Role of Homologous Recombination Genes in Repair of Alkylation Base Damage by Candida albicans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. DNA Extraction and Analysis and Cell Transformation
2.3. Sensitivity to DNA-Damaging Agents
2.4. Generation of MMS-Induced Heat Labile Breaks
2.5. Calculations of Chromosome Fragments Sizes from Closely Spaced Single-Strand Breaks
3. Results
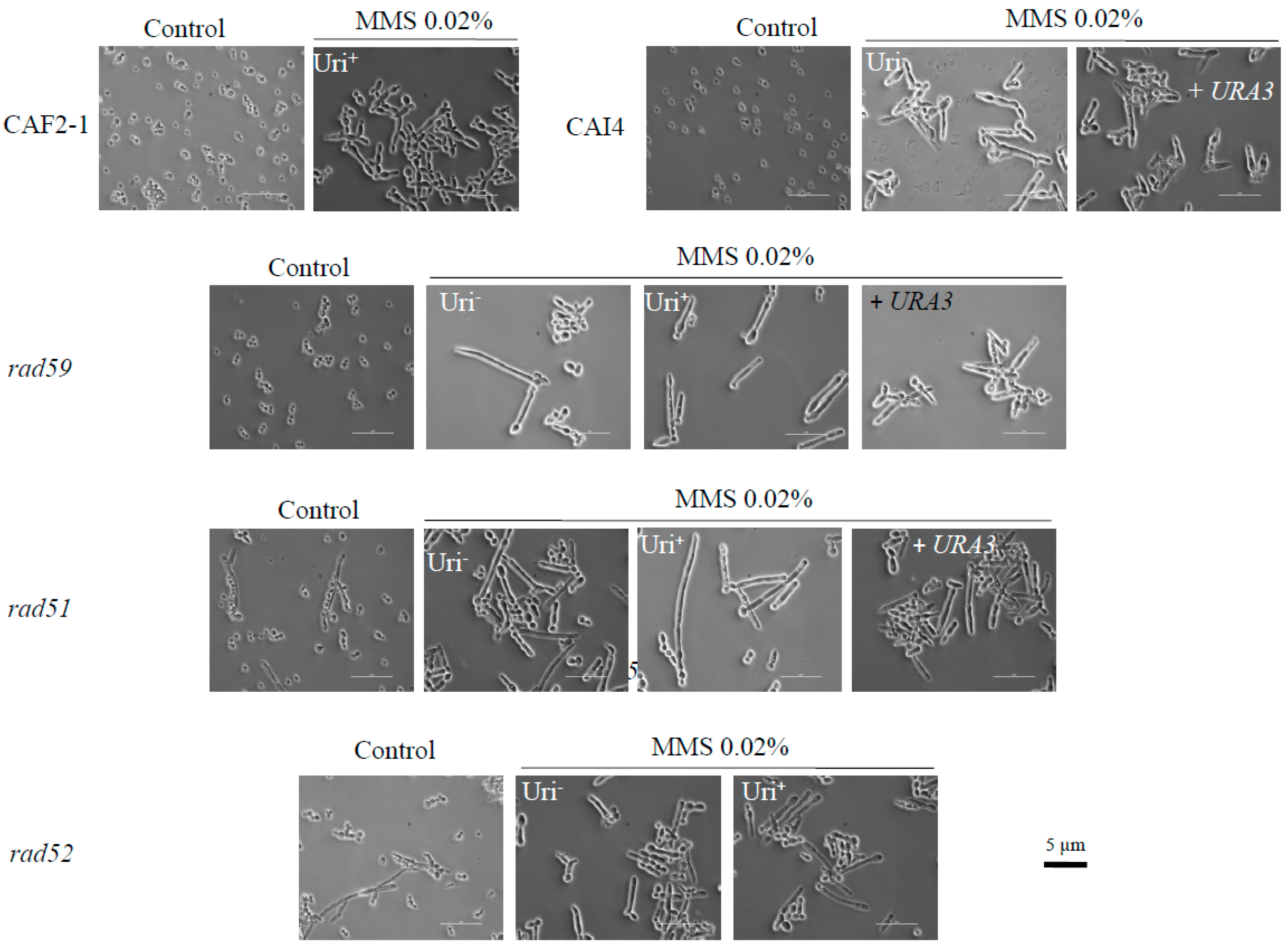
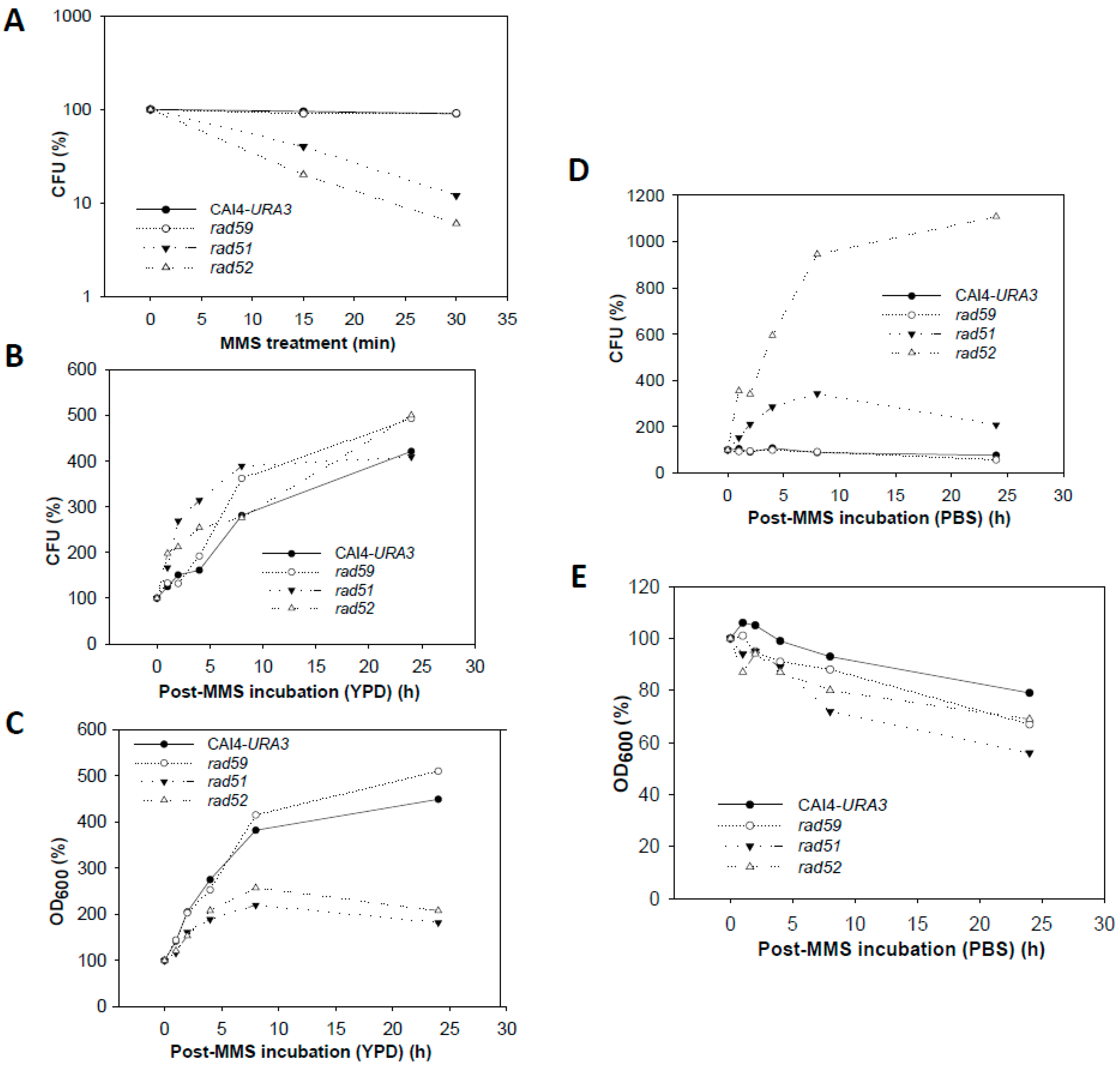
3.1. The Role of HR Genes in Growth Polarization in Response to MMS
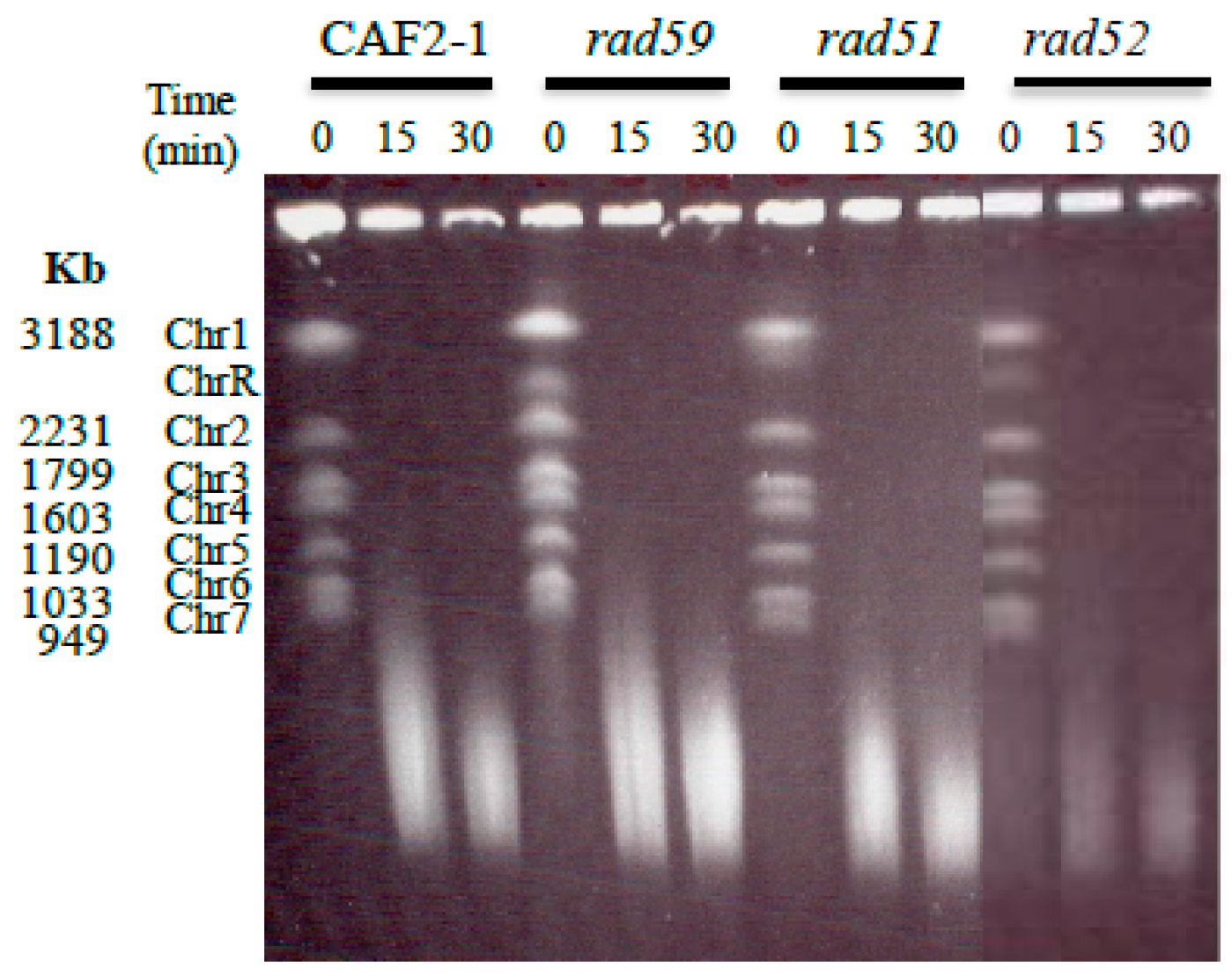
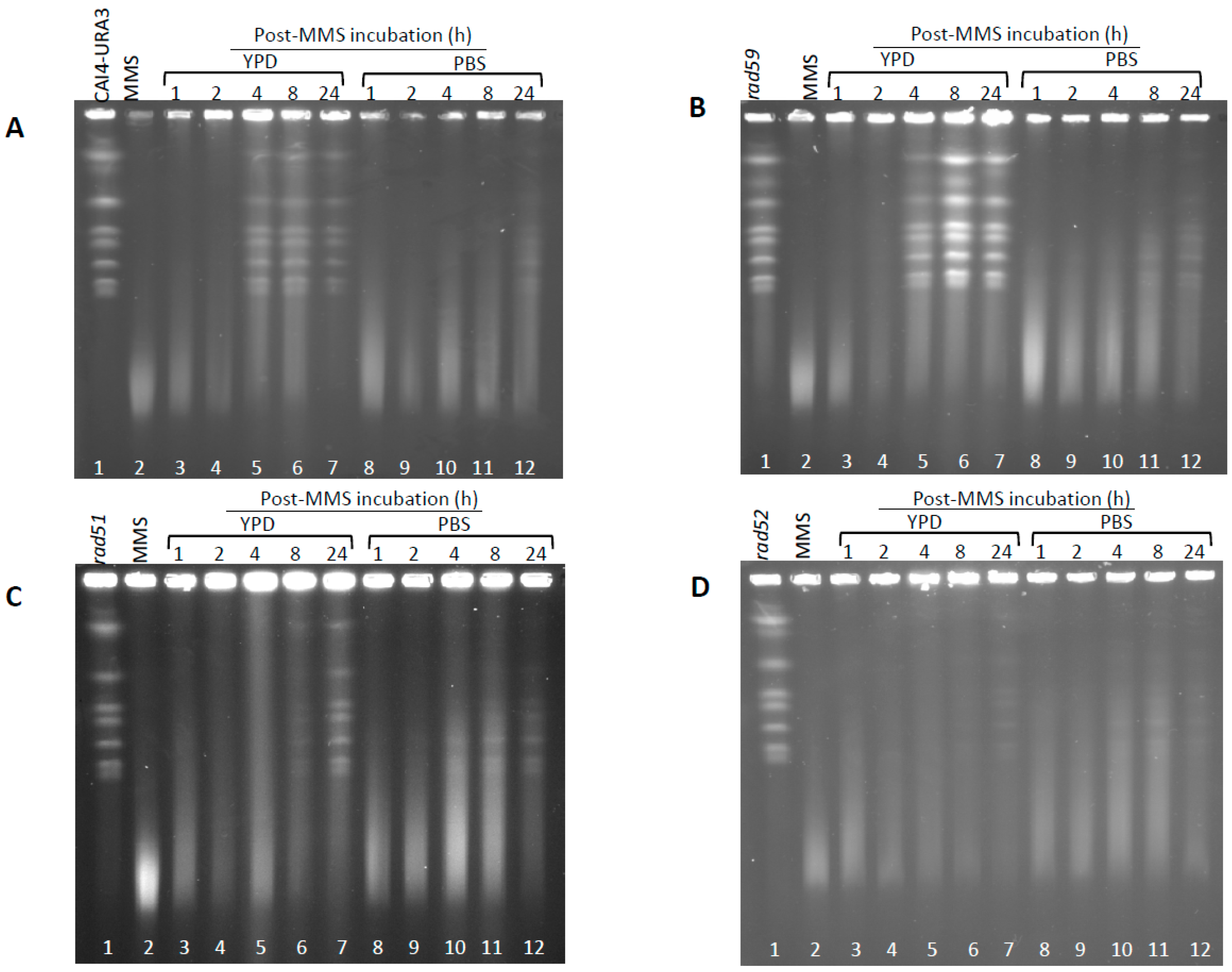
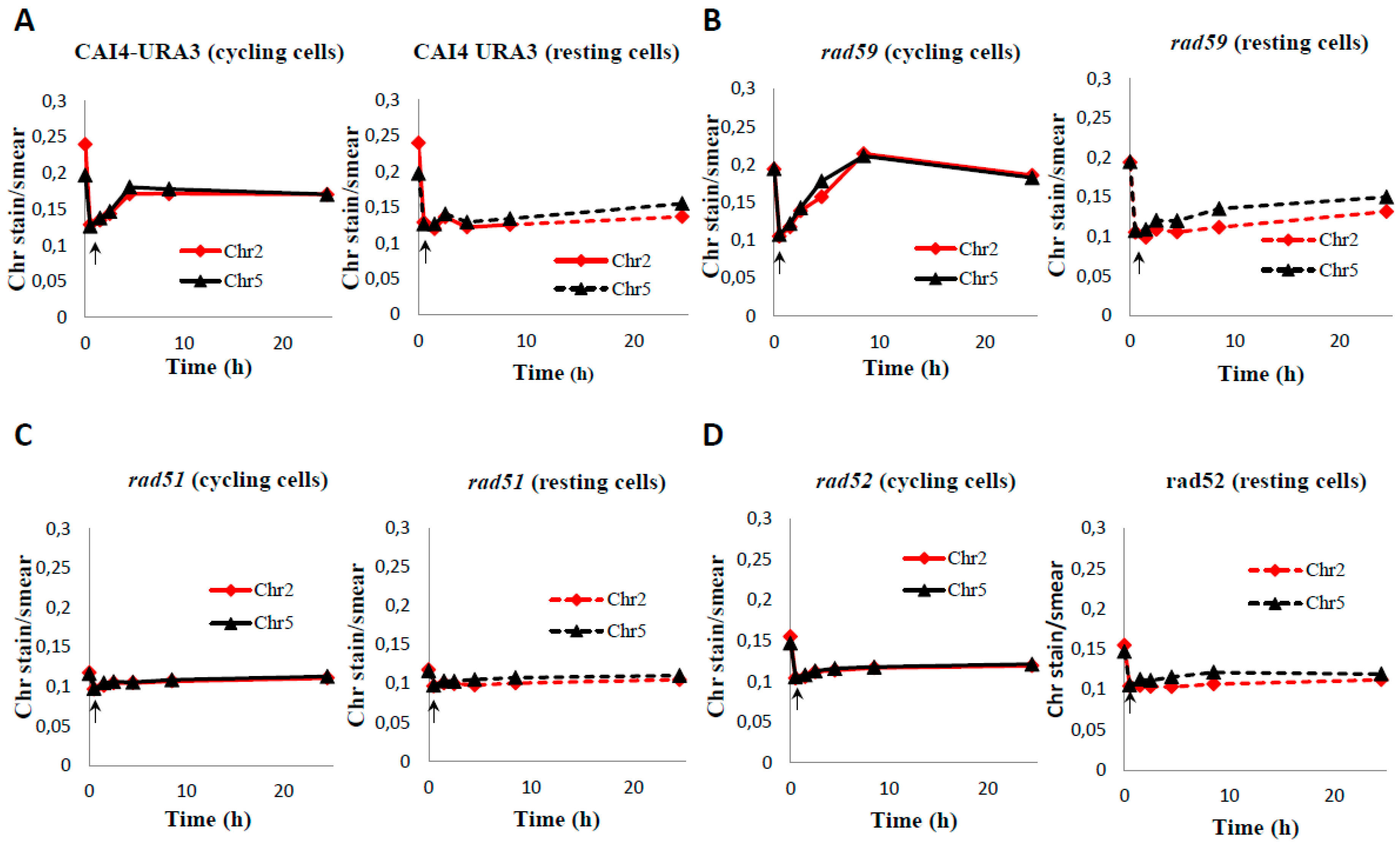
3.2. Generation and Repair of MMS-Induced Heat Labile Lesions on Growing C. albicans Wild Type Cells
3.3. Role of HR on the Repair of HDB
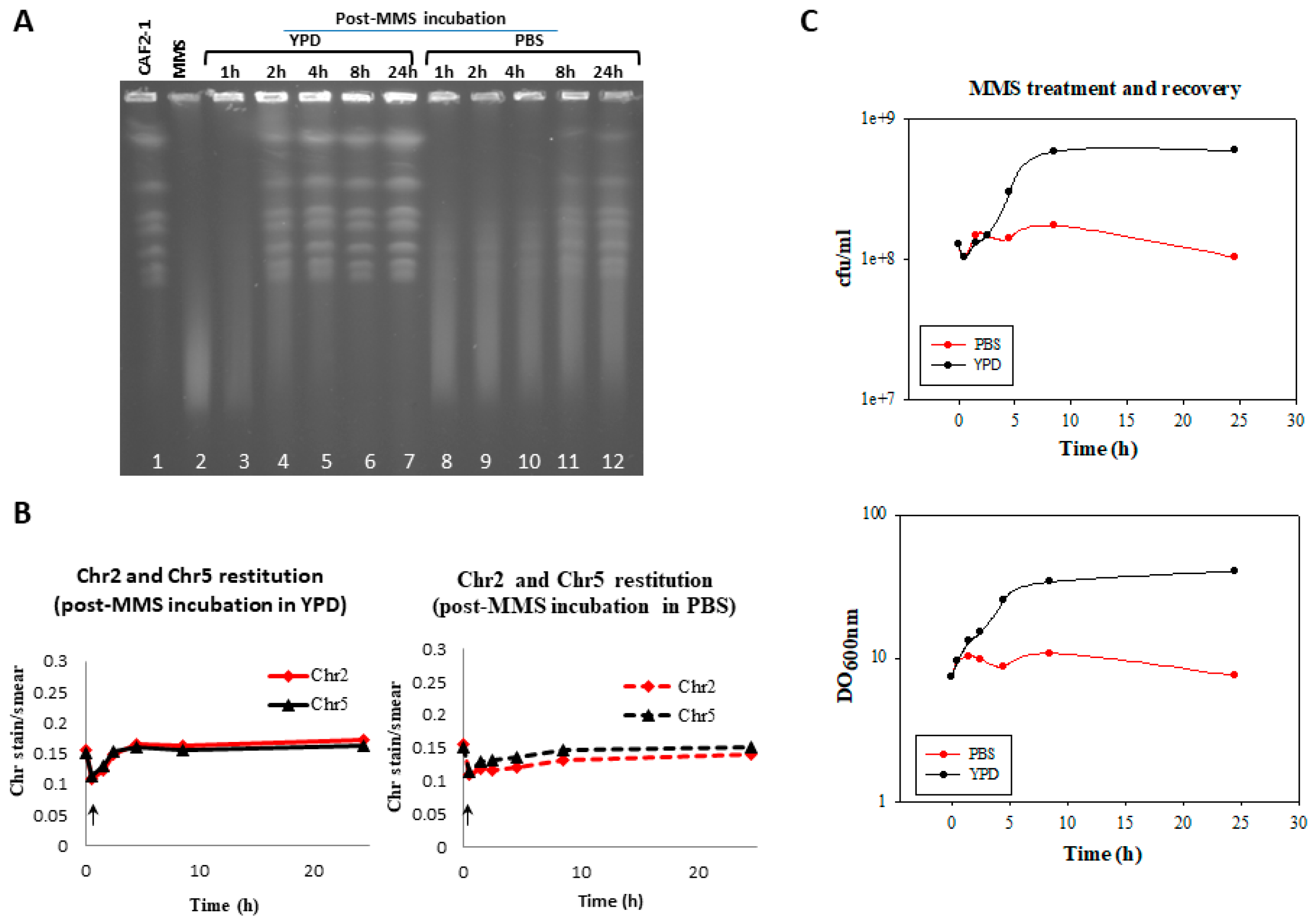
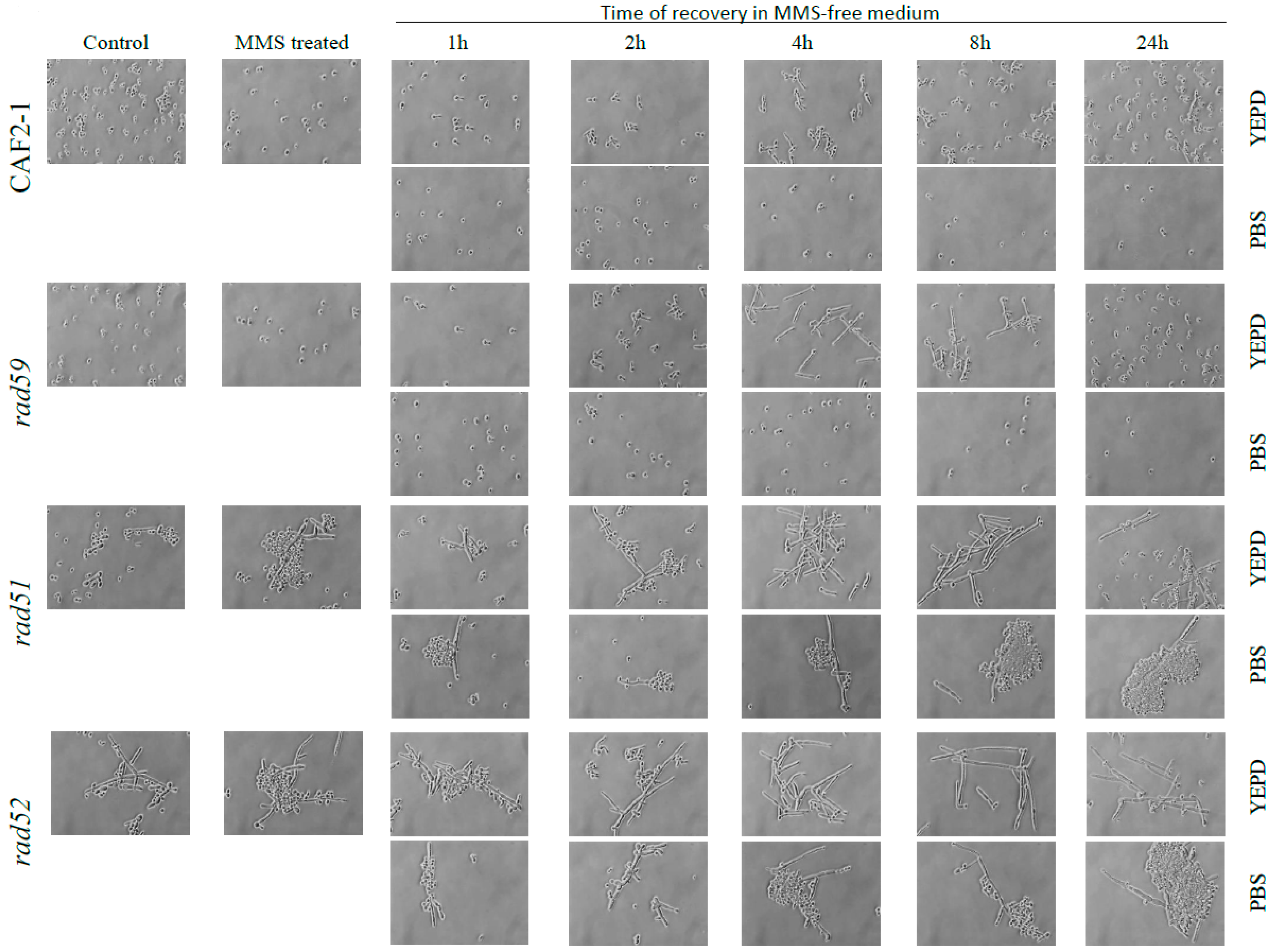
3.4. Effect of the MMS Treatment on Cell Number and Morphology during the Post-Incubation Recovery of Mutants Populations
4. Discussion
4.1. Role of HR in Repair of HDB by Proliferating C. Albicans Cells
4.2. Role of HR in Repair of HDB by Resting C. albicans Cells
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boiteux, S.; Jinks-Robertson, S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 2013, 193, 1025–1064. [Google Scholar] [CrossRef] [PubMed]
