Structural Characteristics and Driving Factors of the Planktonic Eukaryotic Community in the Danjiangkou Reservoir, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Physicochemical Factor Determination in the Danjiangkou Reservoir
2.2. Genomic DNA Extraction from Planktonic Eukaryotes
2.3. Eukaryotic PCR Amplification and High-Throughput Sequencing
2.4. Analysis of Planktonic Eukaryote High-Throughput Data
2.5. Analysis of Experimental Data
3. Results and Analysis
3.1. Analysis of Physicochemical Properties in the Danjiangkou Reservoir
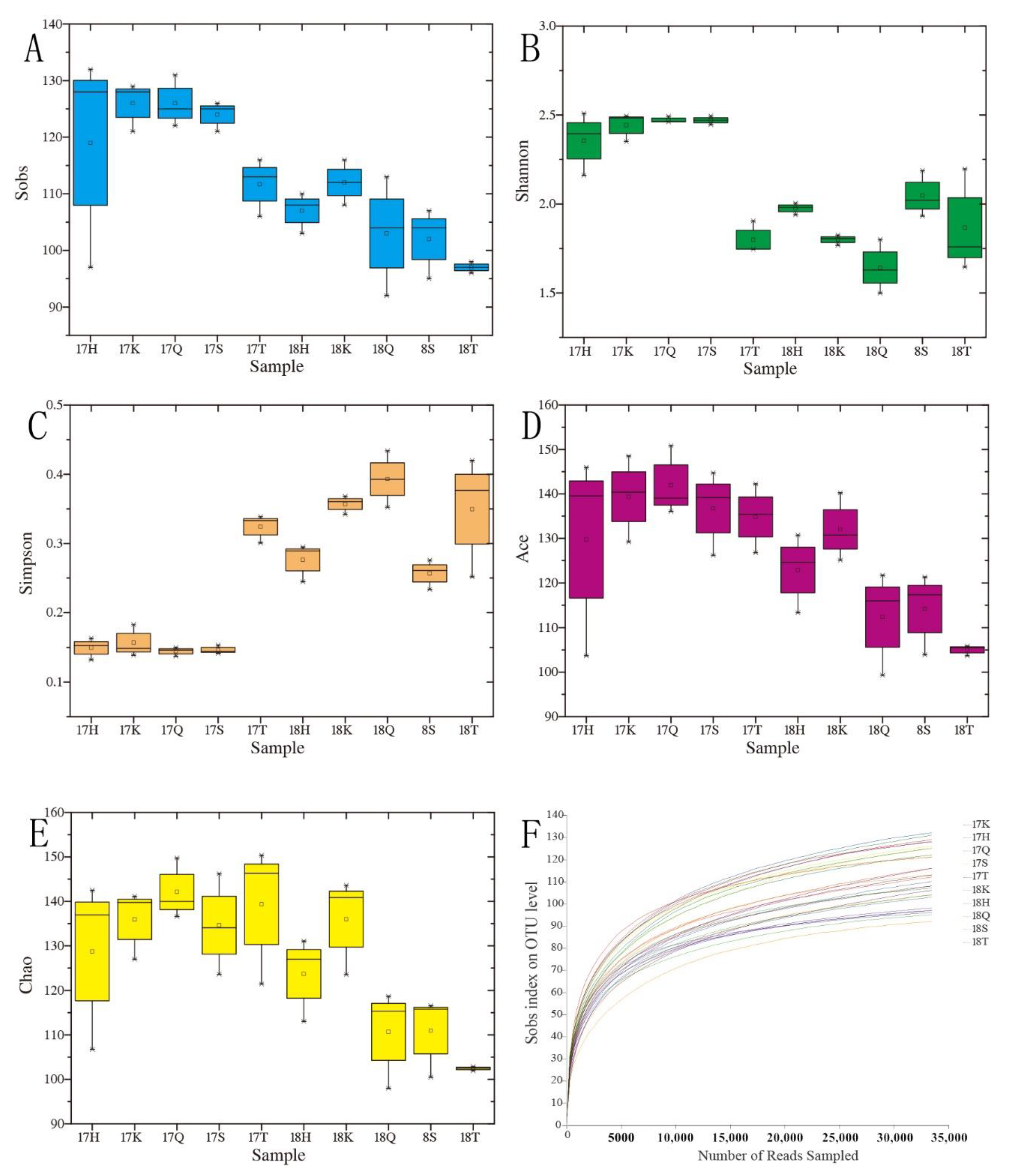
3.2. Analysis of the Alpha Diversity of Planktonic Eukaryotes
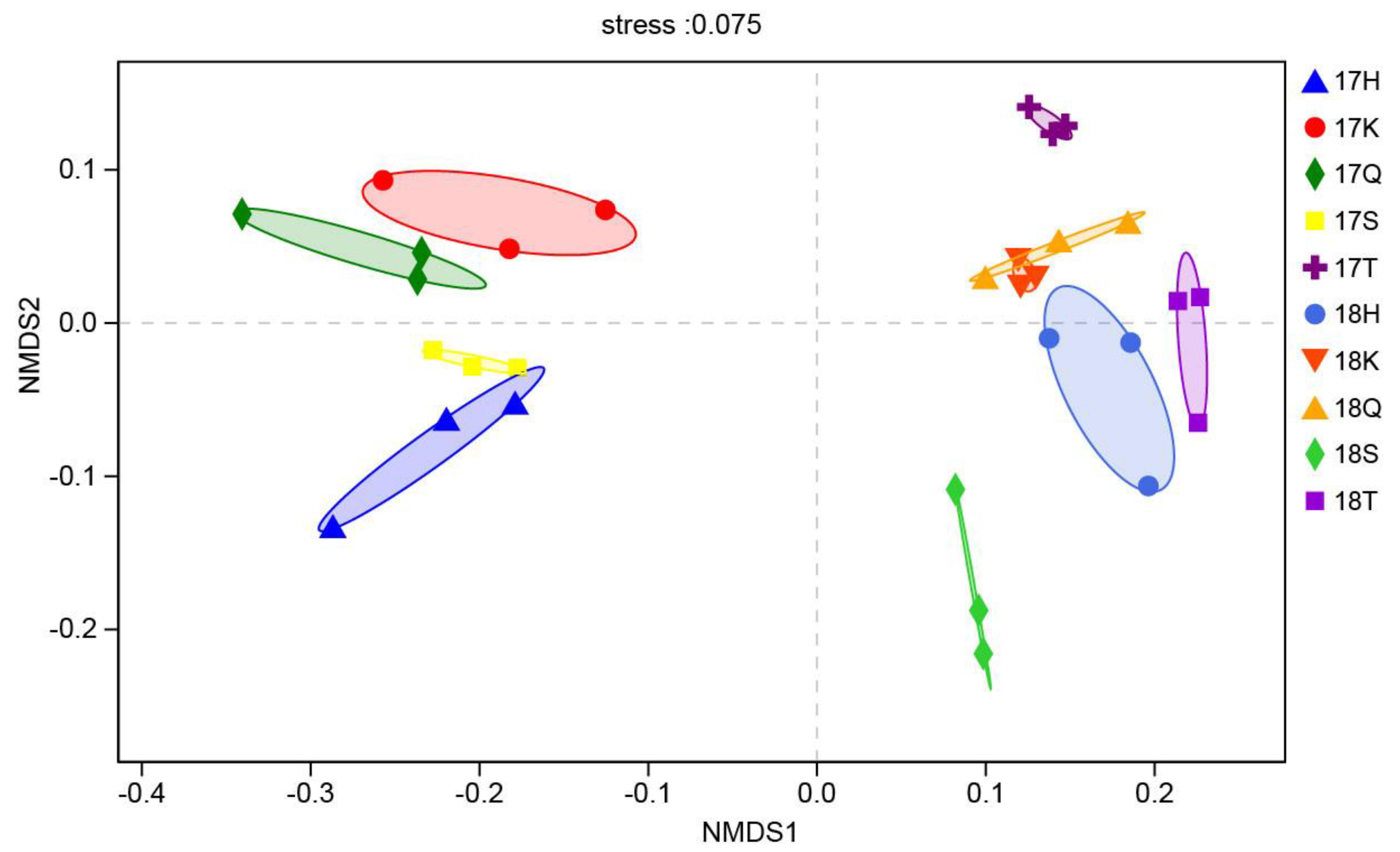
3.3. Analysis of the Beta Diversity of Planktonic Eukaryotes
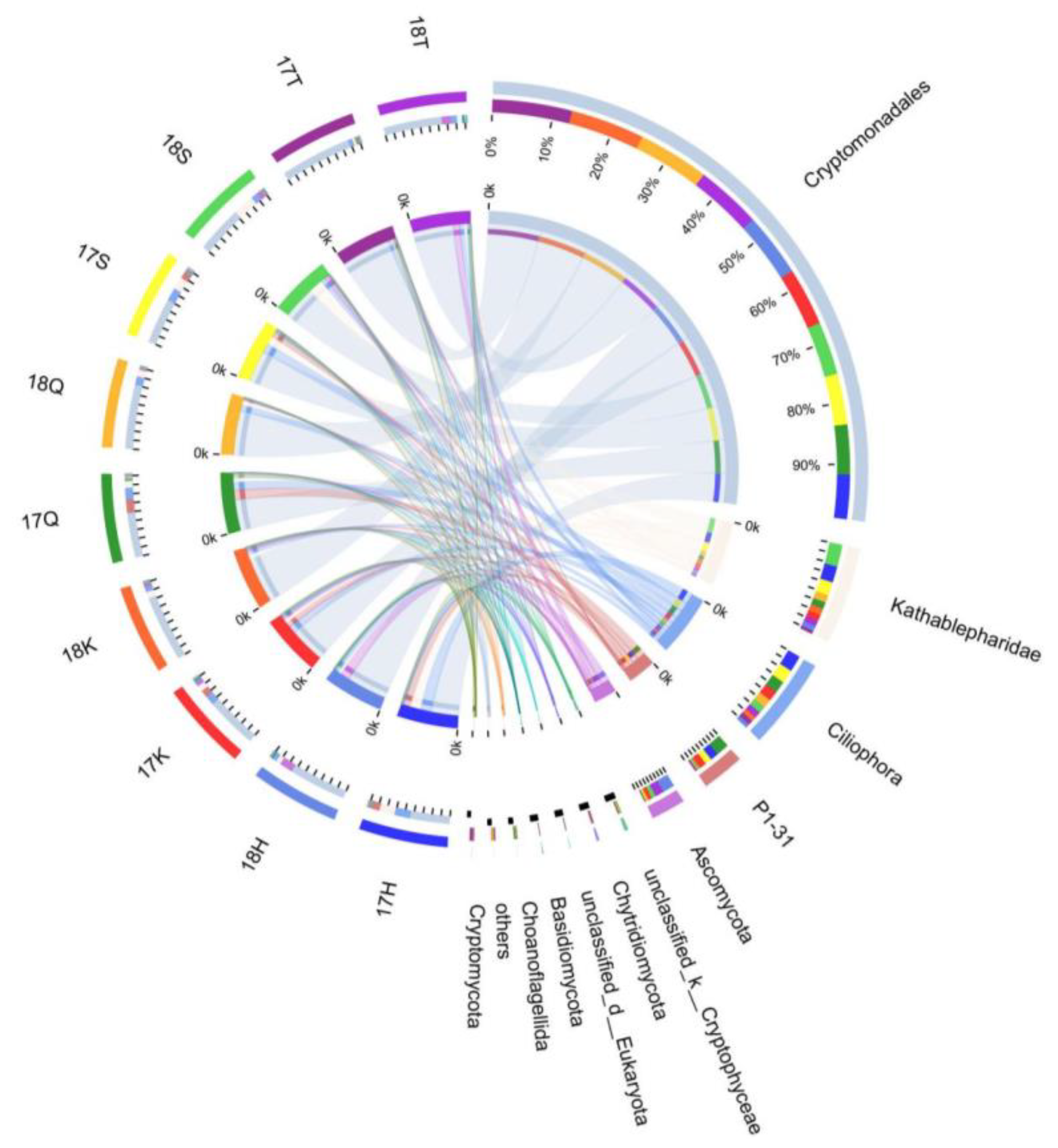
3.4. Taxonomic Composition and Analysis of the Differences in Planktonic Eukaryotes
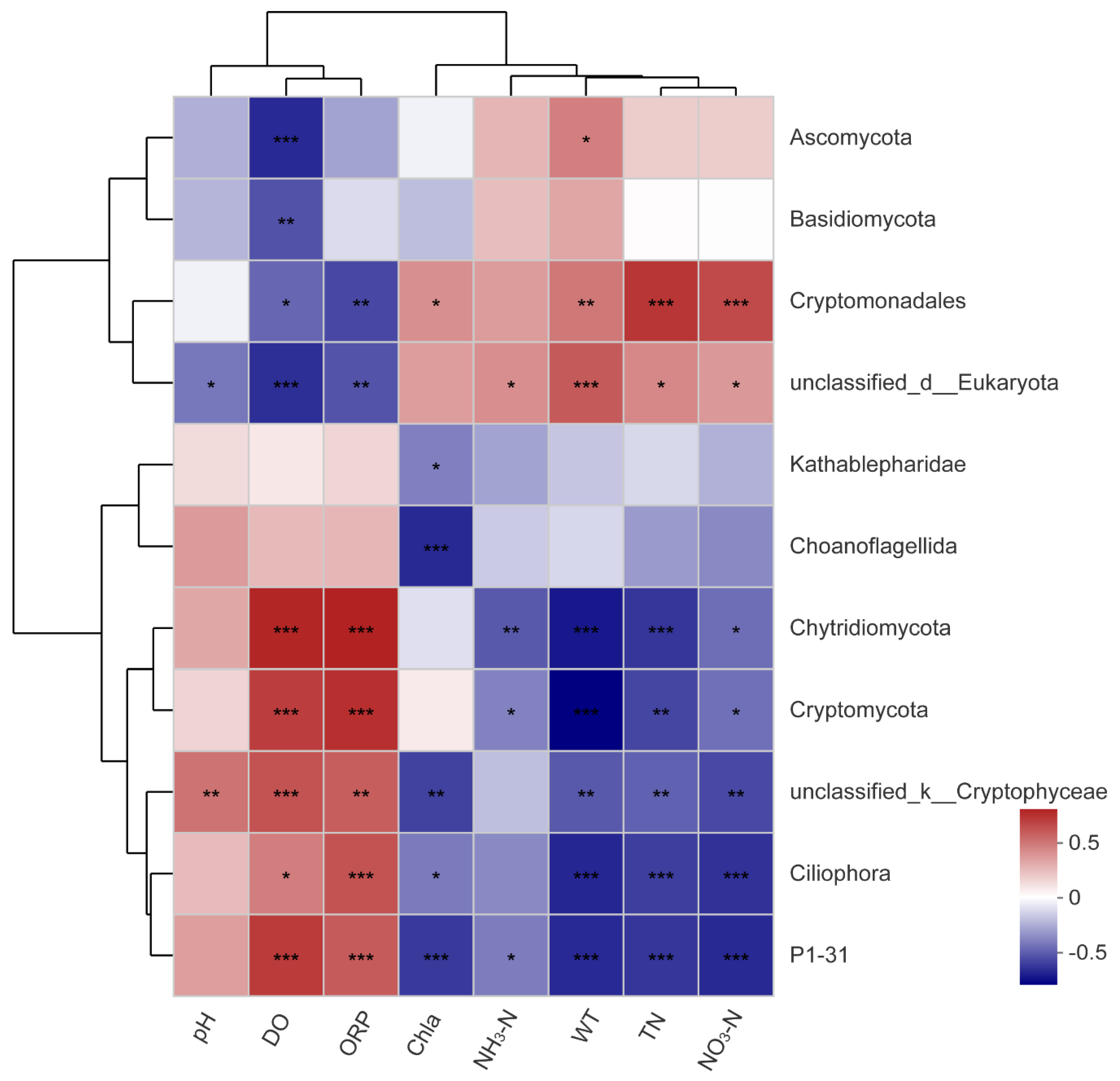
3.5. Analysis of Correlations between Planktonic Eukaryotic Taxa and the Environment
3.6. Network Analysis of Planktonic Eukaryotes
4. Discussion
4.1. Composition of the Planktonic Eukaryotic Community in the Danjiangkou Reservoir
4.2. Relationships between Eukaryotes and Environmental Factors
4.3. Changes in Planktonic Eukaryotic Community Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jiang, Y.; Xu, H.; Zhu, M. Use of planktonic protists for assessing water quality in Jiaozhou Bay, northern China. Protistology 2012, 7, 34–41. [Google Scholar]
- Filker, S.; Sommaruga, R.; Vila, I.; Stoeck, T. Microbial eukaryote plankton communities of high-mountain lakes from three continents exhibit strong biogeographic patterns. Mol. Ecol. 2016, 25, 2286–2301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foissner, W. Biogeography and dispersal of microorganisms: A review emphasizing protists. Acta Protozoologica 2006, 45, 111–136. [Google Scholar]
- Zubkov, M.; Tarran, G.A. High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nat. Cell Biol. 2008, 455, 224–226. [Google Scholar] [CrossRef]
- Caron, D.A. Inorganic nutrients, bacteria, and the microbial loop. Microb. Ecol. 1994, 28, 295–298. [Google Scholar] [CrossRef]
