What Does the Future Hold for Yellow Fever Virus? (II)
Abstract
:1. Introduction, a Re-Emerging Arboviral Haemorrhagic Fever
2. Molecular Biology of Yellow Fever Virus: State of Play
2.1. A Highly Structured and Slowly Evolving Positive Single-Stranded RNA Genome
2.1.1. Promoters
2.1.2. Enhancers
2.1.3. A Genome That Evolves Relatively Slowly
2.2. Structure and Replication of the Viral Particle
3. How to Mitigate and Manage YFV Infections?
3.1. Virus Tracking: Diagnostic Tools Inventory
3.1.1. Molecular YFV Diagnostics
3.1.2. Serological YFV Diagnostics
3.1.3. Changing YFV Diagnostics in Times of Mass Vaccination Campaigns
3.1.4. Limited Information about Genetic YFV Diversity
3.2. Infection Prevention in Endemic and At-Risk Areas
3.2.1. Vaccination Policies
3.2.2. Vector-Control Plans: Past, Present and Future Strategies
3.3. Patient Care: Perspectives for Treatments against Yellow Fever Virus Infection
4. Discussion
Funding
Conflicts of Interest
References
- Monath, T.P. Treatment of yellow fever. Antivir. Res. 2008, 78, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.G.S. Yellow fever vaccine. In Vaccines, 6th ed.; Elsevier: New York, NY, USA, 2012; pp. 870–968. [Google Scholar]
- Beeuwkes, H. Clinical manifestations of yellow fever in the West African native as observed during four extensive epidemics of the disease in the Gold Coast and Nigeria. Trans. R. