Can Genetic Factors Compromise the Success of Dental Implants? A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Methodology
2.1. Standardized Criteria and Type of Study
2.2. Registry Protocol
2.3. Eligibility Criteria
2.4. Inclusion/Exclusion Criteria and Cohort Size
2.4.1. Inclusion Criteria
2.4.2. Exclusion Criteria
2.5. Search Strategy
2.6. Data Collection Process
2.7. Items of Extracted Data
2.8. Evaluation of the Study Quality and Risk of Bias
2.9. Measurements and Statistical Analysis
2.10. Anticipated Outcome
2.10.1. Primary Outcome
2.10.2. Risk of Bias of Quantitative Data
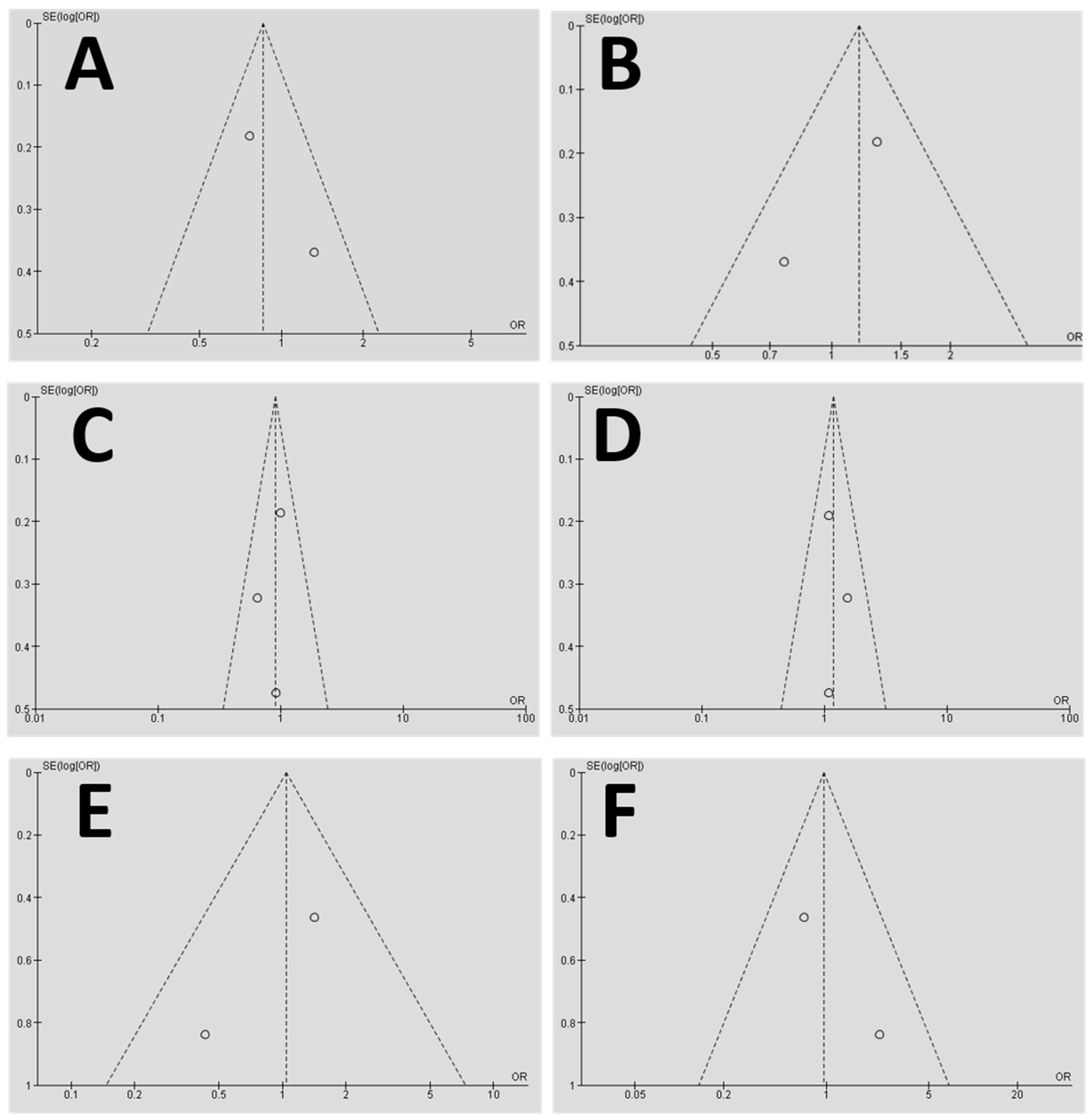
2.10.3. Additional Analysis
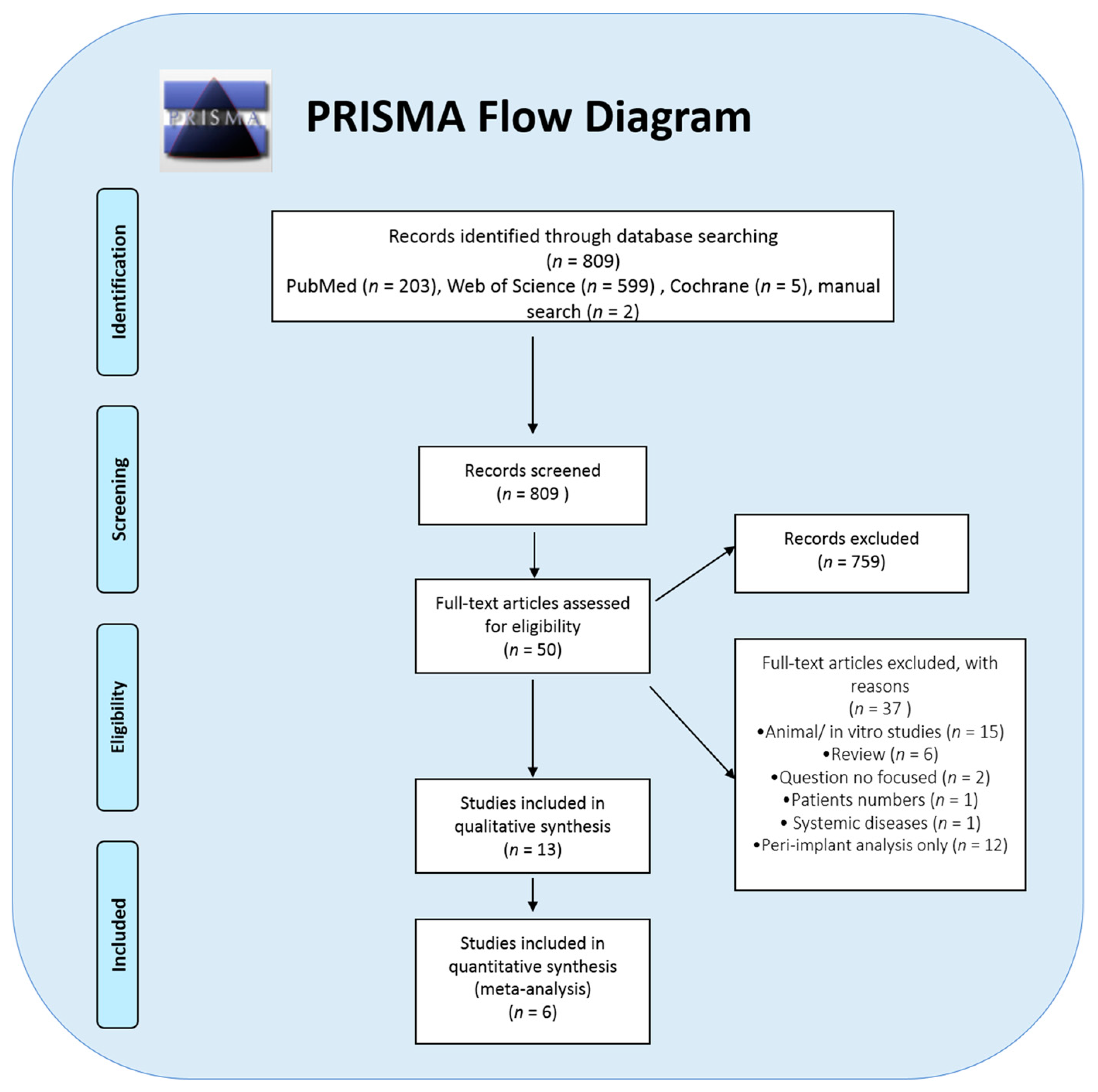
3. Results
3.1. Clinical Parameters
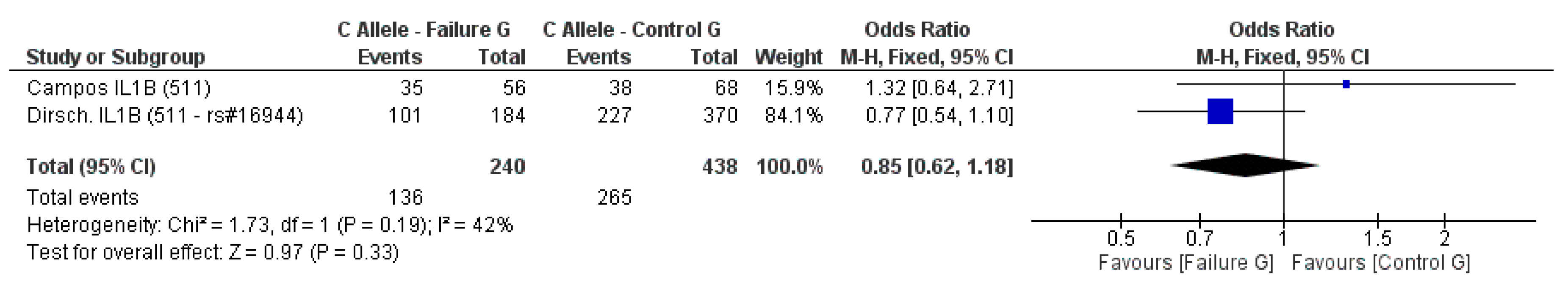
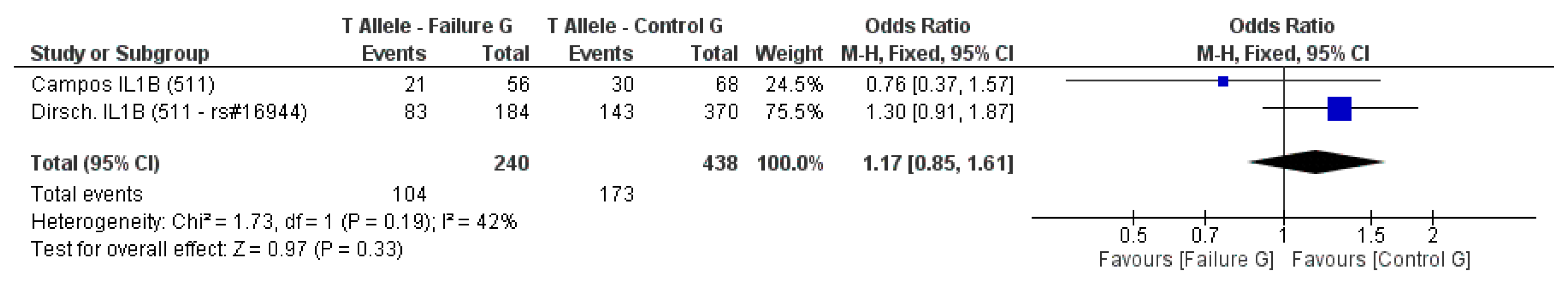
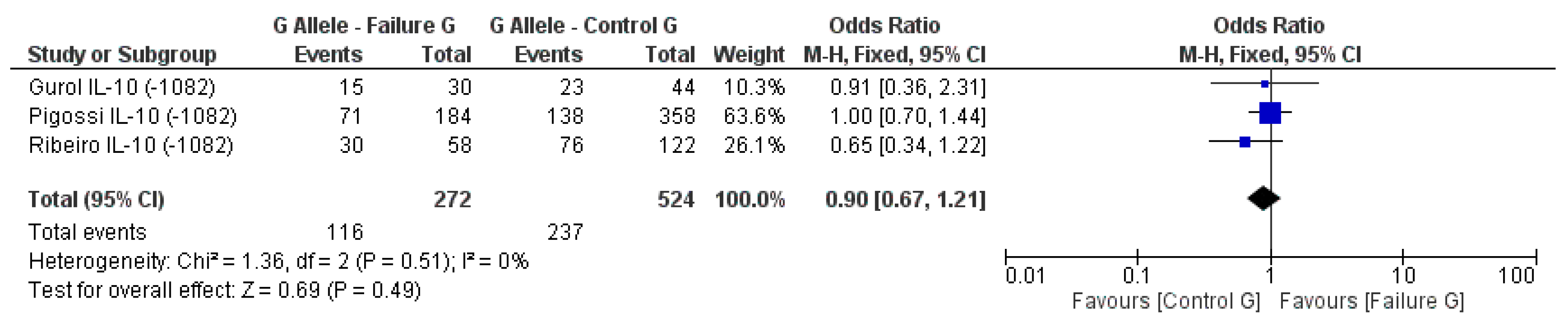
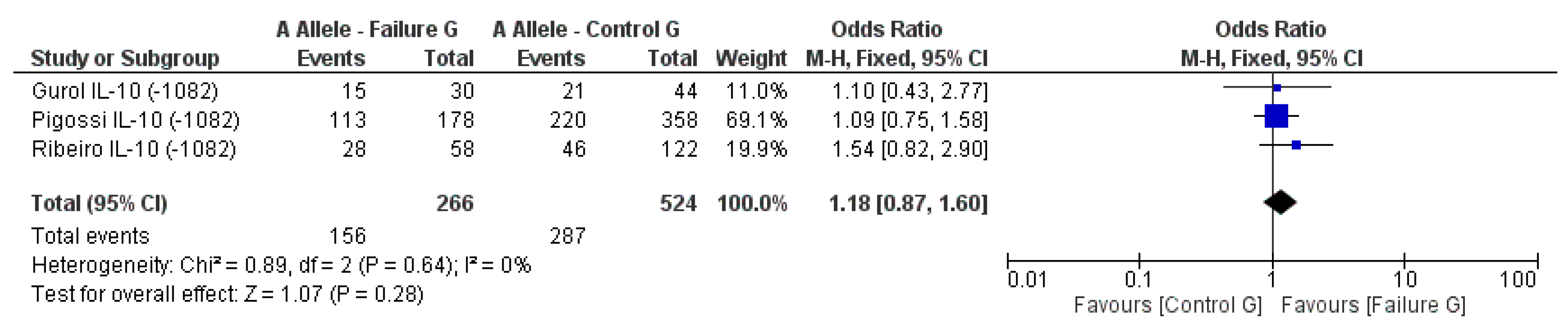
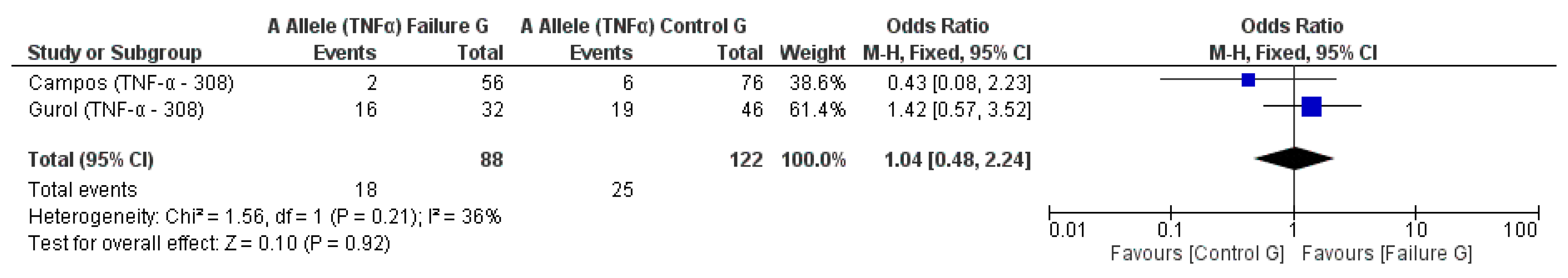
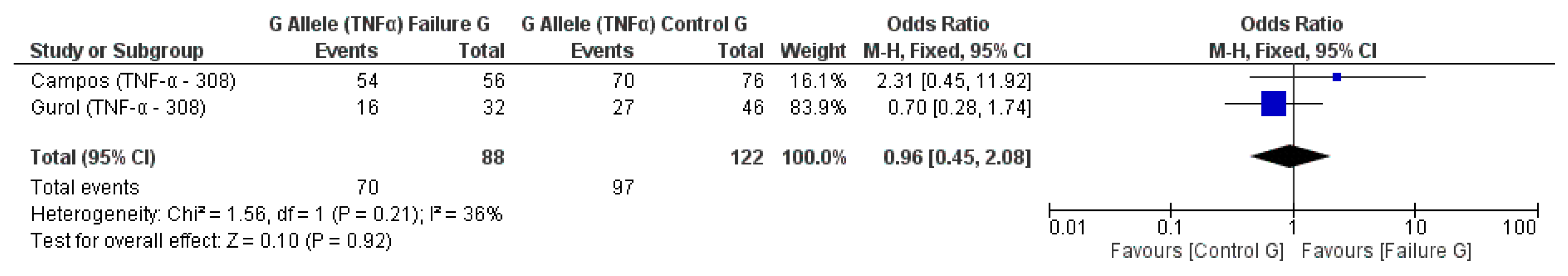
3.2. Meta-Analysis Outcome
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Campos, M.I.; Godoy dos Santos, M.C.; Trevilatto, P.C.; Scarel-Caminaga, R.M.; Bezerra, F.J.; Line, S.R. Interleukin-2 and interleukin-6 gene promoter polymorphisms, and early failure of dental implants. Implant Dent. 2005, 14, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; McInnes, R.; Willard, H. Genes in population. In Genetics in Medicine, 6th ed.; W.S. Company: Philadelphia, PA, USA, 2001; p. 540. [Google Scholar]
- Costa-Junior, F.R.; Alvim-Pereira, C.C.; Alvim-Pereira, F.; Trevilatto, P.C.; de Souza, A.P.; Santos, M.C. Influence of MMP-8 promoter polymorphism in early osseointegrated implant failure. Clin. Oral Investig. 2013, 17, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Dirschnabel, A.J.; Alvim-Pereira, F.; Alvim-Pereira, C.C.; Bernardino, J.F.; Rosa, E.A.; Trevilatto, P.C. Analysis of the association of IL1B(C-511T) polymorphism with dental implant loss and the clusterization phenomenon. Clin. Oral Implants Res. 2011, 22, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [PubMed] [Green Version]
- Ribeiro, R.; Melo, R.; Tortamano Neto, P.; Vajgel, A.; Souza, P.R.; Cimoes, R. Polymorphisms of IL-10 (-1082) and RANKL (-438) genes and the failure of dental implants. Int. J. Dent. 2017, 2017, 3901368. [Google Scholar] [CrossRef] [PubMed]
- Alvim-Pereira, F.; Montes, C.C.; Thome, G.; Olandoski, M.; Trevilatto, P.C. Analysis of association of clinical aspects and vitamin D receptor gene polymorphism with dental implant loss. Clin. Oral Implants Res. 2008, 19, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, M.C.; Campos, M.I.; Souza, A.P.; Scarel-Caminaga, R.M.; Mazzonetto, R.; Line, S.R. Analysis of the transforming growth factor-β1 gene promoter polymorphisms in early osseointegrated implant failure. Implant Dent. 2004, 13, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Mombelli, A.; Cionca, N. Systemic diseases affecting osseointegration therapy. Clin. Oral Implants Res. 2006, 17 (Suppl. 2), 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrektsson, T.; Dahlin, C.; Jemt, T.; Sennerby, L.; Turri, A.; Wennerberg, A. Is marginal bone loss around oral implants the result of a provoked foreign body reaction? Clin. Implant Dent. Relat. Res. 2014, 16, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Gaetti-Jardim, E.C.; Santiago-Junior, J.F.; Goiato, M.C.; Pellizer, E.P.; Magro-Filho, O.; Jardim, E.G. Dental implants in patients with osteoporosis: A clinical reality? J. Craniofac. Surg. 2011, 22, 1111–1113. [Google Scholar] [CrossRef] [PubMed]