- Bunse, C.; Pinhassi, J. Marine Bacterioplankton Seasonal Succession Dynamics. Trends Microbiol. 2017, 25, 494–505. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.; López-García, P.; Deschamps, P.; Moreira, D.; Restoux, G.; Bertolino, P.; Jardillier, L. Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME J. 2015, 9, 1941–1953. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Li, G.; Wang, C.; Jing, Y.; Zhu, Y.; Zhang, S.; Liu, Y. Community dynamics of prokaryotic and eukaryotic microbes in an estuary reservoir. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef]
- Gao, W.; Chen, Z.; Li, Y.; Pan, Y.; Zhu, J.; Guo, S.; Hu, L.; Huang, J. Bioassessment of a Drinking Water Reservoir Using Plankton: High Throughput Sequencing vs. Traditional Morphological Method. Water 2018, 10, 82. [Google Scholar] [CrossRef] [Green Version]
- Keck, F.; Millet, L.; Debroas, D.; Etienne, D.; Galop, D.; Rius, D.; Domaizon, I. Assessing the response of micro-eukaryotic diversity to the Great Acceleration using lake sedimentary DNA. Nat. Commun. 2020, 11, 3831. [Google Scholar] [CrossRef]
- McInnes, A.S.; Laczka, O.F.; Baker, K.G.; Larsson, M.E.; Robinson, C.M.; Clark, J.S.; Laiolo, L.; Alvarez, M.; Laverock, B.; Kremer, C.T.; et al. Live cell analysis at sea reveals divergent thermal performance between photosynthetic ocean microbial eukaryote populations. ISME J. 2019, 13, 1374–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Der Staay, S.Y.M.-V.; De Wachter, R.; Vaulot, D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nat. Cell Biol. 2001, 409, 607–610. [Google Scholar] [CrossRef]
- López-García, P.; Rodríguez-Valera, F.; Pedrós-Alió, C.; Moreira, D. Unexpected diversity of small eukaryotes in deep-sea antarctic plankton. Nature 2001, 409, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Nagarkar, M.; Countway, P.D.; Du Yoo, Y.; Daniels, E.; Poulton, N.; Palenik, B. Temporal dynamics of eukaryotic microbial diversity at a coastal Pacific site. ISME J. 2018, 12, 2278–2291. [Google Scholar] [CrossRef]
- Santoferrara, L.F.; Grattepanche, J.D.; Katz, L.A.; McManus, B. Patterns and processes in microbial biogeography:do molecules and morphologies give the same answers quest. ISME J. 2016, 10, 1779–1790. [Google Scholar] [CrossRef]
- Needham, D.M.; Fichot, E.B.; Wang, E.; Berdjeb, L.; Cram, J.A.; Fichot, C.G.; A Fuhrman, J. Dynamics and interactions of highly resolved marine plankton via automated high-frequency sampling. ISME J. 2018, 12, 2417–2432. [Google Scholar] [CrossRef] [Green Version]
- Wurzbacher, C.M.; Bärlocher, F.; Grossart, H.-P. Fungi in lake ecosystems. Aquat. Microb. Ecol. 2010, 59, 125–149. [Google Scholar] [CrossRef] [Green Version]
