Antibiotic Resistance Genes and Potentially Pathogenic Bacteria in the Central Adriatic Sea: Are They Connected to Urban Wastewater Inputs?
Abstract
:1. Introduction
2. Materials and Methods
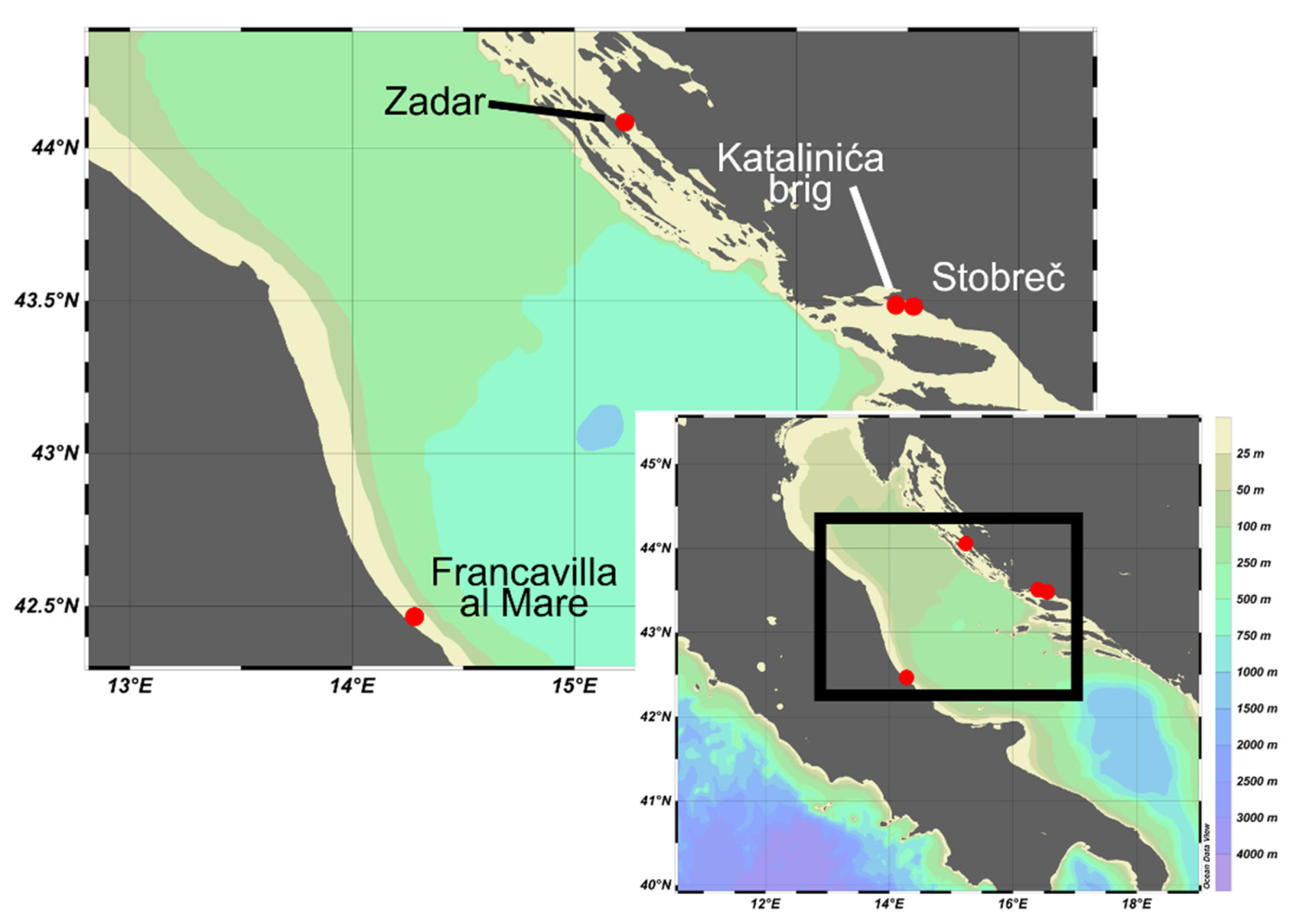
2.1. Study Area and Sampling
2.2. Microbiological Analyses
2.3. DNA Extraction
2.4. Microbial Community Characterization by High-Throughput Amplicon Sequencing
2.5. Quantification of Antibiotic Resistance Genes (ARGs)
2.6. Statistical Analyses
3. Results
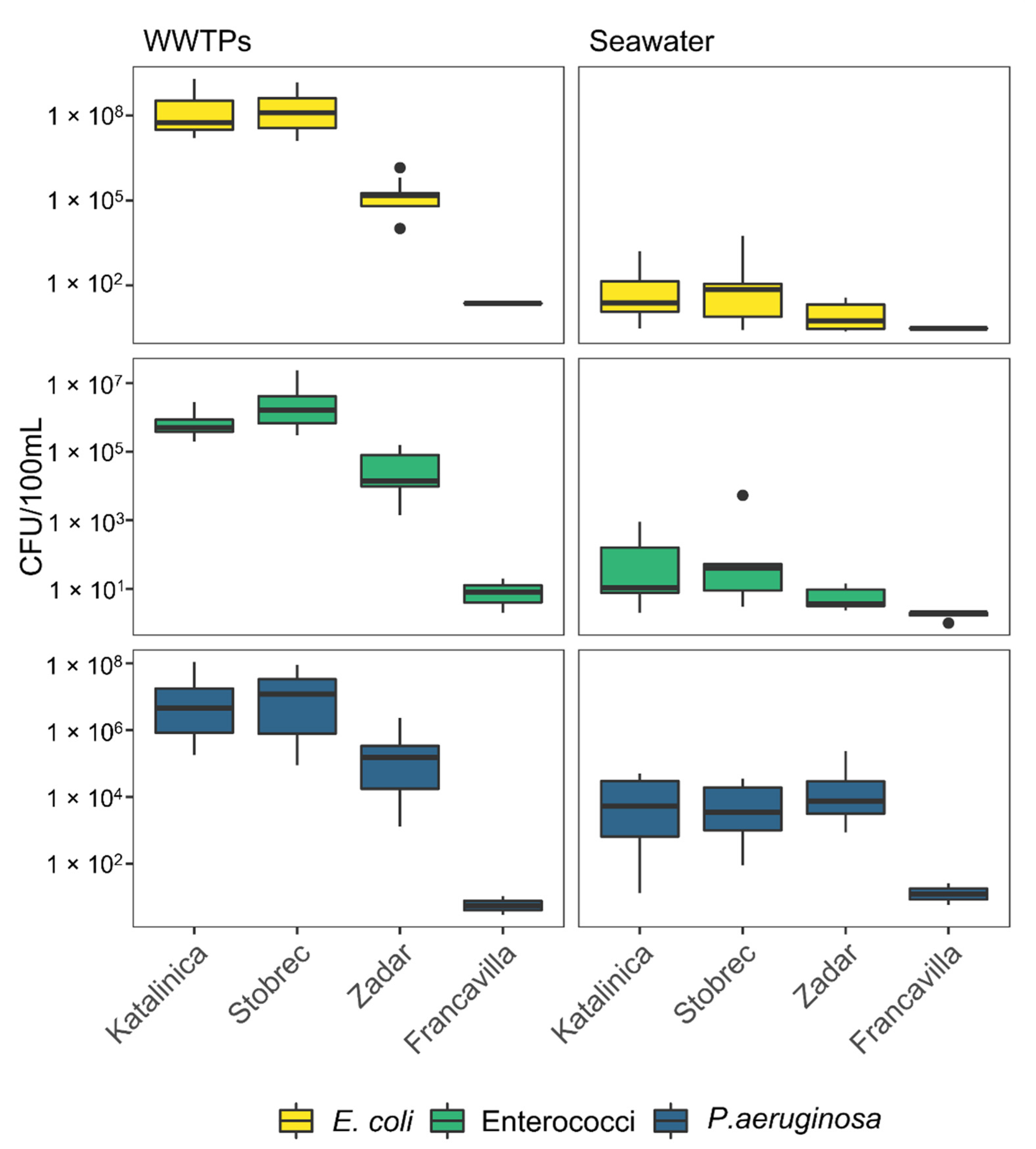
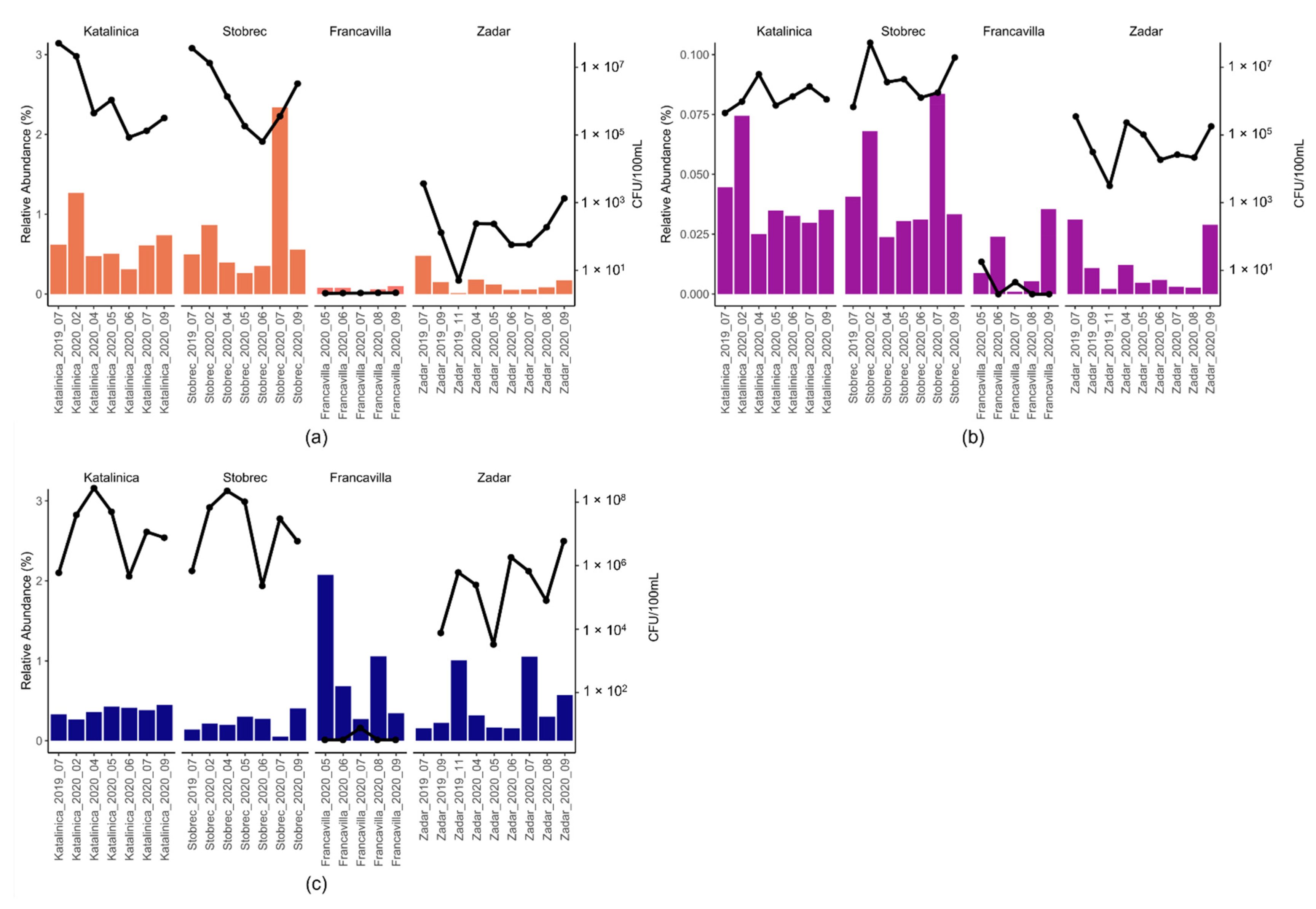
3.1. Enumeration of FIB and P. aeruginosa as Assessed by Microbiological Analyses
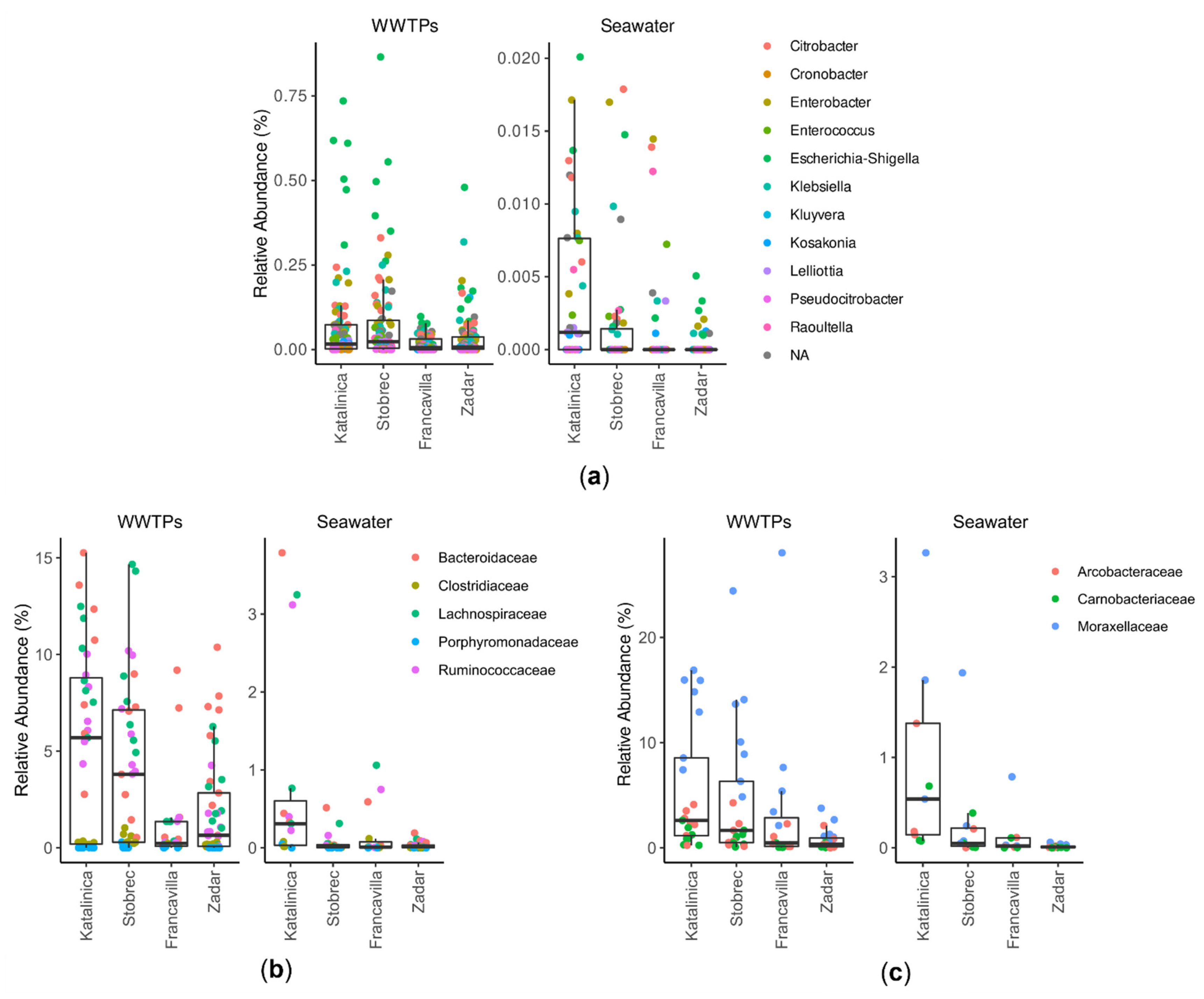
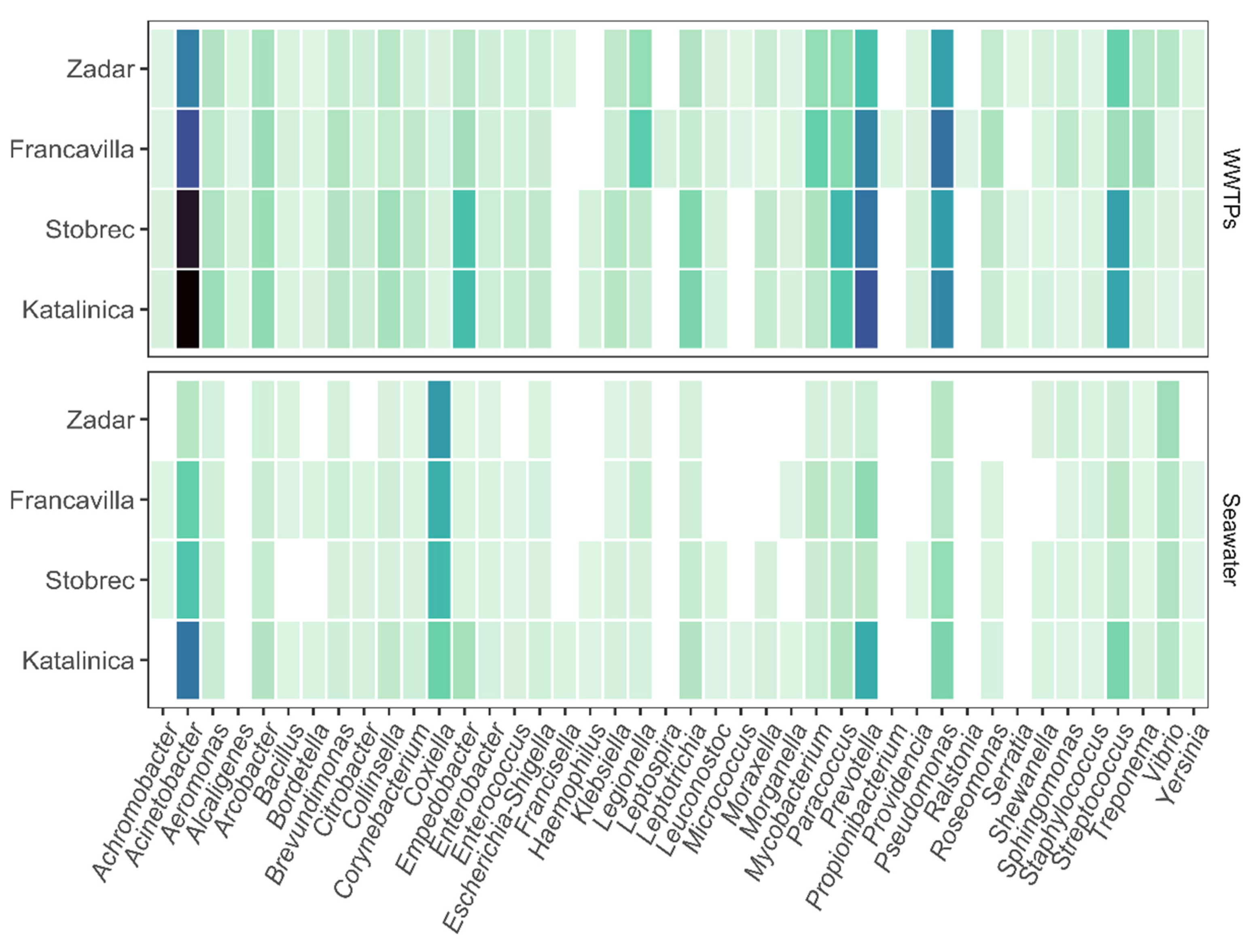
3.2. Bacterial Community Structure
3.3. Effect of WWTPs on Marine Community Structure
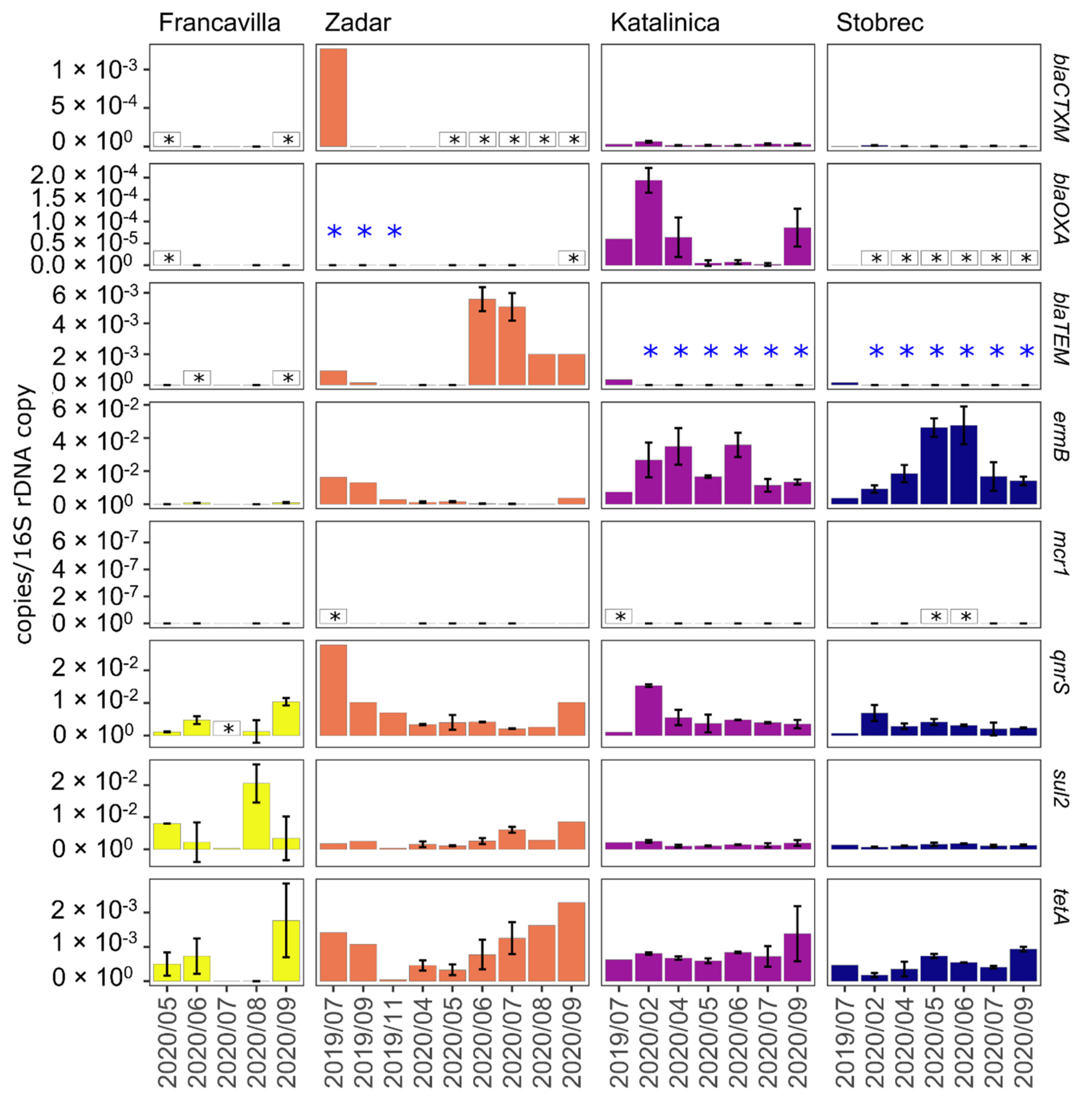
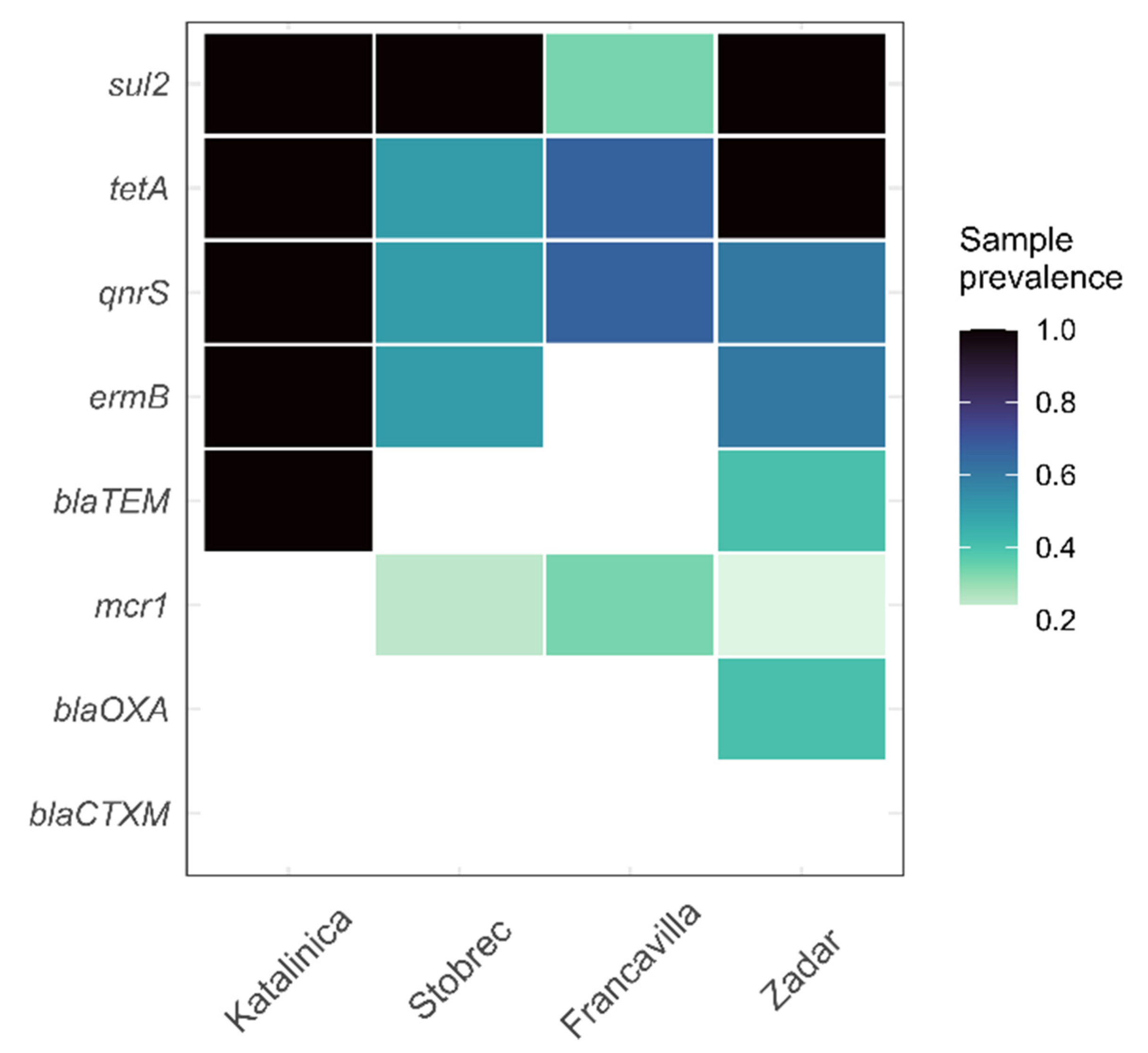
3.4. ARGs in Treated Sewage and Receiving Marine Water
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medlicott, K.; Wester, A.; Gordon, B.; Montgomery, M.; Tayler, E.; Sutherland, D.; Schmoll, O.; De-Souza, M.; Koo-Oshima, S.; Da-Balogh, K.; et al. Technical Brief on Water, Sanitation, Hygiene and Wastewater Management to Prevent Infections and Reduce the Spread of Antimicrobial Resistance. WHO/FAO/OIE Recommendation Report. 2020. Available online: https://www.who.int/publications/i/item/9789240006416 (accessed on 29 July 2021).
- Bruschi, A.; Lisi, I.; De Angelis, R.; Querin, S.; Cossarini, G.; Di Biagio, V.; Salon, S.; Solidoro, C.; Fassina, D.; Ancona, S.; et al. Indexes for the Assessment of Bacterial Pollution in Bathing Waters from Point Sources: The Northern Adriatic Sea CADEAU Service. J. Env. Manag. 2021, 293, 112878. [Google Scholar] [CrossRef]
- Nogales, B.; Lanfranconi, M.P.; Piña-Villalonga, J.M.; Bosch, R. Anthropogenic Perturbations in Marine Microbial Communities. FEMS Microbiol. Rev. 2011, 35, 275–298. [Google Scholar] [CrossRef] [Green Version]
- European Environment Agency. Towards a Cleaner Mediterranean: A Decade of Progress. Joint Map Report. In EEA Reports; Publications Office of the European Union: Copenhagen, Denmark, 2020. [Google Scholar]
- Court of Justice of the European Union. Sentenza Della Corte. Inadempimento Di Uno Stato—Raccolta E Trattamento Delle Acque Reflue Urbane—Direttiva 91/271/CEE—Articoli 3, 4 e 10. Judgment of the Court N. ECLI:EU:C:2018:358, 31 May 2018. Available on line: https://eur-lex.europa.eu/legal-content/IT/TXT/?uri=CELEX%3A62017CJ0251 (accessed on 26 September 2021).
- Paliaga, P.; Korlević, M.; Ivančić, I.; Najdek, M. Limited Influence of Primary Treated Sewage Waters on Bacterial Abundance, Production and Community Composition in Coastal Seawaters. Mar. Environ. Res. 2017, 131, 215–226. [Google Scholar] [CrossRef]
- Di Cesare, A.; Corno, G.; Manaia, C.M.; Rizzo, L. Impact of Disinfection Processes on Bacterial Community in Urban Wastewater: Should We Rethink Microbial Assessment Methods? J. Environ. Chem. Eng. 2020, 8, 104393. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic Resistance Genes Identified in Wastewater Treatment Plant Systems—A Review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, L.; Wojnárovits, L.; Takács, E. Occurrence and Fate of Antibiotics, Antibiotic Resistant Genes (ARGs) and Antibiotic Resistant Bacteria (ARB) in Municipal Wastewater Treatment Plant: An Overview. Sci. Total Environ. 2020, 744, 140997. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röderet, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global Monitoring of Antimicrobial Resistance Based on Metagenomics Analyses of Urban Sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Finley, R.L.; Collignon, P.; Larsson, D.G.J.; Mcewen, S.A.; Li, X.Z.; Gaze, W.H.; Reid-Smithet, R.; Timinounial, M.; Graham, D.W.; Topp, E. The Scourge of Antibiotic Resistance: The Important Role of the Environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, R.J.; McClary, J.S. The Flux and Impact of Wastewater Infrastructure Microorganisms on Human and Ecosystem Health. Curr. Opin. Biotechnol. 2019, 57, 145–150. [Google Scholar] [CrossRef]
- Corno, G.; Yang, Y.; Eckert, E.M.; Fontaneto, D.; Fiorentino, A.; Galafassi, S.; Zhang, T.; Di Cesare, A. Effluents of Wastewater Treatment Plants Promote the Rapid Stabilization of the Antibiotic Resistome in Receiving Freshwater Bodies. Water Res. 2019, 158, 72–81. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupo, A.; Coyne, S.; Berendonk, T.U. Origin and Evolution of Antibiotic Resistance: The Common Mechanisms of Emergence and Spread in Water Bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, R.J.; Hesse, E.; Dowling, A.J.; Coyle, N.M.; Feil, E.J.; Gaze, W.H.; Vos, M. Using the Wax Moth Larva Galleria Mellonella Infection Model to Detect Emerging Bacterial Pathogens. PeerJ 2019, 6, e6150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mughini-Gras, L.; Dorado-García, A.; van Duijkeren, E.; van den Bunt, G.; Dierikx, C.M.; Bonten, M.J.; Bootsma, M.C.; Schmitt, H.; Hald, T.; Evers, E.G.; et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet. Health 2019, 3, e357–e369. [Google Scholar] [CrossRef] [Green Version]
- Cabral-Oliveira, J.; Pratas, J.; Mendes, S.; Pardal, M.A. Trace Elements in Edible Rocky Shore Species: Effect of Sewage Discharges and Human Health Risk Implications. Hum. Ecol. Risk Assess. Int. J. 2015, 21, 135–145. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Singer, A.; Ukoumunne, O.C.; Gaze, W.H.; Garside, R. Is It Safe to Go back into the Water? A Systematic Review and Meta-Analysis of the Risk of Acquiring Infections from Recreational Exposure to Seawater. Int. J. Epidemiol. 2018, 47, 572–586. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Garside, R.; Ukoumunne, O.C.; Gaze, W.H. A Cross-Sectional Study on the Prevalence of Illness in Coastal Bathers Compared to Non-Bathers in England and Wales: Findings from the Beach User Health Survey. Water Res. 2020, 176, 115700. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Zhang, L.; Balfour, A.J.; Garside, R.; Hawkey, P.M.; Murray, A.K.; Ukoumunne, O.C.; Gaze, W.H. Exposure to and Colonisation by Antibi-Otic-Resistant E. coli in UK Coastal Water Users: Environmental Surveillance, Exposure Assessment, and Epidemiological Study (Beach Bum Survey). Environ. Int. 2018, 114, 326–333. [Google Scholar] [CrossRef]
