An Alternative to Laboratory Testing: Random Forest-Based Water Quality Prediction Framework for Inland and Nearshore Water Bodies
Abstract
:1. Introduction
- We explored the relationship between different water quality indicators and designed a water quality prediction framework, which uses machine learning methods to predict the target variable water quality indicators by the dependent water quality parameters
- We compared the performance of different kinds of machine learning methods for total nitrogen (TN) prediction in the inland river and the best performing model was selected to be the core algorithm of the prediction framework
- We discussed the feasibility of the proposed framework for water quality inversion with water surface reflection data acquired by remote sensing. The GEE platform was used to solve the problem of big data calculation and to map the salinity distribution of nearshore water bodies.
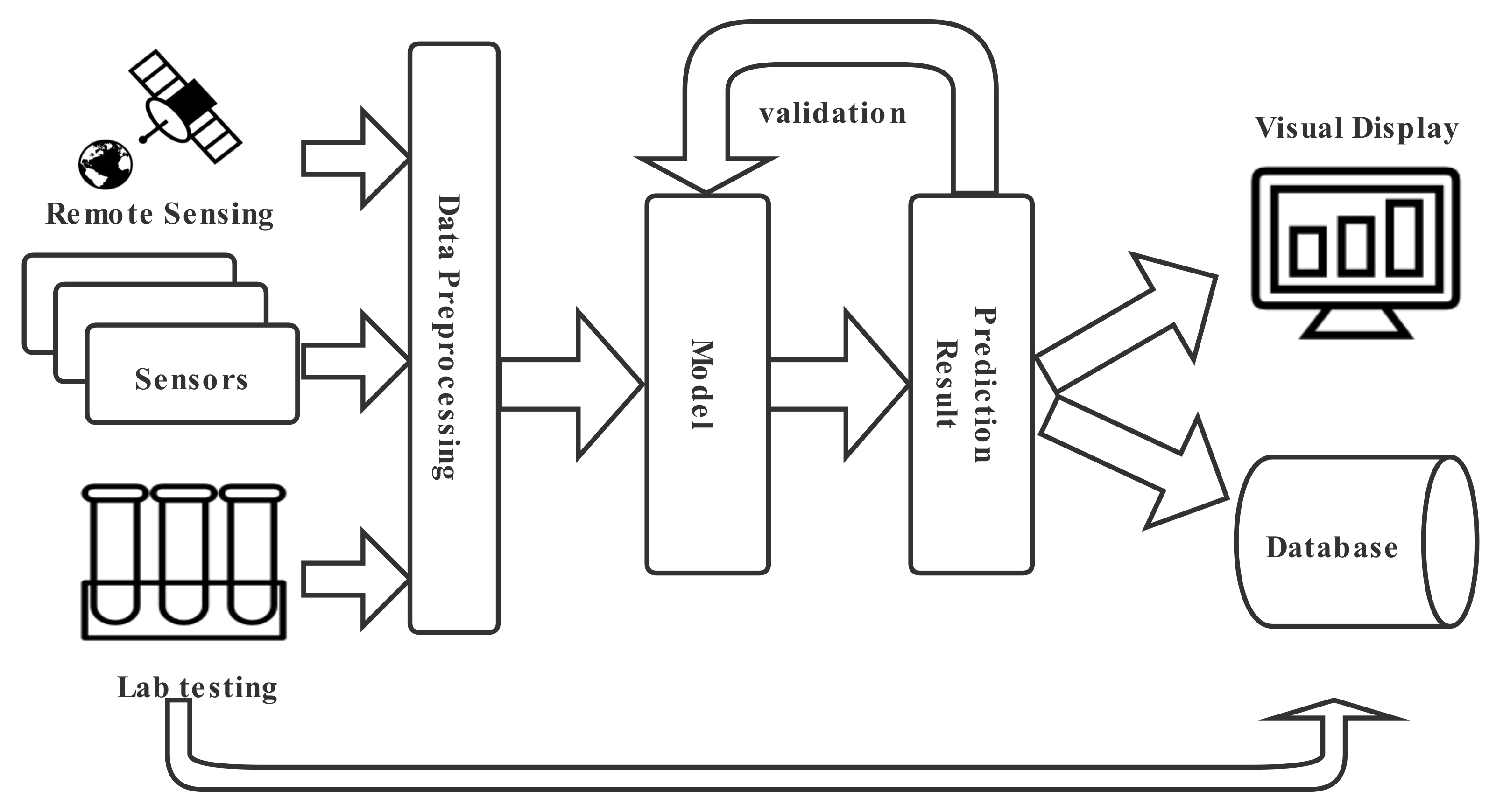
2. Framework for Inland and Nearshore Water Quality Prediction
2.1. Data Acquisition and Pre-Processing
2.2. Decision Tree and Random Forest Model
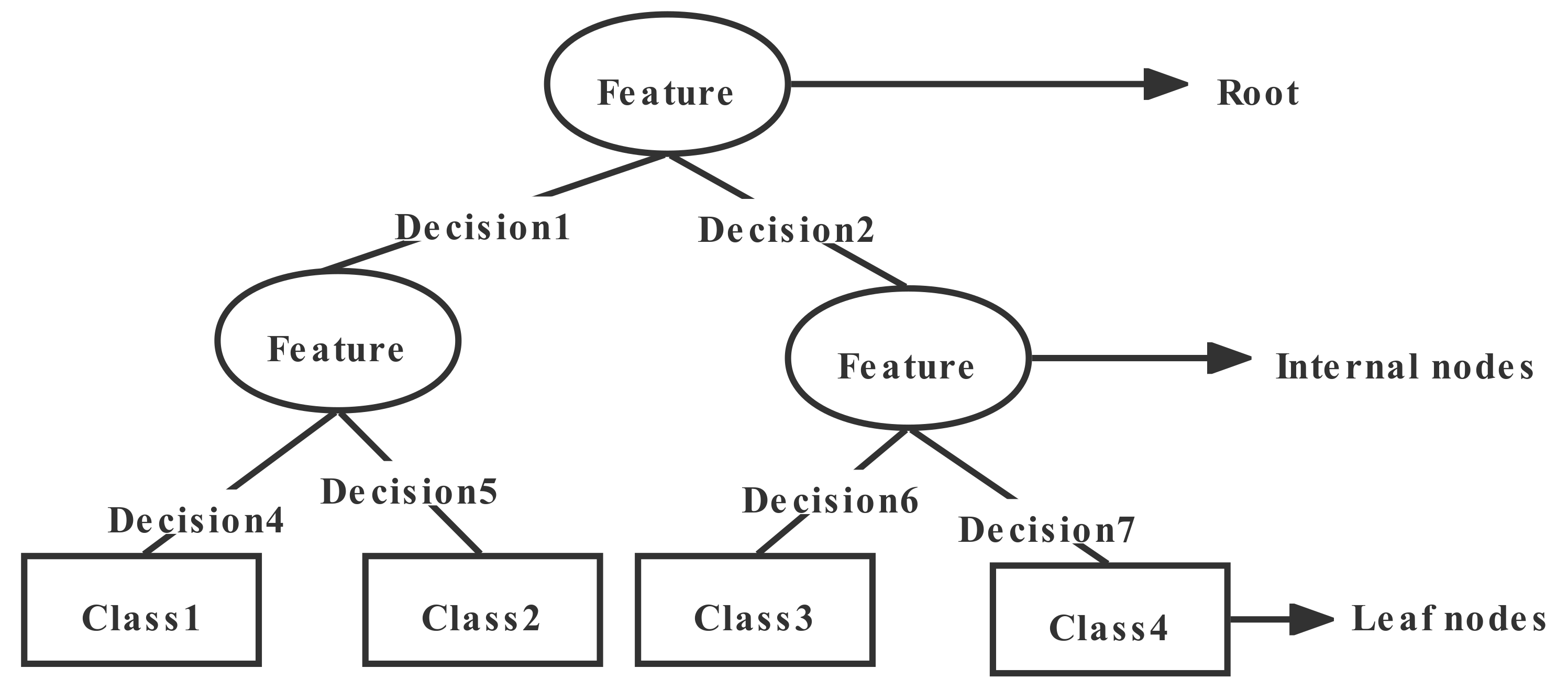
2.2.1. Decision Tree
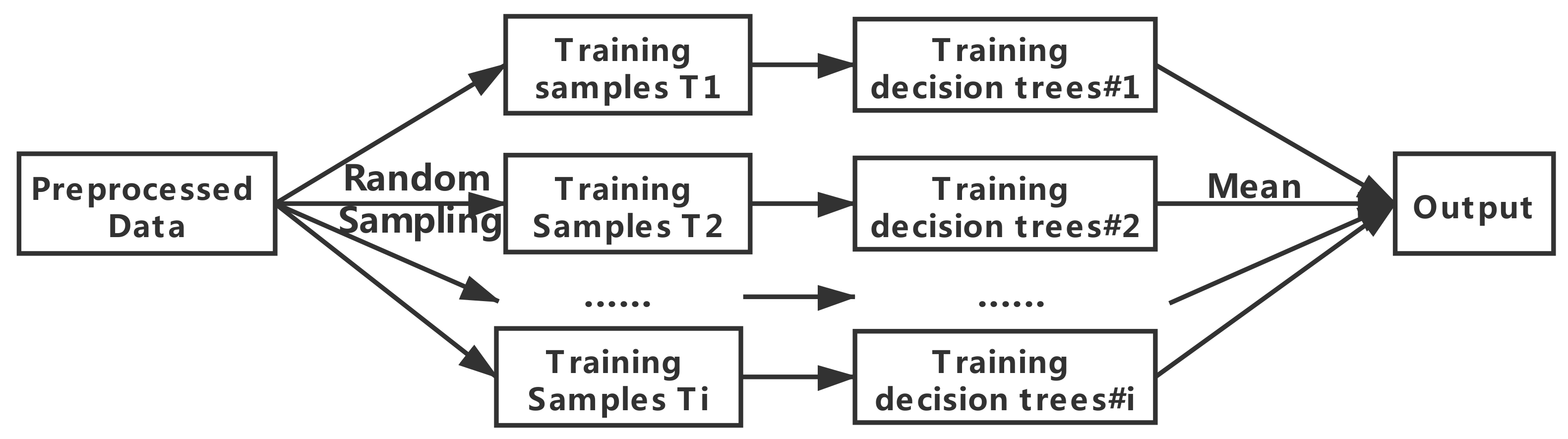
2.2.2. Random Forest
- Sampling: From the training set T, K sets of data sets are generated by Boostrasp sampling with put-back. Each set of data sets is divided into two kinds of sampled data and un-sampled data (out-of-bag data), and each data set will generate a decision tree by training.
- Growth: Each decision tree is trained by training data. At each sub-node, m features are randomly selected from M attributes, and the optimal features are selected based on the Gini metric for full branching growth until no more growth is possible, without pruning.
