-Type Filters" Water 13, no. 23: 3398. https://doi.org/10.3390/w13233398