- Borja, A.; White, M.P.; Berdalet, E.; Bock, N.; Eatock, C.; Kristensen, P.; Leonard, A.; Lloret, J.; Pahl, S.; Parga, M.; et al. Moving toward an Agenda on Ocean Health and Human Health in Europe. Front. Mar. Sci. 2020, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Eckert, E.M.; Di Cesare, A.; Malki, L.S.; Villiger, J.; Pernthaler, J.; Callieri, C.; Bertoni, R.; Corno, G. Seasonality of the Antibiotic Resistance Gene Bla CTX-M in Temperate Lake Maggiore. Hydrobiologia 2019, 843, 143–153. [Google Scholar] [CrossRef]
- WHO. Guidelines for Safe Recreational Water Environments: Coastal and Fresh Waters; World Health Organization: Geneva, Switzerland, 2003; Volume 1. [Google Scholar]
- European Parliament. Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 Concerning the Management of Bathing Water Quality and Repealing Directive 76/160/EEC. 4.3.2006(L64/37). Available online: https://op.europa.eu/en/publication-detail/-/publication/c137dbd0-21c5-492f-81ad-736a1295fc68/language-en (accessed on 26 September 2021).
- Mohammed, R.L.; Echeverry, A.; Stinson, C.M.; Green, M.; Bonilla, T.D.; Hartz, A.; McCorquodale, D.S.; Rogerson, A.; Esiobu, N. Survival Trends of Staphylococcus aureus, Pseudomonas Aeruginosa, and Clostridium Perfringens in a Sandy South Florida Beach. Mar. Pollut. Bull. 2012, 64, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Marino, F.J.; Morinigo, M.A.; Martinez-Manzanares, E.; Borrego, J.J. Microbiological-Epidemiological Study of Selected Marine Beaches in Malaga (Spain). Water Sci. Technol. 1995, 31, 5–9. [Google Scholar] [CrossRef]