- Testing: Using the out-of-bag data to test the accuracy of the model. Because out-of-bag data are not involved in modeling, model effects and generalization capabilities can be tested to some extent. The prediction error of out-of-bag data is picked up to determine the best decision tree in the algorithm and re-modeled.
- Prediction: Using the determined model for new data and prediction, the average of all decision trees prediction results is the final output.
3. Case Studies of Inland and Nearshore Water Quality
3.1. Experiment Description and Settings
3.2. Baseline Methods
3.2.1. SVR
3.2.2. KNN
3.2.3. Ridge Regression
3.2.4. MLP
3.2.5. Gradient Boosted Regression Tree
3.2.6. Bagging
3.3. Evaluation Methodology
- MAE:
- MSE:
- RMSE:
- MAPE:
- :
3.4. Case Studies of Inland Water Quality
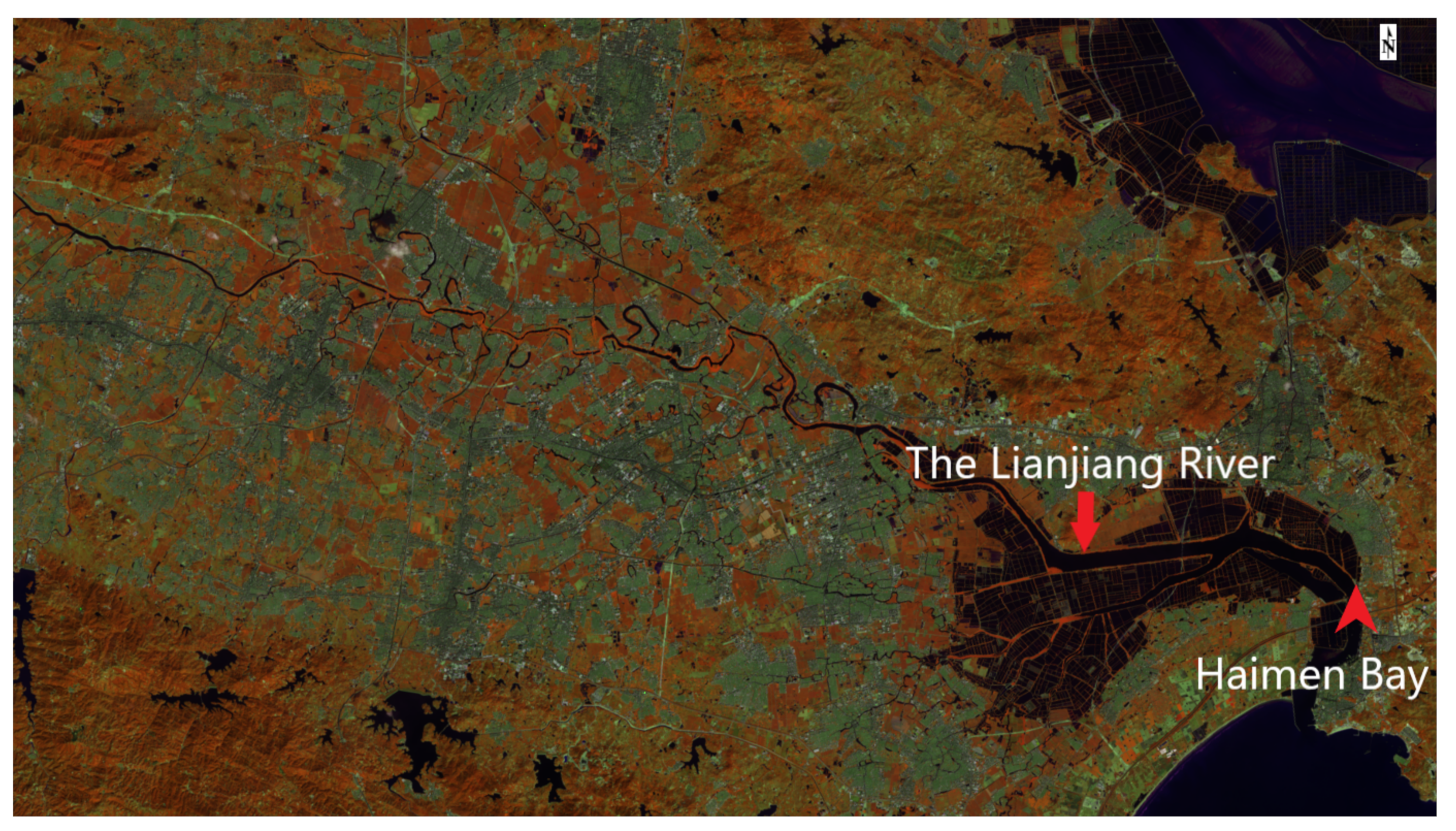
3.4.1. Study Area and Materials
3.4.2. Data Processing
- Data normalizationDifferent evaluation metrics have different magnitudes and units of magnitude [48], for example, the value of conductivity maybe hundreds of times higher than other observed values. Such differences may have an impact on the prediction results. To eliminate the influence of scale between indicators, the Z-score normalization method [49] is used to normalize the filtered data. The normalized data are scaled between 0 and 1, with z-score normalization method is as follows:where denotes the normalized value, X denotes the real monitored value, and represents the minimum value and the maximum value in the set of data.