- Nagano, Y.; Nagahama, T. Fungal diversity in deep-sea extreme environments. Fungal Ecol. 2012, 5, 463–471. [Google Scholar] [CrossRef]
- Taylor, J.D.; Cunliffe, M. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J. 2016, 10, 2118–2128. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef]
- Etan, B.; Ng, C.M.; Nshimyimana, J.P.; Eloh, L.-L.; Gin, K.Y.-H.; Thompson, J.R. Next-generation sequencing (NGS) for assessment of microbial water quality: Current progress, challenges, and future opportunities. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Reich, M.; Wichels, A.; Panzer, K.; Krause, E.; Giménez, L.; Gerdts, G. Impacts of a reduction in seawater pH mimicking ocean acidification on the structure and diversity of mycoplankton communities. Aquat. Microb. Ecol. 2017, 79, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.-J.; He, W.; Liu, W.-X.; Qin, N.; Ouyang, H.-L.; Wang, Q.-M.; Kong, X.-Z.; He, Q.-S.; Yang, C.; Yang, B.; et al. The seasonal and spatial variations of phytoplankton community and their correlation with environmental factors in a large eutrophic Chinese lake (Lake Chaohu). Ecol. Indic. 2014, 40, 58–67. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Q.; Yang, T.; Zhang, L.; Li, X.; Chen, J. River health assessment: Proposing a comprehensive model based on physical habitat, chemical condition and biotic structure. Ecol. Indic. 2019, 103, 446–460. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Yin, X.; Zhao, X.; Guo, R.; Zheng, B. Development and evaluation of a Planktonic Integrity Index (PII) for Jingpo Lake, China. J. Limnol. 2019, 78, 135–146. [Google Scholar] [CrossRef]
- Zhang, Y.; Ban, X.; Li, E.; Wang, Z.; Xiao, F. Evaluating ecological health in the middle-lower reaches of the Hanjiang River with cascade reservoirs using the Planktonic index of biotic integrity (P-IBI). Ecol. Indic. 2020, 114. [Google Scholar] [CrossRef]
- Li, S.; Xu, Z.; Cheng, X.; Zhang, Q. Dissolved trace elements and heavy metals in the Danjiangkou Reservoir, China. Environ. Earth Sci. 2007, 55, 977–983. [Google Scholar] [CrossRef]
- Tropical and Sub-Tropical Reservoir Limnology in China. Lake Chad 2012, 91, 370. [CrossRef]
- Chen, Z.; Yuan, J.; Sun, F.; Zhang, F.; Chen, Y.; Ding, C.; Shi, J.; Li, Y.; Yao, L. Planktonic fungal community structures and their relationship to water quality in the Danjiangkou Reservoir, China. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Dang, C.; Liu, W.; Lin, Y.; Zheng, M.; Jiang, H.; Chen, Q.; Ni, J. Dominant role of ammonia-oxidizing bacteria in nitrification due to ammonia accumulation in sediments of Danjiangkou reservoir, China. Appl. Microbiol. Biotechnol. 2018, 102, 3399–3410. [Google Scholar] [CrossRef]