- Pougnet, R.; Pougnet, L.; Allio, I.; Lucas, D.; Dewitte, J.D.; Loddé, B. Maritime Environment Health Risks Related to Pathogenic Microorganisms in Seawater. Int. Marit. Health 2018, 69, 35–45. [Google Scholar] [CrossRef]
- Di Cesare, A.; Pasquaroli, S.; Vignaroli, C.; Paroncini, P.; Luna, G.M.; Manso, E.; Biavasco, F. The Marine Environment as a Reservoir of Enterococci Carrying Resistance and Virulence Genes Strongly Associated with Clinical Strains. Environ. Microbiol. Rep. 2014, 6, 184–190. [Google Scholar] [CrossRef]
- Perini, L.; Quero, G.M.; Serrano García, E.; Luna, G.M. Distribution of Escherichia Coli in a Coastal Lagoon (Venice, Italy): Temporal Patterns, Genetic Diversity and the Role of Tidal Forcing. Water Res. 2015, 87, 155–165. [Google Scholar] [CrossRef]
- Luna, G.M.; Quero, G.M.; Perini, L. Next Generation Sequencing Reveals Distinct Fecal Pollution Signatures in Aquatic Sediments across Gradients of Anthropogenic Influence. Adv. Oceanogr. Limnol. 2016, 7, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Susmel, S.; Figueredo, F.; Aissa, S.B.; Scognamiglio, V.; Antonacci, A.; Girolametti, F.; Annibaldi, A.; Fonti, V.; Celussi, M.; Manna, V.; et al. A new cross-cutting vision for the quality of the Adriatic Sea under the development of the Interreg project AdSWiM. Transition from WasteWater Treatments (WWT) plant to Water Resource Recovery Facilities (WRRFs) in dialogue with Universities and Research. Water. in prep, in this issue.
- Kvesić, M.; Vojković, M.; Kekez, T.; Maravić, A.; Andričević, R. Spatial and Temporal Vertical Distribution of Chlorophyll in Relation to Submarine Wastewater Effluent Discharges. Water 2021, 13, 2016. [Google Scholar] [CrossRef]
- De Bentzmann, S.; Plésiat, P. The Pseudomonas Aeruginosa Opportunistic Pathogen and Human Infections. Environ. Microbiol. 2011, 13, 1655–1665. [Google Scholar] [CrossRef]
- López-Causapé, C.; Cabot, G.; del Barrio-Tofiño, E.; Oliver, A. The Versatile Mutational Resistome of Pseudomonas Aeruginosa. Front. Microbiol. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging Broad-Spectrum Resistance in Pseudomonas Aeruginosa and Acinetobacter Baumannii: Mechanisms and Epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celussi, M.; Quero, G.M.; Zoccarato, L.; Franzo, A.; Corinaldesi, C.; Rastelli, E.; Lo Martire, M.; Galand, P.E.; Ghiglione, J.F.; Chiggiato, J.; et al. Planktonic Prokaryote and Protist Communities in a Submarine Canyon System in the Ligurian Sea (NW Mediterranean). Prog. Oceanogr. 2018, 168, 210–221. [Google Scholar] [CrossRef]
- 16S Metagenomic Sequencing Library Preparation. 2013. Available online: https://emea.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 29 July 2021).
- Morgan, M.; Anders, S.; Lawrence, M.; Aboyoun, P.; Pages, H.; Gentleman, R. ShortRead: A Bioconductor Package for Input, Quality Assessment and Exploration of High-Throughput Sequence Data. Bioinformatics 2009, 25, 2607–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Im-Proved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. J. Stat. Softw. 2016, 77, 1–3. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Kandlikar, G.S.; Gold, Z.J.; Cowen, M.C.; Meyer, R.S.; Freise, A.; Kraft, N.J.B.; Moberg-Parker, J.; Sprague, J.; Kushner, D.J.; Curd, E.E. Ranacapa: An R Package and Shiny Web App to Explore Environmental DNA Data with Exploratory Statistics and Interactive Visualizations. F1000Research 2018, 7, 1734. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Di Cesare, A.; Eckert, E.M.; Teruggi, A.; Fontaneto, D.; Bertoni, R.; Callieri, C.; Corno, G. Constitutive Presence of Antibiotic Resistance Genes within the Bacterial Community of a Large Subalpine Lake. Mol. Ecol. 2015, 24, 3888–3900. [Google Scholar] [CrossRef]
- Di Cesare, A.; Luna, G.M.; Vignaroli, C.; Pasquaroli, S.; Tota, S.; Paroncini, P.; Biavasco, F. Aquaculture Can Promote the Presence and Spread of Antibiotic-Resistant Enterococci in Marine Sediments. PLoS ONE 2013, 8, e62838. [Google Scholar]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; Pascale, G.D.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Zhang, H.; Miranda, L.; Lin, S. Serious Overestimation in Quantitative PCR by Circular (Supercoiled) Plasmid Standard: Microalgal pcnaas the Model Gene. PLoS ONE 2010, 5, e9545. [Google Scholar] [CrossRef] [Green Version]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the Detection of Tetracycline Resistant Genes. Mol. Cell Probes. 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Pei, R.; Kim, S.C.; Carlson, K.H.; Pruden, A. Effect of River Landscape on the Sediment Concentrations of Antibiotics and Corre-sponding Antibiotic Resistance Genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- Marti, E.; Balcázar, J.L. Real-Time PCR Assays for Quantification of qnr Genes in Environmental Water Samples and Chicken Feces. Appl. Environ. Microbiol. 2013, 79, 1743–1745. [Google Scholar] [CrossRef] [Green Version]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of Antibiotic Resistance Genes and Bacterial Community Composition in a River Influenced by a Wastewater Treatment Plant. PLoS ONE 2013, 8, e78906. [Google Scholar]
- Bontron, S.; Poirel, L.; Nordmann, P. Real-Time PCR for Detection of Plasmid-Mediated Polymyxin Resistance (mcr-1) from Cultured Bacteria and Stools. J. Antimicrob. Chemother. 2016, 71, 2318–2320. [Google Scholar] [CrossRef] [Green Version]
- Bibbal, D.; Dupouy, V.; Ferré, J.P.; Toutain, P.L.; Fayet, O.; Prère, M.F.; Bousquet-Mélou, A. Impact of Three Ampicillin Dosage Regimens on Selection of Ampicillin Resistance in Enterobacteriaceae and Excretion of blaTEM Genes in Swine Feces. Appl. Environ. Microbiol. 2007, 73, 4785–4790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subirats, J.; Royo, E.; Balcázar, J.L.; Borrego, C.M. Real-Time PCR Assays for the Detection and Quantification of Carbapenemase Genes (bla KPC, bla NDM, and bla OXA-48) in Environmental Samples. Environ. Sci. Pollut. Res. 2017, 24, 6710–6714. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Di Cesare, A.; Fontaneto, D.; Doppelbauer, J.; Corno, G. Fitness and Recovery of Bacterial Communities and Antibiotic Resistance Genes in Urban Wastewaters Exposed to Classical Disinfection Treatments. Environ. Sci. Technol. 2016, 50, 10153–10161. [Google Scholar] [CrossRef] [PubMed]
- Zuur, A.F.; Ieno, E.N.; Elphick, C.S. A Protocol for Data Exploration to Avoid Common Statistical Problems. Methods Ecol. Evol. 2010, 1, 3–14. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S.P. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]
- Steinberger, A. Seq-Scripts Release v. 1.0. 2016. Available online: https://zenodo.org/record/1458243#.YZ4Po9BByUk (accessed on 29 July 2021).