- Correlation AnalysisTo find water parameters that are correlated with TN and can be used as independent variables in predicting the TN process, a correlation analysis was performed to extract possible relationships between the parameters. Pearson’s correlation coefficient was used to measure the correlation between water quality indicators. The formula is as follows:where is the correlation coefficient of the random variables X and Y, is the covariance of the random variables X and Y, E is the mathematical expectation or mean, D is the variance, and is the standard deviation.The correlation coefficients between other variables and total nitrogen obtained by Equation (11) are shown in Table 2. As can be seen in the table, the correlation coefficients of DO, pH, and Tud with TN are small, indicating the low probability of their linear correlation with TN. We also tested the variables with small correlations in the experiment and found that removing these variables would negatively affect the predicted results. One possible explanation is that there may be a nonlinear relationship between these variables and TN. For example, pH affects algal growth and thus changes TN concentrations, while algal growth affects DO concentrations [50]. Although the linear correlation between DO and TN is the lowest in the table, DO is directly related to the life and death of aquatic organisms. Proper DO helps these organisms to survive, while when algal overgrowth occurs, the DO of the water column decreases, organisms die, and microbial decomposition leads to higher TN in the water. Based on this consideration we retained and used these variables. TP was not used as an independent variable because the detection of TP is similar to TN, which requires laboratory analysis.
- Data ValidationAccording to the characteristics of machine learning algorithms, we randomly divide the pre-processed 1917 sets of data into training and testing sets, i.e., 90% of the data are used as training set to train the model and 10% are used as testing set to validate the model and calculate the metrics.
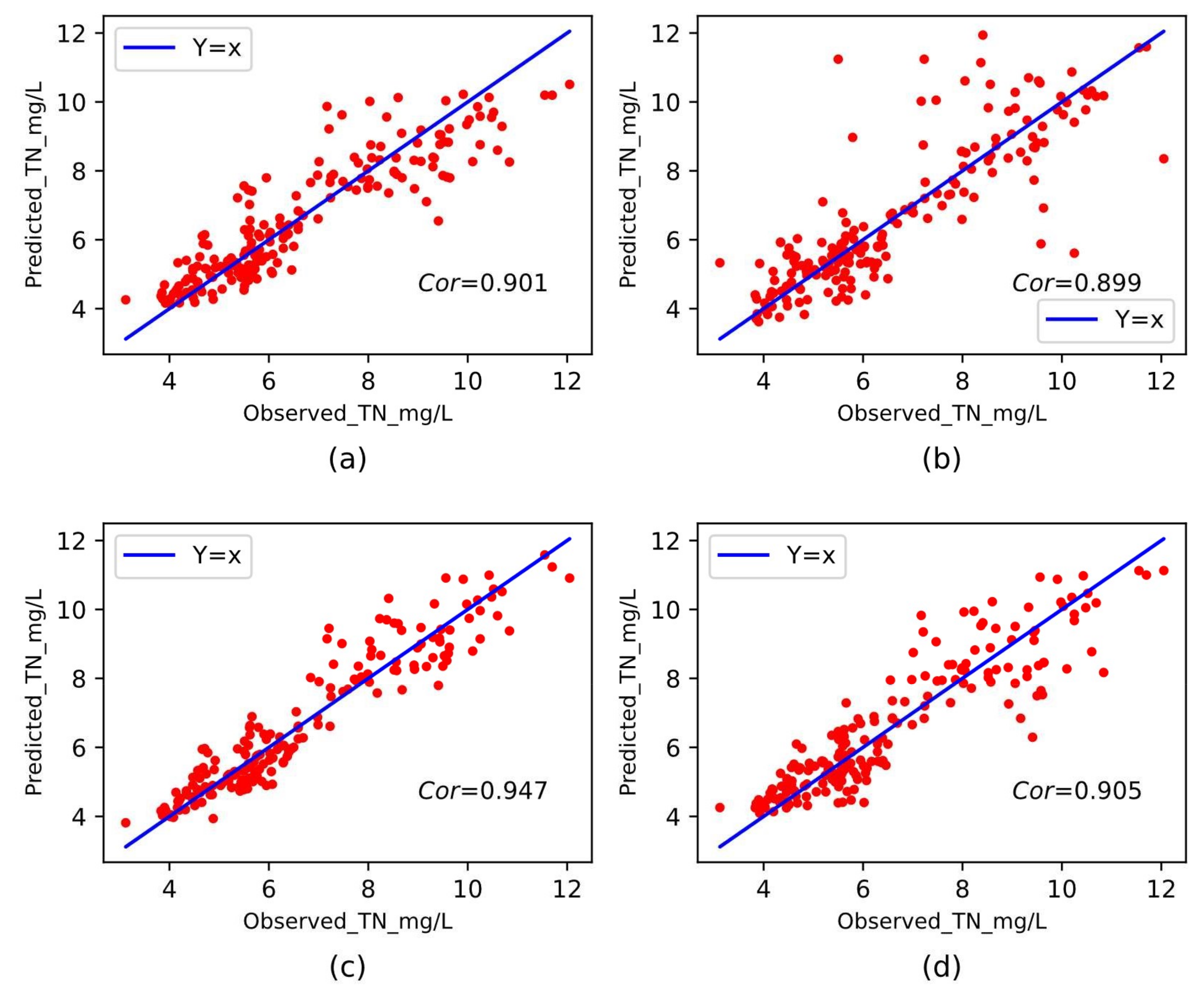
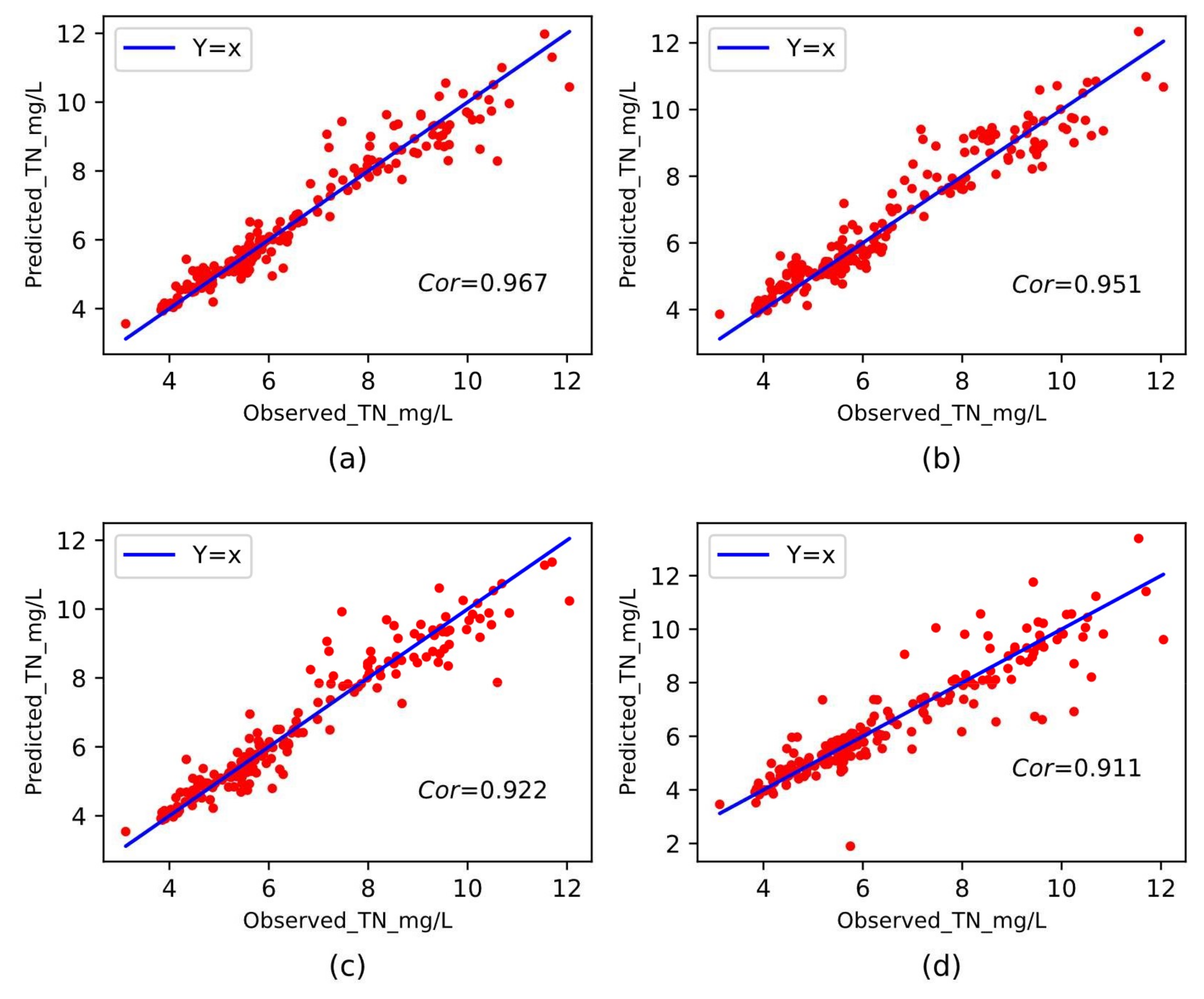
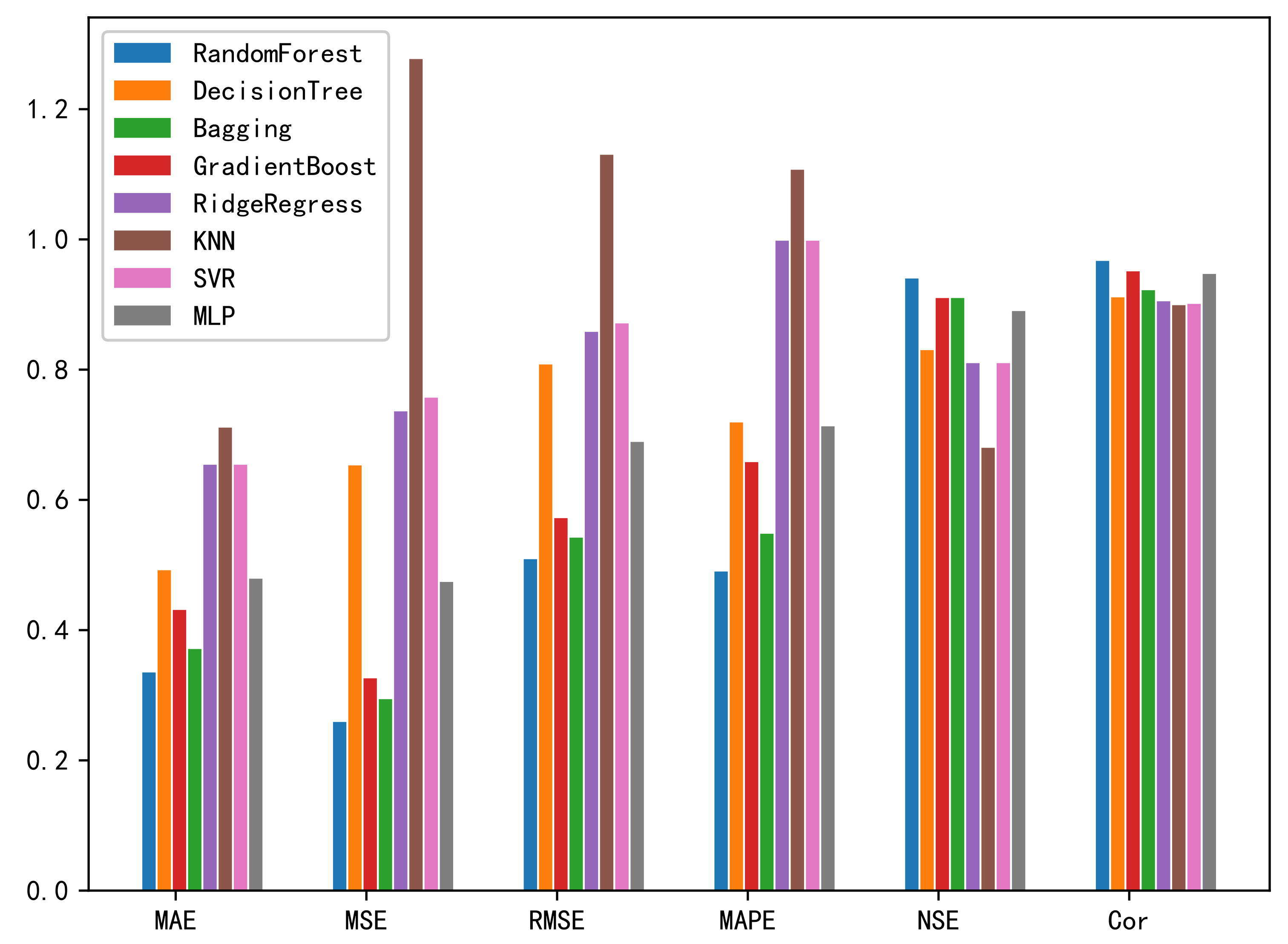
3.4.3. Performance Comparison with Different Baseline Methods
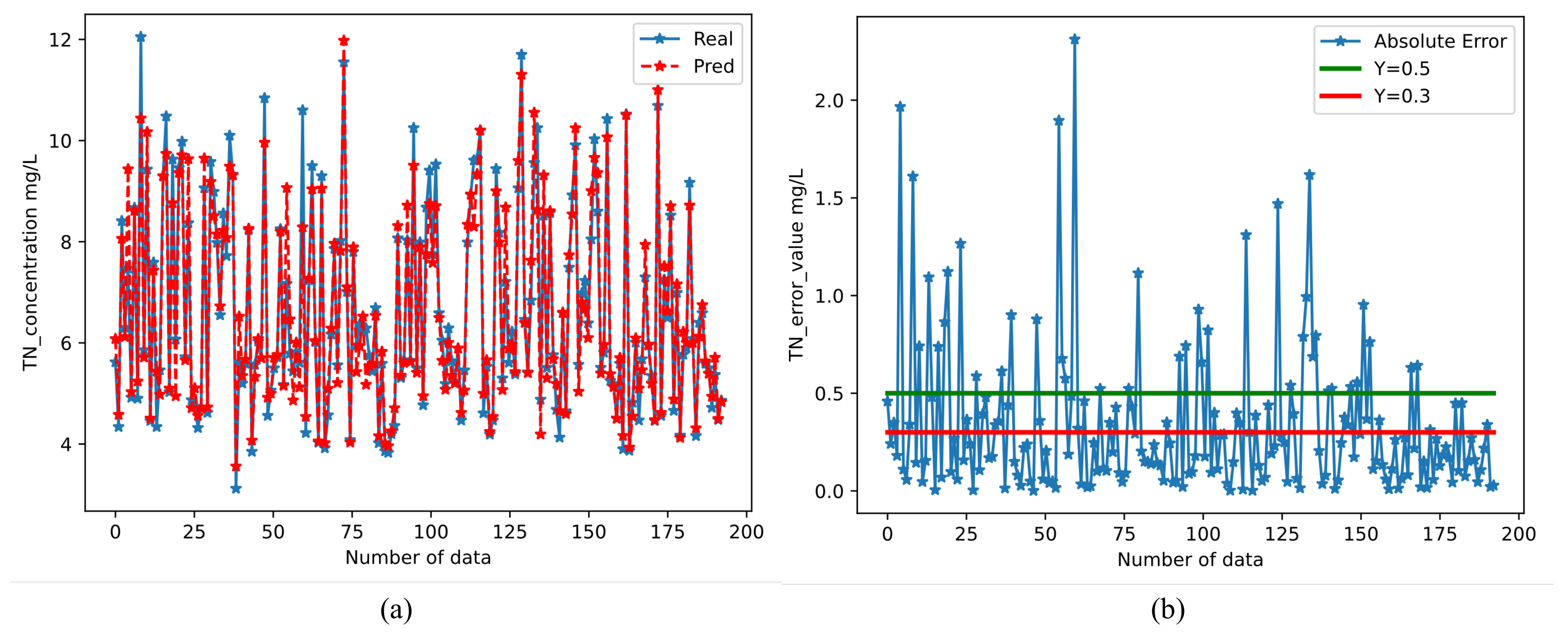
3.4.4. Prediction Results of The Framework for TN
3.5. Case Studies of Nearshore Water Quality
3.5.1. Data Description and Processing
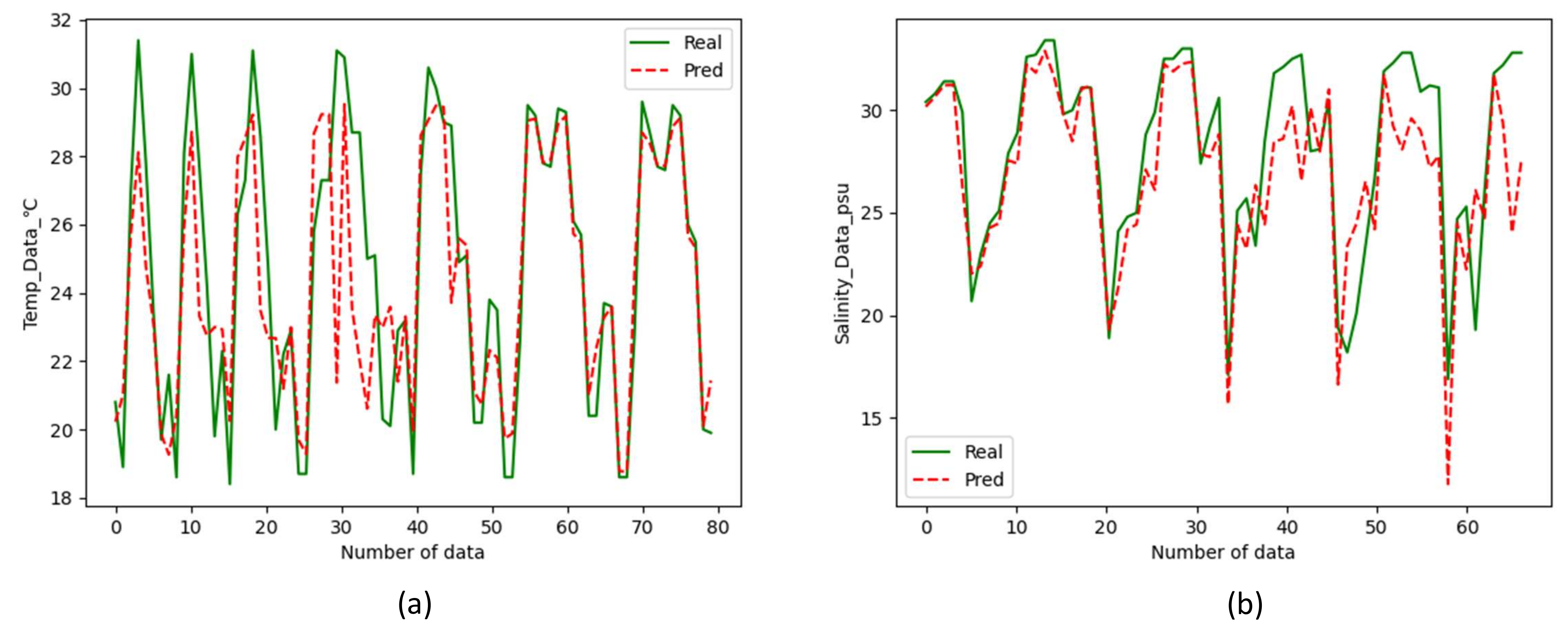
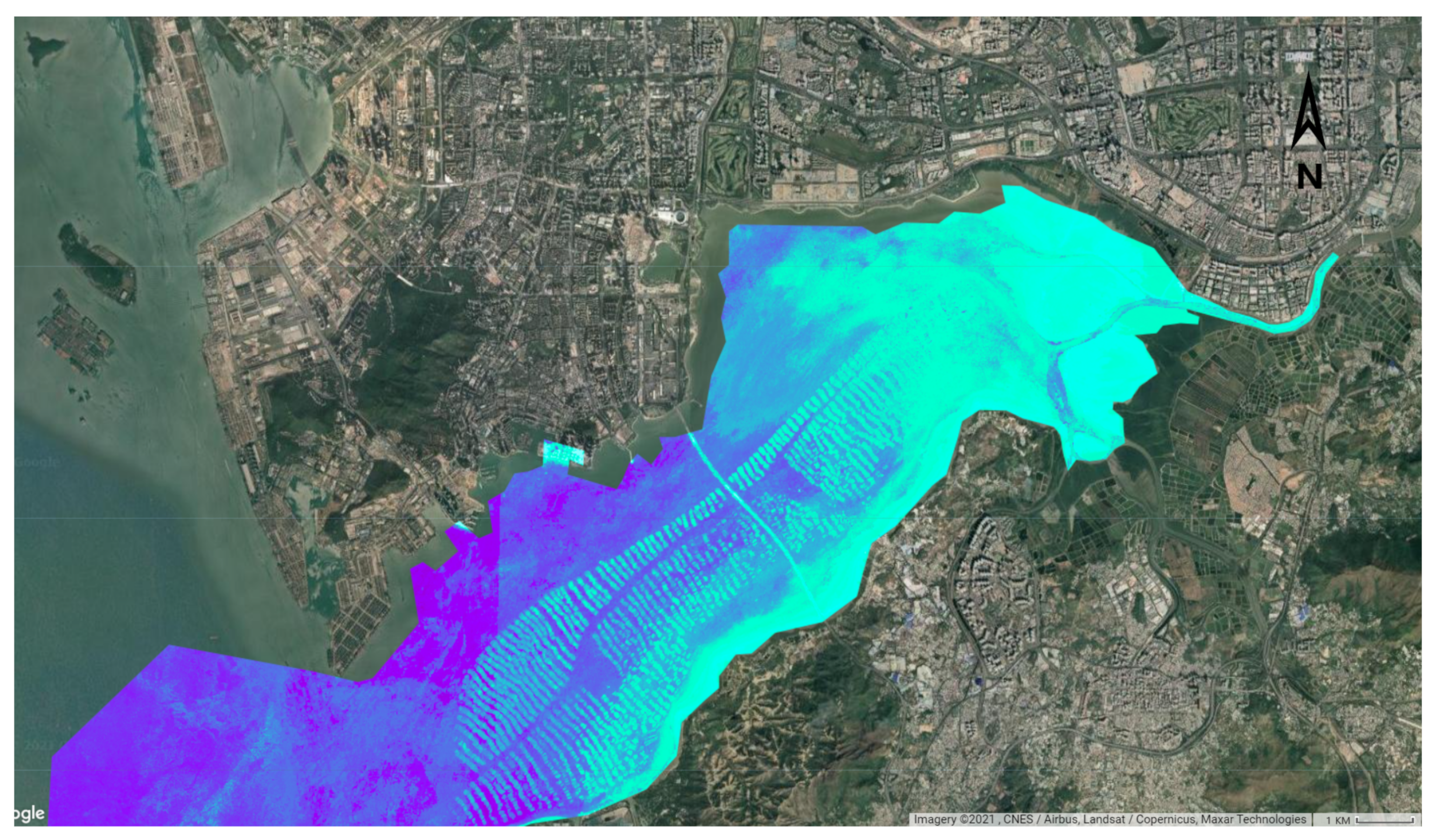
3.5.2. Results of Inversion for Nearshore Water Quality Using Sentinel-2 Data
3.6. Discussion
4. Conclusions
- Machine learning methods and neural network methods can effectively predict the TN in rivers(with an accuracy of 95.1%). Thus, the water quality prediction framework we designed can be used as a soft alternative to sensors in cases where monitoring requirements are less stringent. Research on rivers can provide a real-time and rapid prediction of water quality, which provides a reference basis for river water quality monitoring work, and also provides a decision aid for river management.