- State Environmental Protection Administration. Monitoring and Analysis Methods of Water Waste Water, 4th ed.; Environmental Science Press: Beijing, China, 2002. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Gu, S.; Tan, X.; Zhang, Q. Water quality in the upper Han River basin, China: The impacts of land use/land cover in riparian buffer zone. J. Hazard. Mater. 2009, 165, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, L.; Zhang, H. Spatio-Temporal Variations and Source Apportionment of Water Pollution in Danjiangkou Reservoir Basin, Central China. Water 2015, 7, 2591–2611. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Shi, X.; Liu, G.; Xu, H.; Nie, P. An approach to analyzing taxonomic patterns of protozoan communities for monitoring water quality in Songhua River, northeast China. Hydrobiologia 2009, 638, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Penna, A.; Casabianca, S.; Guerra, A.F.; Vernesi, C.; Scardi, M. Analysis of phytoplankton assemblage structure in the Mediterranean Sea based on high-throughput sequencing of partial 18S rRNA sequences. Mar. Genom. 2017, 36, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Singh, P.; Liu, Y.; Pan, S.Q.; Wang, G. Diversity and biochemical features of culturable fungi from the coastal waters of Southern China. AMB Express 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Kagami, M.; Amano, Y.; Ishii, N. Community Structure of Planktonic Fungi and the Impact of Parasitic Chytrids on Phytoplankton in Lake Inba, Japan. Microb. Ecol. 2011, 63, 358–368. [Google Scholar] [CrossRef]
- Khomich, M.; Kauserud, H.; Logares, R.; Rasconi, S.; Andersen, T. Planktonic protistan communities in lakes along a large-scale environmental gradient. FEMS Microbiol. Ecol. 2016, 93. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Bock, C.; Li, H.R.; Padisák, J.; Krienitz, L. Molecular and microscopic diversity of planktonic eukaryotes in the oligotrophic Lake Stechlin (Germany). Hydrobiologia 2010, 661, 133–143. [Google Scholar] [CrossRef]
- Grujcic, V.; Nuy, J.K.; Salcher, M.M.; Shabarova, T.; Kasalicky, V.; Boenigk, J.; Jensen, M.; Simek, K. Cryptophyta as major bacterivores in freshwater summer plankton. ISME J. 2018, 12, 1668–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.Y.; Li, J.Z.; Liang, Z.A.; Yin, W.; Hu, L.Q.; Shi, J.W.; Wang, Y. Planktonic Algae and Epilithic Algae in Typical Rivers of Danjingkou Reservoir. Adv. Mater. Res. 2011, 356, 59–62. [Google Scholar] [CrossRef]
- Callieri, C. Picophytoplankton in Freshwater Ecosystems: The Importance of Small-Sized Phototrophs. Freshw. Rev. 2008, 1, 1–28. [Google Scholar] [CrossRef]
- Lepère, C.; Isabelle, D.; Taïb, N.; Mangot, J.-F.; Bronner, G.; Delphine, B.; Dider, D. Geographic distance and ecosystem sizedetermine the distribution of smallest protists in lacustrine ecosystems. FEMS Microbiol. Ecol. 2013, 85, 85–94. [Google Scholar]
- Cudowski, A.; Pietryczuk, A.; Hauschild, T. Aquatic fungi in relation to the physical and chemical parameters of water quality in the Augustów Canal. Fungal Ecol. 2015, 13, 193–204. [Google Scholar] [CrossRef]
- Diaz, R.J.; Rosenberg, R. Spreading Dead Zones and Consequences for Marine Ecosystems. Science 2008, 321, 926–929. [Google Scholar] [CrossRef]
- Liu, T.; Ni Zhang, A.; Wang, J.; Liu, S.; Jiang, X.; Dang, C.; Ma, T.; Liu, S.; Chen, Q.; Xie, S.; et al. Integrated biogeography of planktonic and sedimentary bacterial communities in the Yangtze River. Microbiome 2018, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zohary, T.; Flaim, G.; Sommer, U. Temperature and the size of freshwater phytoplankton. Hydrobiologia 2020, 1. [Google Scholar] [CrossRef]
- Wan, X.; Yang, T.; Zhang, Q.; Wang, W.; Wang, Y. Joint effects of habitat indexes and physic-chemical factors for freshwater basin of semi-arid area on plankton integrity—A case study of the Wei River Basin, China. Ecol. Indic. 2020, 120. [Google Scholar] [CrossRef]
- Edberg, S.; Rice, E.; Karlin, R.; Allen, M. Escherichia coli: The best biological drinking water indicator for public health protection. J. Appl. Microbiol. 2000, 88, 106S–116S. [Google Scholar] [CrossRef]
- Parmar, T.K.; Rawtani, D.; Agrawal, Y.K. Bioindicators: The natural indicator of environmental pollution. Front. Life Sci. 2016, 9, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Gaedke, U.; Seifried, A.; Adrian, R. Biomass Size Spectra and Plankton Diversity in a Shallow Eutrophic Lake. Int. Rev. Hydrobiol. 2004, 89, 1–20. [Google Scholar] [CrossRef]