- Goiato, M.C.; dos Santos, D.M.; Santiago, J.F.J.; Moreno, A.; Pellizzer, E.P. Longevity of dental implants in type IV bone: A systematic review. Int. J. Oral Maxillofac. Surg. 2014, 43, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- De Medeiros, F.; Kudo, G.A.H.; Leme, B.G.; Saraiva, P.P.; Verri, F.R.; Honorio, H.M.; Pellizzer, E.P.; Santiago Junior, J.F. Dental implants in patients with osteoporosis: A systematic review with meta-analysis. Int. J. Oral Maxillofac. Surg. 2017. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.; Hirsch, J.M.; Lekholm, U.; Thomsen, P. Biological factors contributing to failures of osseointegrated oral implants. (I). Success criteria and epidemiology. Eur. J. Oral Sci. 1998, 106, 527–551. [Google Scholar] [CrossRef] [PubMed]
- Gurol, C.; Kazazoglu, E.; Dabakoglu, B.; Korachi, M. A comparative study of the role of cytokine polymorphisms interleukin-10 and tumor necrosis factor alpha in susceptibility to implant failure and chronic periodontitis. Int. J. Oral Maxillofac. Implants 2011, 26, 955–960. [Google Scholar] [PubMed]
- Takashiba, S.; Naruishi, K. Gene polymorphisms in periodontal health and disease. Periodontology 2000 2006, 40, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Pigossi, S.C.; Alvim-Pereira, F.; Alvim-Pereira, C.C.; Trevilatto, P.C.; Scarel-Caminaga, R.M. Association of interleukin 4 gene polymorphisms with dental implant loss. Implant Dent. 2014, 23, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Vaz, P.; Gallas, M.M.; Braga, A.C.; Sampaio-Fernandes, J.C.; Felino, A.; Tavares, P. IL1 gene polymorphisms and unsuccessful dental implants. Clin. Oral Implants Res. 2012, 23, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Cavalla, F.; Araujo-Pires, A.C.; Biguetti, C.C.; Garlet, G.P. Cytokine networks regulating inflammantion and immune defense in the oral cavity. Curr. Oral Health Rep. 2014, 1, 104–113. [Google Scholar] [CrossRef]
- Campos, M.I.; dos Santos, M.C.; Trevilatto, P.C.; Scarel-Caminaga, R.M.; Bezerra, F.J.; Line, S.R. Early failure of dental implants and TNF-alpha (G-308A) gene polymorphism. Implant Dent. 2004, 13, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Montes, C.C.; Alvim-Pereira, F.; de Castilhos, B.B.; Sakurai, M.L.; Olandoski, M.; Trevilatto, P.C. Analysis of the association of IL1B (C+3954T) and IL1RN (intron 2) polymorphisms with dental implant loss in a Brazilian population. Clin. Oral Implants Res. 2009, 20, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.; Green, S. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0; The Cochrane Collaboration: London, UK, 2011; Available online: www.cochrane-handbook.org (accessed on 5 September 2018).
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Santiago Junior, J.F.; de Souza Batista, V.E.; Verri, F.R.; Honorio, H.M.; de Mello, C.C.; Almeida, D.A.d.; Pellizzer, E.P. Platform-switching implants and bone preservation: A systematic review and meta-analysis. Int. J. Oral Maxillofac. Surg. 2016, 45, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Lopes, L.F.d.P.; da Silva, V.F.; Santiago, J.F., Jr.; Panzarini, S.R.; Pellizzer, E.P. Placement of dental implants in the maxillary tuberosity: A systematic review. Int. J. Oral Maxillofac. Surg. 2015, 44, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Lemos, C.A.; Verri, F.R.; de Souza Batista, V.E.; Santiago Junior, J.F.; Mello, C.C.; Pellizzer, E.P. Complete overdentures retained by mini implants: A systematic review. J. Dent. 2017, 57, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Lemos, C.A.; de Souza Batista, V.E.; Almeida, D.A.; Santiago Junior, J.F.; Verri, F.R.; Pellizzer, E.P. Evaluation of cement-retained versus screw-retained implant-supported restorations for marginal bone loss: A systematic review and meta-analysis. J. Prosthet. Dent. 2015, 115, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Lemos, C.A.A.; Verri, F.R.; Bonfante, E.A.; Santiago Junior, J.F.; Pellizzer, E.P. Comparison of external and internal implant-abutment connections for implant supported prostheses. A systematic review and meta-analysis. J. Dent. 2017, 70, 14–22. [Google Scholar] [CrossRef] [PubMed]
- NHMRC. National Health and Medical Research Council. How to Use the Evidence: Assessment and Application of Scientific Evidence. Available online: https://www.nhmrc.gov.au/_files_nhmrc/publications/attachments/cp69.pdf (accessed on 5 September 2018).
- Higgins, J.P.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef] [PubMed]
- Atieh, M.A.; Ibrahim, H.M.; Atieh, A.H. Platform switching for marginal bone preservation around dental implants: A systematic review and meta-analysis. J. Periodontol. 2010, 81, 1350–1366. [Google Scholar] [CrossRef] [PubMed]
- Annibali, S.; Bignozzi, I.; Sammartino, G.; La Monaca, G.; Cristalli, M.P. Horizontal and vertical ridge augmentation in localized alveolar deficient sites: A retrospective case series. Implant Dent. 2012, 21, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.I.; Santos, M.C.; Trevilatto, P.C.; Scarel-Caminaga, R.M.; Bezerra, F.J.; Line, S.R. Evaluation of the relationship between interleukin-1 gene cluster polymorphisms and early implant failure in non-smoking patients. Clin. Oral Implants Res. 2005, 16, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Pigossi, S.C.; Alvim-Pereira, F.; Montes, C.C.; Finoti, L.S.; Secolin, R.; Trevilatto, P.C.; Scarel-Caminaga, R.M. Genetic association study between interleukin 10 gene and dental implant loss. Arch. Oral Biol. 2012, 57, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- De Souza Batista, V.E.; Santiago Junior, J.F.; de Faria Almeida, D.A.; de Toledo Piza Lopes, L.F.; Verri, F.R.; Pellizzer, E.P. The effect of offset implant configuration on bone stress distribution: A systematic review. J. Prosthodont.-Implant Esthet. Reconstr. Dent. 2015, 24, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Goiato, M.C.; Pellizzer, E.P.; Moreno, A.; Gennari-Filho, H.; dos Santos, D.M.; Santiago, J.F., Jr.; dos Santos, E.G. Implants in the zygomatic bone for maxillary prosthetic rehabilitation: A systematic review. Int. J. Oral Maxillofac. Surg. 2014, 43, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Santiago Junior, J.F.; Verri, F.R.; de Faria Almeida, D.A.; de Souza Batista, V.E.; Araujo Lemos, C.A.; Pellizzer, E.P. Finite element analysis on influence of implant surface treatments, connection and bone types. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 63, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Pietruski, J.K.; Pietruska, M.D.; Stokowska, W.; Pattarelli, G.M. Serum levels of interleukin-1 (IL-1), interleukin-6 (IL-6) and interleukin-8 (IL-8) in patients treated with dental implants. Rocz. Akad. Med. Bialymstoku 2001, 46, 28–37. [Google Scholar]
- Panagakos, F.S.; Aboyoussef, H.; Dondero, R.; Jandinski, J.J. Detection and measurement of inflammatory cytokines in implant crevicular fluid: A pilot study. Int. J. Oral Maxillofac. Implants 1996, 11, 794–799. [Google Scholar] [PubMed]
- Salcetti, J.M.; Moriarty, J.D.; Cooper, L.F.; Smith, F.W.; Collins, J.G.; Socransky, S.S.; Offenbacher, S. The clinical, microbial, and host response characteristics of the failing implant. Int. J. Oral Maxillofac. Implants 1997, 12, 32–42. [Google Scholar] [PubMed]
- Duarte, P.M.; Serrao, C.R.; Miranda, T.S.; Zanatta, L.C.; Bastos, M.F.; Faveri, M.; Figueiredo, L.C.; Feres, M. Could cytokine levels in the peri-implant crevicular fluid be used to distinguish between healthy implants and implants with peri-implantitis? A systematic review. J. Periodontal Res. 2016, 51, 689–698. [Google Scholar] [CrossRef] [PubMed]
| Selected Study | Type of Study | Study Place | Analyzed Variable | Results (Association of Genetic Factors on Implant Failure) |
|---|---|---|---|---|
| Alvim-Pereira et al. 2008 [7] | Prospective | Brazil | Vitamin D Receptor (rs731236) * | n.s. |
| Campos et al. 2005a [1] | Prospective | Brazil | IL-2 (T330G) IL-6 (G174C) * | n.s. |
| Campos et al. 2005b [33] | Prospective | Brazil | IL-1A (−889) IL-1B (3953) IL-1B (−511C/T) IL-RN (intron 2) * | n.s. |
| Campos et al. 2004 [20] | Prospective | Brazil | TNF-α (−308) * | n.s. |
| Costa-Jr et al. 2013 [3] | Prospective, Multicentric | Brazil | MMP-8 (C799T) * | Significant association of MMP-8 with dental implant failure (p = 0.0011) |
| Dirschnabel et al. 2011 [4] | Prospective | Brazil (S) ** | IL1B (−511C/T) * | n.s. |
| Dos Santos et al. 2004 [8] | Prospective | Brazil (SE & NE) ** | Growth factor-β1 (C509T, G800A) | n.s. |
| Gurol et al. 2011 [15] | Prospective | Turkey | IL-10 (−1082A/G, 819, 592); TNF-α (308) | n.s. for IL-10 and TNF-α alleles |
| Montes et al. 2009 [21] | Prospective | Brazil | IL-1B (3954); IL-1RN (intron 2) | n.s. for genotype and allele frequencies of IL1B and IL1RN $ |
| Pigossi et al. 2012 [34] | Prospective | Brazil | IL-10 (−1082A, −819, −519) | n.s. for dental implants loss with genotypes (p > 0.05) |
| Pigossi et al. 2014 [17] | Prospective | Brazil | IL-4 (−590C/T; 33C/T) | Significant association of IL-4 C allele with implant loss (p = 0.0236, OR = 1.61, CI = 1.1–2.4). |
| Ribeiro et al. 2017 [6] | Retrospective | Brazil | IL-10 (−1082A/G) RANKL (−438A/G) | n.s. for IL-10 and RANKL alleles |
| Vaz et al. 2012 [18] | Prospective | Portugal | IL1A (−889) IL1B (3953) | Significant association of IL-1A and IL-1B alleles with dental implant failure |
| Selected Studies | No. Patient | Groups | Ave. Age (years) | Implants | Trade Mark | Periodontal Evaluation (Partially Edentulous Patients) |