- Random forest-based water quality prediction framework can be applied to the inversion of ocean temperature (accuracy reaches 95.35%) and salinity(accuracy reaches 92.94%). Through the reflectance of the water body to light bands, the trained model can invert the water body without the help of water quality data.
- GEE platform is friendly for remote sensing calculation. GEE provides satellite data resources including Landsat series and Sentinel series, and product resources developed with these data. Coupled with its powerful computing power, it can easily solve the problem of large-scale remote sensing calculations.
5. Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Son, G.; Kim, D.; Kim, Y.D.; Lyu, S.; Kim, S. A Forecasting Method for Harmful Algal Bloom (HAB)-Prone Regions Allowing Preemptive Countermeasures Based Only on Acoustic Doppler Current Profiler Measurements in a Large River. Water 2020, 12, 3488. [Google Scholar] [CrossRef]
- Singh, J.; Yadav, P.; Pal, A.K.; Mishra, V. Water pollutants: Origin and status. In Sensors in Water Pollutants Monitoring: Role of Material; Springer: Berlin/Heidelberg, Germany, 2020; pp. 5–20. [Google Scholar]
- Jiang, J.; Tang, S.; Han, D.; Fu, G.; Solomatine, D.; Zheng, Y. A comprehensive review on the design and optimization of surface water quality monitoring networks. Environ. Model. Softw. 2020, 132, 104792. [Google Scholar] [CrossRef]
- Park, J.; Kim, K.T.; Lee, W.H. Recent advances in information and communications technology (ICT) and sensor technology for monitoring water quality. Water 2020, 12, 510. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhang, F.; Ge, X.; Zhang, X.; Chan, N.; Qi, Y. Measurement of total nitrogen concentration in surface water using hyperspectral band observation method. Water 2020, 12, 1842. [Google Scholar] [CrossRef]
- Di Trapani, A.; Corbari, C.; Mancini, M. Effect of the Three Gorges Dam on Total Suspended Sediments from MODIS and Landsat Satellite Data. Water 2020, 12, 3259. [Google Scholar] [CrossRef]
- Zhao, C.; Chen, L.; Zhong, G.; Wu, Q.; Liu, J.; Liu, X. A portable analytical system for rapid on-site determination of total nitrogen in water. Water Res. 2021, 202, 117410. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, D.; Yang, Q.; Pan, S.; Gai, Y.; Cheng, W.; Liu, X.; Tang, S. Variations of Water Transparency and Impact Factors in the Bohai and Yellow Seas from Satellite Observations. Remote Sens. 2021, 13, 514. [Google Scholar] [CrossRef]
- Ho, J.Y.; Afan, H.A.; El-Shafie, A.H.; Koting, S.B.; Mohd, N.S.; Jaafar, W.Z.B.; Sai, H.L.; Malek, M.A.; Ahmed, A.N.; Mohtar, W.H.M.W.; et al. Towards a time and cost effective approach to water quality index class prediction. J. Hydrol. 2019, 575, 148–165. [Google Scholar] [CrossRef]
- Robertson, D.M.; Hubbard, L.E.; Lorenz, D.L.; Sullivan, D.J. A surrogate regression approach for computing continuous loads for the tributary nutrient and sediment monitoring program on the Great Lakes. J. Great Lakes Res. 2018, 44, 26–42. [Google Scholar] [CrossRef]