- Chen, M.; Chen, F.; Yu, Y.; Ji, J.; Kong, F. Genetic Diversity of Eukaryotic Microorganisms in Lake Taihu, a Large Shallow Subtropical Lake in China. Microb. Ecol. 2008, 56, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Breuer, F.; Janz, P.; Farrelly, E.; Ebke, K.-P.; Breuer, F.; Janz, P.; Farrelly, E.; Ebke, K.-P. Environmental and structural factors influencing algal communities in small streams and ditches in central Germany. J. Freshw. Ecol. 2016, 32, 65–83. [Google Scholar] [CrossRef] [Green Version]
- Belykh, O.I.; Ekaterina, G.; Sorokovikova, T.; Saphonova, A.; Tikhonova, I.V. Autotrophic picoplankton of Lake Baikal: Composition, abundance and structure. Hydrobiologia 2006, 568, 9–17. [Google Scholar] [CrossRef]
- Barkmann, S. The significance of photoautotrophic picoplankton in the eutrophic Lake Belau (Bornhöveder Seenkette, North Germany). Limnologica 2000, 30, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Soballe, D.M.; Kimmel, B.L. A Large-Scale Comparison of Factors Influencing Phytoplankton Abundance in Rivers, Lakes, and Impoundments. Ecology 1987, 68, 1943–1954. [Google Scholar] [CrossRef]
- Schallenberg, M.; Burns, C.W. Phytoplankton biomass and productivity in two oligotrophic lakes of short hydraulic res idence time. New Zeal. J. Mar. 1997, 31, 119–134. [Google Scholar] [CrossRef] [Green Version]
- Lindström, E.S.; Bergström, A.K. Influence of inlet bacteria on bacterioplankton assemblage composition in lakes of different hydraulic retention time. Limnol. Oceanogr. 2004, 49, 125–136. [Google Scholar] [CrossRef]
- Lv, H.; Yang, J.; Liu, L. Temporal pattern prevails over spatial variability in phytoplankton communities from a subtropical water supply reservoir. Oceanol. Hydrobiol. Stud. 2013, 42, 420–430. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, H.; Yang, J.; Liu, M.; Huang, B. Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 2018, 12, 2263–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, T.; Baranyi, C.; Reckendorfer, W.; Schiemer, F. The impact of surface water exchange on the nutrient and particle dynamics in side-arms along the River Danube, Austria. Sci. Total. Environ. 2004, 328, 207–218. [Google Scholar] [CrossRef] [PubMed]
- GoŁdyn, R.; Szela̧g-Wasielewska, E. Changes in the phytoseston of a river-lake system in Drawieński National Park. Oceanol. Hydrobiol. Stud. 2004, 33, 17–28. [Google Scholar]