- Budzowska, M.; Kanaar, R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009, 53, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Westmoreland, J.W.; Gordenin, D.A.; Resnick, M.A. Alkylation base damage is converted into repairable double-strand breaks and complex intermediates in G2 cells lacking AP endonuclease. PLoS Genet. 2011, 7, e1002059. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, B.; Bates, P.A.; Paik, J.; Jacobs, S.C.; Lindahl, T. Repair of alkylated DNA: Recent advances. DNA Repair 2007, 6, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Resnick, M.A.; Gordenin, D.A. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008, 36, 1836–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legrand, M.; Chan, C.L.; Jauert, P.A.; Kirkpatrick, D.T. Analysis of base excision and nucleotide excision repair in Candida albicans. Microbiology 2008, 154, 2446–2456. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, M.D.; Pittman, D.L. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006, 19, 1580–1594. [Google Scholar] [CrossRef] [PubMed]
- Valenti, A.; Napoli, A.; Ferrara, M.C.; Nadal, M.; Rossi, M.; Ciaramella, M. Selective degradation of reverse gyrase and DNA fragmentation induced by alkylating agent in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2006, 34, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Andersson, A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 1972, 11, 3618–3623. [Google Scholar] [CrossRef] [PubMed]
- Lundin, E.; North, M.; Erixon, K.; Walters, K.; Jenssen, D.; Goldman, A.S.; Helleday, T. Methyl methane sulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005, 33, 3799–3811. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Panduri, V.; Sterling, J.F.; Van Houten, B.; Gordenin, D.A.; Resnick, M.A. The transition of closely opposed lesions to double-strand breaks during long-patch base excision repair is prevented by the coordinated action of DNA polymerase delta and Rad27/Fen1. Mol. Cell. Biol. 2009, 29, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Rothstein, R.; Lisby, M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 2014, 198, 795–835. [Google Scholar] [CrossRef] [PubMed]
- Begley, T.J.; Rosenbach, A.S.; Ideker, T.; Samson, L.D. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer Res. 2002, 1, 103–112. [Google Scholar] [PubMed]
- Vázquez, M.V.; Rojas, V.; Tercero, J.A. sMultiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair 2008, 7, 1693–1704. [Google Scholar] [CrossRef] [PubMed]
- Tercero, J.A.; Diffley, J.F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 2001, 412, 553–557. [Google Scholar] [CrossRef] [PubMed]
- González-Prieto, R.; Muñoz-Cabello, A.M.; Cabello-Lobato, M.J.; Prado, F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013, 32, 1307–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legrand, M.; Chan, C.L.; Jauert, P.A.; Kirkpatrick, D.T. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot. Cell 2007, 6, 2194–2205. [Google Scholar] [CrossRef] [PubMed]
- Drablos, F.; Feyzi, E.; Aas, P.A.; Vaagbø, C.B.; Kavli, B.; Bratlie, M.S.; Peña-Diaz, J.; Otterlei, M.; Slupphaug, G.; Krokan, H.E. Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair 2004, 3, 1389–1407. [Google Scholar] [CrossRef] [PubMed]
- Larson, K.; Sahm, J.; Shenkar, R.; Strauss, B. Methylation-induced blocks to in vitro DNA replication. Mutat. Res. 1985, 150, 77–84. [Google Scholar] [CrossRef]
- García-Prieto, F.; Gómez-Raja, J.; Andaluz, E.; Calderone, R.; Larriba, G. Role of the homologous recombination genes RAD51 and RAD59 in the resistance of Candida albicans to UV light, radiomimetic and anti-tumor compounds and oxidizing agents. Fungal Genet. Biol. 2010, 47, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Bellido, A.; Andaluz, E.; Gómez-Raja, J.; Álvarez-Barrientos, A.; Larriba, G. Genetic interactions among homologous recombination mutants in Candida albicans. Fungal Genet. Biol. 2015, 74, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Andaluz, E.; Ciudad, T.; Gómez-Raja, J.; Calderone, R.; Larriba, G. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 2006, 59, 1452–1472. [Google Scholar] [CrossRef] [PubMed]