- Jones, A.S.; Stevens, D.K.; Horsburgh, J.S.; Mesner, N.O. Surrogate Measures for Providing High Frequency Estimates of Total Suspended Solids and Total Phosphorus Concentrations 1. JAWRA J. Am. Water Resour. Assoc. 2011, 47, 239–253. [Google Scholar] [CrossRef] [Green Version]
- Kuefner, W.; Ossyssek, S.; Geist, J.; Raeder, U. The silicification value: A novel diatom-based indicator to assess climate change in freshwater habitats. Diatom Res. 2020, 35, 1–16. [Google Scholar] [CrossRef]
- Shah, M.I.; Javed, M.F.; Abunama, T. Proposed formulation of surface water quality and modelling using gene expression, machine learning, and regression techniques. Environ. Sci. Pollut. Res. 2021, 28, 13202–13220. [Google Scholar] [CrossRef]
- Abba, S.; Hadi, S.J.; Sammen, S.S.; Salih, S.Q.; Abdulkadir, R.; Pham, Q.B.; Yaseen, Z.M. Evolutionary computational intelligence algorithm coupled with self-tuning predictive model for water quality index determination. J. Hydrol. 2020, 587, 124974. [Google Scholar] [CrossRef]
- Schenk, L.; Bragg, H. Sediment transport, turbidity, and dissolved oxygen responses to annual streambed drawdowns for downstream fish passage in a flood control reservoir. J. Environ. Manag. 2021, 295, 113068. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.L.; Yang, S.H.; Hsieh, S.L.; Wang, H.J.; Yeh, K.C. Artificial intelligence methodologies applied to prompt pluvial flood estimation and prediction. Water 2020, 12, 3552. [Google Scholar] [CrossRef]
- Yaseen, Z.M.; El-Shafie, A.; Jaafar, O.; Afan, H.A.; Sayl, K.N. Artificial intelligence based models for stream-flow forecasting: 2000–2015. J. Hydrol. 2015, 530, 829–844. [Google Scholar] [CrossRef]
- Rahmati, O.; Choubin, B.; Fathabadi, A.; Coulon, F.; Soltani, E.; Shahabi, H.; Mollaefar, E.; Tiefenbacher, J.; Cipullo, S.; Ahmad, B.B.; et al. Predicting uncertainty of machine learning models for modelling nitrate pollution of groundwater using quantile regression and UNEEC methods. Sci. Total Environ. 2019, 688, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Lucius, M.A.; Johnston, K.E.; Eichler, L.W.; Farrell, J.L.; Moriarty, V.W.; Relyea, R.A. Using machine learning to correct for nonphotochemical quenching in high-frequency, in vivo fluorometer data. Limnol. Oceanogr. Methods 2020, 18, 477–494. [Google Scholar] [CrossRef]
- Shen, L.Q.; Amatulli, G.; Sethi, T.; Raymond, P.; Domisch, S. Estimating nitrogen and phosphorus concentrations in streams and rivers, within a machine learning framework. Sci. Data 2020, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mateo Pérez, V.; Mesa Fernández, J.M.; Villanueva Balsera, J.; Alonso Álvarez, C. A Random Forest Model for the Prediction of FOG Content in Inlet Wastewater from Urban WWTPs. Water 2021, 13, 1237. [Google Scholar] [CrossRef]
- Chen, Y.; Song, L.; Liu, Y.; Yang, L.; Li, D. A review of the artificial neural network models for water quality prediction. Appl. Sci. 2020, 10, 5776. [Google Scholar] [CrossRef]