- Kim, D.Y.; Countway, P.D.; Jones, A.C.; Schnetzer, A.; Yamashita, W.; Tung, C.; Caron, D.A. Monthly to interannual variability of microbial eukaryote assemblages at four depths in the eastern North Pacific. ISME J. 2014, 8, 515–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Pan, Y.; Yang, J.; Chen, H.; Holohan, B.; Vaudrey, J.; Lin, S.; McManus, G.B. The diversity and biogeography of abundant and rare intertidal marine microeukaryotes explained by environment and dispersal limitation. Environ. Microbiol. 2017, 20, 462–476. [Google Scholar] [CrossRef]
- Meadow, J.F.; Zabinski, C.A. Spatial heterogeneity of eukaryotic microbial communities in an unstudied geothermal diatomaceous biological soil crust: Yellowstone National Park, WY, USA. FEMS Microbiol. Ecol. 2012, 82, 182–191. [Google Scholar] [CrossRef] [Green Version]
- Lefranc, M.; Thénot, A.; Lepère, C.; Debroas, D. Genetic Diversity of Small Eukaryotes in Lakes Differing by Their Trophic Status. Appl. Environ. Microbiol. 2005, 71, 5935–5942. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Xu, G.; Ding, C.-Y.; Zheng, B.-H.; Chen, Y.; Han, H.; Li, Y.; Shi, J.-W.; Hu, L.-Q. Illumina MiSeq sequencing and network analysis the distribution and co-occurrence of bacterioplankton in Danjiangkou Reservoir, China. Arch. Microbiol. 2020, 202, 859–873. [Google Scholar] [CrossRef]
- Stoecker, D.K.; Lavrentyev, P.J. Mixotrophic Plankton in the Polar Seas: A Pan-Arctic Review. Front. Mar. Sci. 2018, 5. [Google Scholar] [CrossRef]
- Stoecker, D.K.; Hansen, P.J.; Caron, D.A.; Mitra, A. Mixotrophy in the Marine Plankton. Annu. Rev. Mar. Sci. 2017, 9, 311–335. [Google Scholar] [CrossRef] [Green Version]
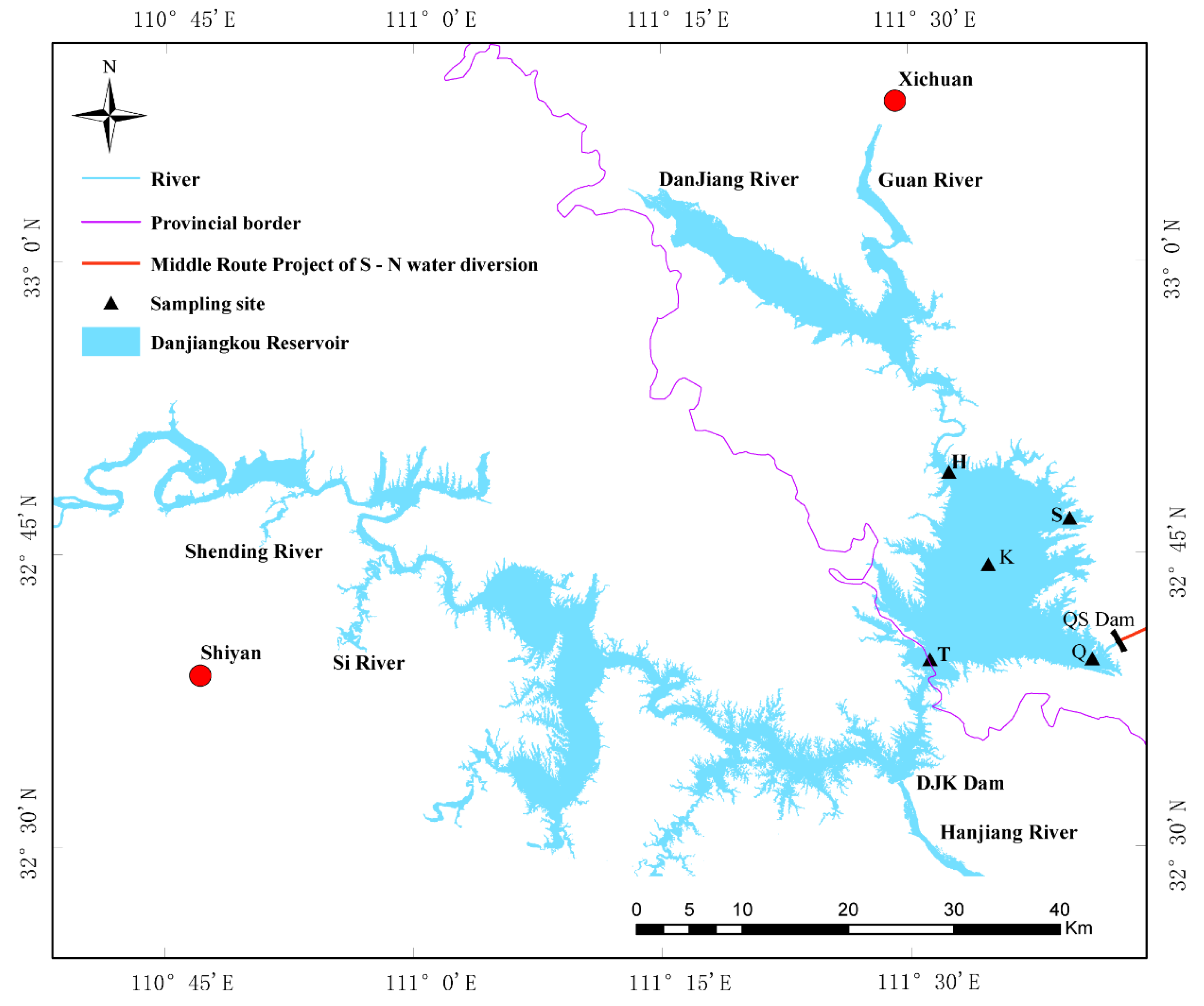




| Sample | Abbreviation | 2017 Abbreviation | 2018 Abbreviation | Latitude | Longitude |
|---|---|---|---|---|---|
| Kuxin | K | 17K | 18K | 32°45′06″ N | 111°34′45″ E |
| Qushou | Q | 17Q | 18Q | 32°40′03″ N | 111°37′47″ E |
| Songgang | S | 17S | 18S | 32°46′06″ N | 111°36′40″ E |
| Heijizui | H | 17H | 18H | 32°48′37″ N | 111°33′17″ E |
| Taizishan | T | 17T | 18T | 32°40′42″ N | 111°32′01″ E |
| Sample | Level /m | WT /°C | pH | DO /mg·L−1 | ORP /mv | TP /mg·L−1 | TN /mg·L−1 | NO3-N /mg·L−1 | NH3-N /mg·L−1 | Chla /mg·m3 | Water Quality Classification |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 17Q | 156.00 ± 1.00 | 16.90 ± 0.10 b | 8.45 ± 0.01 c | 9.79 ± 0.02 i | 335.00 ± 1.00 f | 0.02 ± 0.01 d | 0.90 ± 0.02 b | 0.70 ± 0.01 c | 0.05 ± 0.00 d | 0.60 ± 0.01 de | Class II |
| 17K | 156.33 ± 1.15 | 16.53 ± 0.15 a | 8.49 ± 0.01 c | 8.70 ± 0.10 g | 346.00 ± 1.00 h | 0.01 ± 0.00 a | 0.95 ± 0.03 c | 0.76 ± 0.04 d | 0.03 ± 0.00 b | 0.50 ± 0.01 c | Class I |