- Bärtsch, S.; Kang, L.E.; Symington, L.S. RAD51 is required for the repair of plasmid double-stranded gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 2000, 20, 1194–1205. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, C.; Fink, G.R. Guide to Yeast Genetics and Molecular Biology; Academic Press: San Diego, CA, USA, 1991. [Google Scholar]
- Gillum, A.M.; Tsay, E.Y.; Kirsch, D.R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984, 198, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Fonzi, W.A.; Irwin, M.Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics 1993, 134, 717–728. [Google Scholar]
- Ciudad, T.; Andaluz, E.; Steinberg-Neifach, O.; Lue, N.F.; Gow, N.A.; Calderone, R.A.; Larriba, G. Homologous recombination in Candida albicans: Role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 2004, 53, 1177–1194. [Google Scholar] [CrossRef] [PubMed]
- Andaluz, E.; Gómez-Raja, J.; Hermosa, B.; Ciudad, T.; Rustchenko, E.; Calderone, R.; Larriba, G. Loss and fragmentation of chromosome 5 are major events linked to the adaptation of rad52-DeltaDelta strains of Candida albicans to sorbose. Fungal Genet. Biol. 2007, 44, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Andaluz, E.; Bellido, A.; Gómez-Raja, J.; Selmecki, A.; Bouchonville, K.; Calderone, R.; Berman, J.; Larriba, G. Rad52 function prevents chromosome loss and truncation in Candida albicans. Mol. Microbiol. 