- Newton, R.J.; Bootsma, M.J.; Morrison, H.G.; Sogin, M.L.; McLellan, S.L. A Microbial Signature Approach to Identify Fecal Pollution in the Waters Off an Urbanized Coast of Lake Michigan. Microb. Ecol. 2013, 65, 1011–1023. [Google Scholar] [CrossRef]
- WHO. Microbial Fact Sheets. In World Health Organization Guidelines for Drinking-Water Quality (WHO GDWQ); WHO: Geneva, Switzerland, 2011. [Google Scholar]
- Janda, J.M. Shewanella: A Marine Pathogen as an Emerging Cause of Human Disease. Clin. Microbiol. Newsl. 2014, 36, 25–29. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Bürgmann, H.; Frigon, D.; Gaze, W.H.; Manaia, C.M.; Pruden, A.; Singer, A.C.; Smets, B.F.; Zhang, T. Water and Sanitation: An Essential Battlefront in the War on Antimicrobial Resistance. FEMS Microbiol. Ecol. 2018, 94, fiy101. [Google Scholar] [CrossRef]
- Fiorentino, A.; Ferro, G.; Alferez, M.C.; Polo-López, M.I.; Fernández-Ibañez, P.; Rizzo, L. Inactivation and Regrowth of Multidrug Resistant Bacteria in Urban Wastewater after Disinfection by Solar-Driven and Chlorination Processes. J. Photochem. Photobiol. B Biol. 2015, 148, 43–50. [Google Scholar] [CrossRef]
- Lazarova, V.; Janex, M.L.; Fiksdal, L.; Oberg, C.; Barcina, I.; Pommepuy, M. Advanced Wastewater Disinfection Technologies: Short and Long Term Efficiency. Water Sci. Technol. 1998, 38, 109–117. [Google Scholar] [CrossRef]
- Wade, T.J.; Sams, E.A.; Beach, M.J.; Collier, S.A.; Dufour, A.P. The Incidence and Health Burden of Earaches Attributable to Recrea-Tional Swimming in Natural Waters: A Prospective Cohort Study. Environ. Health 2013, 12, 67. [Google Scholar] [CrossRef] [Green Version]
- Bonadonna, L.; Briancesco, R.; Coccia, A.M.; Semproni, M.; Stewardson, D. Occurrence of Potential Bacterial Pathogens in Coastal Areas of the Adriatic Sea. Environ Monit. Assess. 2002, 77, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, C.; Celussi, M.; Russell, H.; Del Negro, P. Phenotypic and Genetic Diversity of Coexisting Listonella Anguillarum, Vibrio Harveyi and Vibrio Chagassi Recovered from Skin Haemorrhages of Diseased Sand Smelt, Atherina Boyeri, in the Gulf of Trieste (NE Adriatic Sea). Lett. Appl. Microbiol. 2012, 54, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Bacosa, H.P.; Liu, Z.; Erdner, D.L. Natural Sunlight Shapes Crude Oil-Degrading Bacterial Communities in Northern Gulf of Mexico Surface Waters. Front. Microbiol. 2015, 6, 1325. [Google Scholar] [CrossRef] [Green Version]
- Weitz, J.S.; Stock, C.A.; Wilhelm, S.W.; Bourouiba, L.; Coleman, M.L.; Buchan, A.; Follows, M.J.; Fuhrman, J.A.; Jover, L.F.; Lennon, J.T.; et al. A Multitrophic Model to Quantify the Effects of Marine Viruses on Microbial Food Webs and Ecosystem Processes. ISME J. 2015, 9, 1352–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varela, A.R.; Manageiro, V.; Ferreira, E.; Guimarães, M.A.; da Costa, P.M.; Caniça, M.; Manaia, C.M. Molecular Evidence of the Close Relat-Edness of Clinical, Gull and Wastewater Isolates of Quinolone-Resistant Escherichia Coli. J. Glob. Antimicrob. Resist. 2015, 3, 286–289. [Google Scholar] [CrossRef]
- Abgottspon, H.; Nüesch-Inderbinen, M.T.; Zurfluh, K.; Althaus, D.; Hächler, H.; Stephan, R. Enterobacteriaceae with Extend-ed-Spectrum-and pAmpC-Type β-Lactamase-Encoding Genes Isolated from Freshwater Fish from Two Lakes in Switzerland. Antimicrob. Agents Chemother. 2014, 58, 2482–2484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Huang, L.; Méndez-García, C.; Kuang, J.; Hua, Z.; Liu, J.; Shu, W.S. Microbial Communities, Processes and Functions in Acid Mine Drainage Ecosystems. Curr. Opin. Biotechnol. 2016, 38, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhang, M.; Zhang, S.; Yu, X. Can Chlorination Co-Select Antibiotic-Resistance Genes? Chemosphere 2016, 156, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.T.; Yuan, Q.B.; Yang, J. Distinguishing Effects of Ultraviolet Exposure and Chlorination on the Horizontal Transfer of Antibiotic Resistance Genes in Municipal Wastewater. Environ. Sci. Technol. 2015, 49, 5771–5778. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, A.; Dai, T.; Li, F.; Xie, H.; Chen, L.; Wen, D. Cell-Free DNA: A Neglected SOURCE for Antibiotic Resistance Genes Spreading from WWTPs. Environ. Sci. Technol. 2018, 52, 248–257. [Google Scholar] [CrossRef]
- Di Cesare, A.; Pjevac, P.; Eckert, E.; Curkov, N.; Miko Šparica, M.; Corno, G.; Orlić, S. The Role of Metal Contamination in Shaping Microbial Communities in Heavily Polluted Marine Sediments. Environ. Pollut. 2020, 265, 114823. [Google Scholar] [CrossRef]
- Šamanić, I.; Kalinić, H.; Fredotović, Ž.; Dželalija, M.; Bungur, A.M.; Maravić, A. Bacteria Tolerant to Colistin in Coastal Marine Environment: Detection, Microbiome Diversity and Antibiotic Resistance genes’ Repertoire. Chemosphere 2021, 281, 130945. [Google Scholar] [CrossRef]
- Lozano-Leon, A.; Garcia-Omil, C.; Dalama, J.; Rodriguez-Souto, R.; Martinez-Urtaza, J.; Gonzalez-Escalona, N. Detection of Col-istin Resistance mcr-1 Gene in Salmonella Enterica Serovar Rissen Isolated from Mussels, Spain, 2012 to 2016. Eurosurveillance 2019, 24, 1900200. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P. Aquaculture, Exaptation, and the Origin of mcr-Positive Colistin Resistance. Antimicrob. Agents Chemother. 2018, 62, e01903-18. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonti, V.; Di Cesare, A.; Šangulin, J.; Del Negro, P.; Celussi, M. Antibiotic Resistance Genes and Potentially Pathogenic Bacteria in the Central Adriatic Sea: Are They Connected to Urban Wastewater Inputs? Water 2021, 13, 3335. https://doi.org/10.3390/w13233335
Fonti V, Di Cesare A, Šangulin J, Del Negro P, Celussi M. Antibiotic Resistance Genes and Potentially Pathogenic Bacteria in the Central Adriatic Sea: Are They Connected to Urban Wastewater Inputs? Water. 2021; 13(23):3335. https://doi.org/10.3390/w13233335
Chicago/Turabian StyleFonti, Viviana, Andrea Di Cesare, Jadranka Šangulin, Paola Del Negro, and Mauro Celussi. 2021. "Antibiotic Resistance Genes and Potentially Pathogenic Bacteria in the Central Adriatic Sea: Are They Connected to Urban Wastewater Inputs?" Water 13, no. 23: 3335. https://doi.org/10.3390/w13233335