- Xu, J.; Wang, K.; Lin, C.; Xiao, L.; Huang, X.; Zhang, Y. FM-GRU: A Time Series Prediction Method for Water Quality Based on seq2seq Framework. Water 2021, 13, 1031. [Google Scholar] [CrossRef]
- Mateo Pérez, V.; Mesa Fernández, J.M.; Ortega Fernández, F.; Villanueva Balsera, J. Gross Solids Content Prediction in Urban WWTPs Using SVM. Water 2021, 13, 442. [Google Scholar] [CrossRef]
- Stajkowski, S.; Zeynoddin, M.; Farghaly, H.; Gharabaghi, B.; Bonakdari, H. A methodology for forecasting dissolved oxygen in urban streams. Water 2020, 12, 2568. [Google Scholar] [CrossRef]
- Tang, X.; Huang, M. Inversion of chlorophyll-a concentration in Donghu Lake based on machine learning algorithm. Water 2021, 13, 1179. [Google Scholar] [CrossRef]
- Song, C.M. Application of convolution neural networks and hydrological images for the estimation of pollutant loads in ungauged watersheds. Water 2021, 13, 239. [Google Scholar] [CrossRef]
- Yu, H.; Yang, L.; Li, D.; Chen, Y. A hybrid intelligent soft computing method for ammonia nitrogen prediction in aquaculture. Inf. Process. Agric. 2021, 8, 64–74. [Google Scholar] [CrossRef]
- Gholizadeh, M.H.; Melesse, A.M.; Reddi, L. A comprehensive review on water quality parameters estimation using remote sensing techniques. Sensors 2016, 16, 1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topp, S.N.; Pavelsky, T.M.; Jensen, D.; Simard, M.; Ross, M.R. Research trends in the use of remote sensing for inland water quality science: Moving towards multidisciplinary applications. Water 2020, 12, 169. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wu, L.; Ren, H.; Liu, Y.; Zheng, Y.; Liu, Y.; Dong, J. Mapping water quality parameters in urban rivers from hyperspectral images using a new self-adapting selection of multiple artificial neural networks. Remote Sens. 2020, 12, 336. [Google Scholar] [CrossRef] [Green Version]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Chen, Y.; Clinton, N.; Wang, J.; Wang, X.; Liu, C.; Gong, P.; Yang, J.; Bai, Y.; Zheng, Y.; et al. Mapping major land cover dynamics in Beijing using all Landsat images in Google Earth Engine. Remote Sens. Environ. 2017, 202, 166–176. [Google Scholar] [CrossRef]
- Goldblatt, R.; You, W.; Hanson, G.; Khandelwal, A.K. Detecting the boundaries of urban areas in india: A dataset for pixel-based image classification in google earth engine. Remote Sens. 2016, 8, 634. [Google Scholar] [CrossRef] [Green Version]
- Talukdar, S.; Singha, P.; Mahato, S.; Pal, S.; Liou, Y.A.; Rahman, A. Land-use land-cover classification by machine learning classifiers for satellite observations—A review. Remote Sens. 2020, 12, 1135. [Google Scholar] [CrossRef] [Green Version]
- Perrone, M.; Scalici, M.; Conti, L.; Moravec, D.; Kropáček, J.; Sighicelli, M.; Lecce, F.; Malavasi, M. Water Mixing Conditions Influence Sentinel-2 Monitoring of Chlorophyll Content in Monomictic Lakes. Remote Sens. 2021, 13, 2699. [Google Scholar] [CrossRef]
- Weigelhofer, G.; Hein, T.; Bondar-Kunze, E. Phosphorus and nitrogen dynamics in riverine systems: Human impacts and management options. Riverine Ecosyst. Manag. 2018, 187. [Google Scholar] [CrossRef]
- Loh, W.Y. Classification and regression trees. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Bangira, T.; Alfieri, S.M.; Menenti, M.; Van Niekerk, A. Comparing thresholding with machine learning classifiers for mapping complex water. Remote Sens. 2019, 11, 1351. [Google Scholar] [CrossRef] [Green Version]
- Peterson, K.T.; Sagan, V.; Sidike, P.; Hasenmueller, E.A.; Sloan, J.J.; Knouft, J.H. Machine learning-based ensemble prediction of water-quality variables using feature-level and decision-level fusion with proximal remote sensing. Photogramm. Eng. Remote Sens. 2019, 85, 269–280. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, C.; Liu, Q.; Xi, B.; Qian, G.; Zhang, D.; Huo, S. Method to predict key factors affecting lake eutrophication–A new approach based on Support Vector Regression model. Int. Biodeterior. Biodegrad. 2015, 102, 308–315. [Google Scholar] [CrossRef]
- Chomboon, K.; Chujai, P.; Teerarassamee, P.; Kerdprasop, K.; Kerdprasop, N. An empirical study of distance metrics for k-nearest neighbor algorithm. In Proceedings of the 3rd International Conference on Industrial Application Engineering, Sanya, China, 15–18 April 2015; pp. 280–285. [Google Scholar]
- McDonald, G.C. Ridge regression. Wiley Interdiscip. Rev. Comput. Stat. 2009, 1, 93–100. [Google Scholar] [CrossRef]
- Chen, Y.R.; Rezapour, A.; Tzeng, W.G. Privacy-preserving ridge regression on distributed data. Inf. Sci. 2018, 451, 34–49. [Google Scholar] [CrossRef]
- Ghorbani, M.A.; Deo, R.C.; Karimi, V.; Kashani, M.H.; Ghorbani, S. Design and implementation of a hybrid MLP-GSA model with multi-layer perceptron-gravitational search algorithm for monthly lake water level forecasting. Stoch. Environ. Res. Risk Assess. 2019, 33, 125–147. [Google Scholar] [CrossRef]
- Schapire, R.E. The boosting approach to machine learning: An overview. In Nonlinear Estimation and Classification; Springer: Berlin/Heidelberg, Germany, 2003; pp. 149–171. [Google Scholar]
- Bühlmann, P.; Yu, B. Analyzing bagging. Ann. Stat. 2002, 30, 927–961. [Google Scholar] [CrossRef]
- Karami, J.; Alimohammadi, A.; Seifouri, T. Water quality analysis using a variable consistency dominance-based rough set approach. Comput. Environ. Urban Syst. 2014, 43, 25–33. [Google Scholar] [CrossRef]
- Antanasijević, D.; Pocajt, V.; Perić-Grujić, A.; Ristić, M. Modelling of dissolved oxygen in the Danube River using artificial neural networks and Monte Carlo Simulation uncertainty analysis. J. Hydrol. 2014, 519, 1895–1907. [Google Scholar] [CrossRef]
- Klose, K.; Cooper, S.D.; Leydecker, A.D.; Kreitler, J. Relationships among catchment land use and concentrations of nutrients, algae, and dissolved oxygen in a southern California river. Freshw. Sci. 2012, 31, 908–927. [Google Scholar] [CrossRef]
- Dinnat, E.P.; Le Vine, D.M.; Boutin, J.; Meissner, T.; Lagerloef, G. Remote sensing of sea surface salinity: Comparison of satellite and in situ observations and impact of retrieval parameters. Remote Sens. 2019, 11, 750. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.H. Ensemble learning. In Machine Learning; Springer: Berlin/Heidelberg, Germany, 2021; pp. 181–210. [Google Scholar]