|---|---|---|---|---|---|---|
| Alvim-Pereira et al. 2008 [7] | 217 | CG: 137 SG: 80 | 51.7 ± 11.3 | CG:1232 SG: 135 | Neodent™ | Gingival Index: 0.64 ± 0.38 (CG), 0.65 ± 0.55 (SG). Plaque Index: 0.14 ± 0.26 (CG), 0.25 ± 0.42 (SG). Calculus Index: 0.08 ± 0.13 (CG), 0.14 ± 0.25 (SG). Probing attachment (mm): 2.68 ± 0.41 (CG), 2.52 ± 0.47 (SG). Clinical attachment (mm): 3.61 ± 0.76 (CG), 3.66 ± 1.10 (SG). Mobility (absence/presence): 98/13 (CG); 60/15 (SG) |
| Campos et al. 2005a [1] | 74 | CG: 40 SG: 34 | 43.8 49.3 | NI | NI | NI |
| Campos et al. 2005b [33] | 72 | CG: 34 SG: 28 | 43.3 52.7 | NI 97 | 3i™/ Conexão™ | NI |
| Campos et al. 2004 [20] | 66 | CG: 38 * SG: 28 | NC | NI | 3™/ Conexão™ | NI |
| Costa-Jr et al. 2013 [3] | 180 | CG:100 ** SG: 80 | >18 | NI | NI | NI |
| Dirschnabel et al. 2011 [4] | 277 | CG:185 * SG: 92 | 53.6 ± 11.1 | NI | NI | Gingival Index: 0.64 ± 0.37 (CG), 0.65 ± 0.53 (SG) Plaque Index: 0.12 ± 0.23 (CG), 0.23 ± 0.41 (SG) Calculus Index: 0.07 ± 0.12 (CG), 0.13 ± 0.24 (SG) Probing attachment (mm): 2.72 ± 0.46 (CG), 2.54 ± 0.47 (SG) Clinical attachment (mm): 3.62 ± 0.85 (CG), 3.66 ± 1.07 (SG) Mobility (absence/presence): 132/19 (CG), 70/15 (SG) |
| Dos Santos et al. 2004 [8] | 68 | CG:40 $ SG: 28 | >18 | NI | 3i™/ Conexão™ | NI |
| Gurol et al. 2011 [15] | 108 | CG: 70 SG: 38 | 25–48 | NI 16 | NI | NI |
| Montes et al. 2009 [21] | 266 | SG: 90 CG: 176 | 51.5 ± 11.5 | 1232 135 | Neodent™ | Gingival Index: 0.63 ± 0.38 (CG) and 0.65 ± 0.53 (SG) Plaque Index: 0.12 ± 0.24 (CG) and 0.24 ± 0.42 (SG) Calculus Index: 0.07 ± 0.12 (CG) and 0.13 ± 0.24 (SG). Probing attachment (mm): 2.72 ± 0.46 (CG) and 2.55 ± 0.47 (SG). Clinical attachment (mm): 3.61 ± 0.85 (CG) and 3.67 ± 1.07 (SG). Dental Mobility 18 (142) (CG); 16 (83) (SG) |
| Pigossi et al. 2012 [34] | 277 | CG: 185 SG: 92 | 53.79 ± 11.3 | NI | Neodent™ | NI |
| Pigossi et al. 2014 [17] | 280 | CG: 186 * SG: 94 | 56.1 ± 11.3 | 1232 135 | Neodent™ | Gingival Index: 0.63 ± 0.38 (CG) and 0.64 ± 0.28 (SG) Plaque Index: 0.12 ± 0.23 (CG) and 0.23 ± 0.41 (SG) Calculus Index: 0.07 ± 0.12 (CG) and 0.13 ± 0.24 (SG). Probing attachment level (mm): 2.72 ± 0.46 (CG) and 2.55 ± 0.47 (SG). Clinical attachment level (mm): 3.61 ± 0.85 (CG) and 3.67 ± 1.07 (SG). Dental Mobility: 19 (12.5) (CG); 16 (18.6) (SG) |
| Ribeiro et al. 2017 [6] | 90 | 1 Group | 54.5 | 245 | Straumann™ | NI |
| Vaz et al. 2012 [18] | 155 | CG: 100 SG: 55 | NI | NI | NI | NI |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago Junior, J.F.; Biguetti, C.C.; Matsumoto, M.A.; Abu Halawa Kudo, G.; Parra da Silva, R.B.; Pinto Saraiva, P.; Fakhouri, W.D. Can Genetic Factors Compromise the Success of Dental Implants? A Systematic Review and Meta-Analysis. Genes 2018, 9, 444. https://doi.org/10.3390/genes9090444
Santiago Junior JF, Biguetti CC, Matsumoto MA, Abu Halawa Kudo G, Parra da Silva RB, Pinto Saraiva P, Fakhouri WD. Can Genetic Factors Compromise the Success of Dental Implants? A Systematic Review and Meta-Analysis. Genes. 2018; 9(9):444. https://doi.org/10.3390/genes9090444
Chicago/Turabian StyleSantiago Junior, Joel Ferreira, Claudia Cristina Biguetti, Mariza Akemi Matsumoto, Guilherme Abu Halawa Kudo, Raquel Barroso Parra da Silva, Patrícia Pinto Saraiva, and Walid D. Fakhouri. 2018. "Can Genetic Factors Compromise the Success of Dental Implants? A Systematic Review and Meta-Analysis" Genes 9, no. 9: 444. https://doi.org/10.3390/genes9090444