| 17S | 156.00 ± 1.00 | 16.65 ± 0.01 a | 8.64 ± 0.01 d | 8.50 ± 0.10 f | 340.00 ± 1.00 g | 0.02 ± 0.01 d | 1.01 ± 0.01 d | 0.51 ± 0.03 a | 0.15 ± 0.01 h | 0.42 ± 0.01 b | Class II |
| 17H | 156.00 ± 1.00 | 16.66 ± 0.02 a | 8.20 ± 0.10 a | 9.63 ± 0.01 h | 335.00 ± 1.00 f | 0.01 ± 0.01 ab | 0.78 ± 0.03 a | 0.66 ± 0.02 b | 0.02 ± 0.00 a | 0.56 ± 0.01 d | Class I |
| 17T | 156.00 ± 1.00 | 17.20 ± 0.10 c | 8.44 ± 0.01 bc | 8.80 ± 0.10 g | 302.00 ± 1.00 e | 0.02 ± 0.01 d | 1.21 ± 0.01 f | 1.08 ± 0.00 h | 0.07 ± 0.01 e | 1.28 ± 0.01 h | Class II |
| 18Q | 155.00 ± 1.00 | 18.00 ± 0.10 d | 8.40 ± 0.10 bc | 5.40 ± 0.10 c | 267.00 ± 1.00 a | 0.01 ± 0.00 bc | 1.59 ± 0.00 h | 1.35 ± 0.00 i | 0.07 ± 0.00 e | 1.15 ± 0.01 g | Class II |
| 18K | 156.00 ± 1.00 | 22.20 ± 0.10 h | 8.40 ± 0.10 bc | 5.20 ± 0.10 b | 266.00 ± 1.00 a | 0.01 ± 0.00 abc | 1.58 ± 0.01 h | 0.98 ± 0.00 f | 0.10 ± 0.00 g | 0.35 ± 0.01 a | Class II |
| 18S | 156.00 ± 1.00 | 21.60 ± 0.10 g | 8.40 ± 0.10 bc | 5.80 ± 0.10 d | 278.00 ± 1.00 c | 0.01 ± 0.00 abc | 1.46 ± 0.00 g | 1.10 ± 0.00 h | 0.04 ± 0.00 c | 0.86 ± 0.01 f | Class II |
| 18H | 156.00 ± 1.00 | 20.00 ± 0.06 f | 8.30 ± 0.06 ab | 7.90 ± 0.10 e | 271.00 ± 0.58 b | 0.02 ± 0.00 cd | 1.17 ± 0.00 f | 1.04 ± 0.00 g | 0.09 ± 0.00 f | 0.62 ± 0.01 e | Class II |
| 18T | 156.00 ± 1.00 | 18.20 ± 0.10 e | 8.20 ± 0.10 a | 4.50 ± 0.10 a | 281.00 ± 1.00 d | 0.01 ± 0.00 abc | 1.09 ± 0.00 e | 0.95 ± 0.00 e | 0.09 ± 0.01 fg | 1.20 ± 0.10 g | Class II |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, B.-H.; Chen, Z.-J.; Li, Y.-Y.; Fohrer, N.; Zhang, Y.; Wu, D.-Y.; Yan, X.-Y.; Li, B.-L. Structural Characteristics and Driving Factors of the Planktonic Eukaryotic Community in the Danjiangkou Reservoir, China. Water 2020, 12, 3499. https://doi.org/10.3390/w12123499
Zheng B-H, Chen Z-J, Li Y-Y, Fohrer N, Zhang Y, Wu D-Y, Yan X-Y, Li B-L. Structural Characteristics and Driving Factors of the Planktonic Eukaryotic Community in the Danjiangkou Reservoir, China. Water. 2020; 12(12):3499. https://doi.org/10.3390/w12123499
Chicago/Turabian StyleZheng, Bao-Hai, Zhao-Jin Chen, Yu-Ying Li, Nicola Fohrer, Yun Zhang, Dong-Yu Wu, Xue-Yan Yan, and Bai-Lian Li. 2020. "Structural Characteristics and Driving Factors of the Planktonic Eukaryotic Community in the Danjiangkou Reservoir, China" Water 12, no. 12: 3499. https://doi.org/10.3390/w12123499