| Temp | pH | DO | Tud | COD | T.D.S | TN | ||
|---|---|---|---|---|---|---|---|---|
| Magnitude | ℃ | - | mg/L | NTU | mg/L | S/cm | mg/L | mg/L |
| MAX | 35.00 | 9.43 | 15.00 | 500.00 | 70.10 | 8895 | 12.12 | 14.20 |
| MIN | 15.70 | 6.15 | 0.09 | 1.00 | 7.00 | 0.00 | 0.01 | 1.90 |
| Mean | 25.51 | 7.34 | 4.78 | 61.65 | 25.22 | 1848 | 2.60 | 6.46 |
| Median | 25.70 | 7.31 | 4.50 | 52.00 | 24.90 | 1271 | 2.19 | 5.77 |
| Mode | 19.60 | 7.23 | 3.55 | 49.00 | 21.60 | 398 | 0.24 | 4.72 |
| SD | 4.78 | 0.40 | 2.46 | 41.84 | 7.63 | 1589 | 2.12 | 2.05 |
| Set | 1917 | 1917 | 1917 | 1917 | 1917 | 1917 | 1917 | 1917 |
| Parameter | Temp | pH | T.D.S | DO | Tud | COD | TP | TN | |
|---|---|---|---|---|---|---|---|---|---|
| Coefficient | −0.75 | 0.12 | 0.3 | 0.012 | −0.05 | 0.63 | 0.47 | 0.81 | 1 |
| MLP | 0.479 | 0.474 | 0.689 | 7.13% | 0.89 | 0.947 |
| SVR | 0.654 | 0.757 | 0.871 | 9.98% | 0.81 | 0.901 |
| KNN | 0.711 | 1.277 | 1.13 | 11.07% | 0.68 | 0.899 |
| RidgeRegress | 0.654 | 0.736 | 0.858 | 9.98% | 0.81 | 0.905 |
| GradientBoost | 0.431 | 0.326 | 0.572 | 6.58% | 0.91 | 0.951 |
| Bagging | 0.371 | 0.294 | 0.542 | 5.48% | 0.91 | 0.922 |
| Decision Tree | 0.492 | 0.653 | 0.808 | 7.19% | 0.83 | 0.911 |
| RFR | 0.335 | 0.259 | 0.509 | 4.90% | 0.94 | 0.967 |
| B11 | B12 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | B8A | |
|---|---|---|---|---|---|---|---|---|---|---|
| Temperature | 0.51 | 0.50 | 0.49 | 0.45 | 0.48 | 0.52 | 0.57 | 0.57 | 0.54 | 0.56 |
| Salinity | −0.65 | −0.65 | −0.62 | −0.68 | −0.74 | −0.76 | −0.69 | −0.71 | −0.68 | −0.68 |
| Temperature | Salinity | |||||
|---|---|---|---|---|---|---|
| MLP | 2.490 | 0.52 | 10.83% | 1.773 | 0.76 | 7.22% |
| SVR | 1.744 | 0.61 | 8.42% | 2.182 | 0.61 | 8.99% |
| KNN | 1.754 | 0.60 | 9.45% | 2.667 | 0.62 | 13.03% |
| RidgeRegress | 2.527 | 0.51 | 10.95% | 2.165 | 0.72 | 8.26% |
| GradientBoot | 1.129 | 0.77 | 5.44% | 1.887 | 0.71 | 7.32% |
| Bagging | 1.337 | 0.76 | 5.83% | 2.130 | 0.65 | 8.47% |
| Decision Tree | 1.620 | 0.57 | 6.27% | 1.782 | 0.76 | 7.23% |
| RFR | 1.107 | 0.82 | 4.65% | 1.755 | 0.77 | 7.06% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Xu, Z.; Kuang, J.; Lin, C.; Xiao, L.; Huang, X.; Zhang, Y. An Alternative to Laboratory Testing: Random Forest-Based Water Quality Prediction Framework for Inland and Nearshore Water Bodies. Water 2021, 13, 3262. https://doi.org/10.3390/w13223262
Xu J, Xu Z, Kuang J, Lin C, Xiao L, Huang X, Zhang Y. An Alternative to Laboratory Testing: Random Forest-Based Water Quality Prediction Framework for Inland and Nearshore Water Bodies. Water. 2021; 13(22):3262. https://doi.org/10.3390/w13223262
Chicago/Turabian StyleXu, Jianlong, Zhuo Xu, Jianjun Kuang, Che Lin, Lianghong Xiao, Xingshan Huang, and Yufeng Zhang. 2021. "An Alternative to Laboratory Testing: Random Forest-Based Water Quality Prediction Framework for Inland and Nearshore Water Bodies" Water 13, no. 22: 3262. https://doi.org/10.3390/w13223262