2011, 79, 1462–1482. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.; Wendland, J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 2003, 42, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.M.; Johnson, A.D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 2005, 4, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.M.; Wang, Y.M.; Zheng, X.D.; Lee, R.T.; Wang, Y. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 2007, 18, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Legrand, M.; Chan, C.L.; Jauert, P.A.; Kirkpatrick, D.T. The contribution of the S-phase checkpoint genes MEC1 and SGS1 to genome stability maintenance in Candida albicans. Fungal Genet. Biol. 2011, 48, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Ciudad, T.; Hickman, M.; Bellido, A.; Berman, J.; Larriba, G. Phenotypic consequences of a spontaneous loss of heterozygosity in a common laboratory strain of Candida albicans. Genetics 2016, 203, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Covo, S.; Westmoreland, J.W.; Gordenin, D.A.; Resnick, M.A. Cohesin is limiting for the suppression of DNA damage-induced recombination between homologous chromosomes. PLoS Genet. 2010, 6, e1001006. [Google Scholar] [CrossRef] [PubMed]
- Redon, C.; Pilch, D.R.; Rogakou, E.P.; Orr, A.H.; Lowndes, N.F.; Bonner, W.M. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 2003, 4, 678–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auerbach, P.A.; Demple, B. Roles of Rev1, Pol zeta, Pol32 and Pol eta in the bypass of chromosomal abasic sites in Saccharomyces cerevisiae. Mutagenesis 2010, 25, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Calvo, J.A.; Samson, L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 2013, 12, 104–120. [Google Scholar] [CrossRef] [PubMed]
| Strains (Old Name) | Genotype | Parental | Reference |
|---|---|---|---|
| Candida albicans | |||
| SC5314 | Wild type | Gillum et al., 1984 [26] | |
| CAF2-1 | Δura3::imm434/URA3 | SC5314 | Fonzi and Irwin, 1993 [27] |
| CAI4 | Δura3::imm434/Δura3::imm434 | CAF2-1 | Fonzi and Irwin, 1993 [27] |
| CAGL4 (TCR2.1) | Δura3::imm434/Δura3::imm434 rad52::hisG/Δrad52::hisG-URA3-hisG | CAI4 | Ciudad et al., 2004 [28] |
| CAGL4.1 (TCR2.1.1) | Δura3::imm434/Δura3::imm434 Δrad52::hisG/Δrad52::hisG | CAGL4 | Ciudad et al., 2004 [28] |
| CAGL17 (BNC1.1) | Δura3::imm434/Δura3::imm434 Δrad59::hisG/Δrad59::hisG-URA3-hisG | CAI4 | García-Prieto et al., 2010 [21] |
| CAGL17.1 (BNC23.1) | Δura3::imm434/Δura3::imm434 Δrad59::hisG/Δrad59::hisG | CAGL17 | Bellido et al., 2015 [22] |
| CAGL19 (JGR5) | Δura3::imm434/Δura3::imm434 Δrad51::hisG/Δrad51::hisG-URA3-hisG | CAI4 | García-Prieto et al., 2010 [21] |
| CAGL19.1 (JGR5A) | Δura3::imm434/Δura3::imm434 Δrad51::hisG/Δrad51::hisG | CAGL19 | García-Prieto et al., 2010 [21] |
| S. cerevisiae | |||
| LSY0695-7D W303 haploid | MATa; ADE2 RAD5 met17-s ade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 | Bärtsch et al., 2000 [24] | |
| W303 diploid | MATa/α; ADE2 RAD5 met17-s/met17-s ade2-1/ade2-1 trp1-1/trp1-1 his3-11,15/his3-11,15 can1-100/can1-100 ura3-1/ura3-1 leu2-3,112/leu2-3,112 | Bellido et al., 2015 [22] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciudad, T.; Bellido, A.; Andaluz, E.; Hermosa, B.; Larriba, G. Role of Homologous Recombination Genes in Repair of Alkylation Base Damage by Candida albicans. Genes 2018, 9, 447. https://doi.org/10.3390/genes9090447
Ciudad T, Bellido A, Andaluz E, Hermosa B, Larriba G. Role of Homologous Recombination Genes in Repair of Alkylation Base Damage by Candida albicans. Genes. 2018; 9(9):447. https://doi.org/10.3390/genes9090447
Chicago/Turabian StyleCiudad, Toni, Alberto Bellido, Encarnación Andaluz, Belén Hermosa, and Germán Larriba. 2018. "Role of Homologous Recombination Genes in Repair of Alkylation Base Damage by Candida albicans" Genes 9, no. 9: 447. https://doi.org/10.3390/genes9090447





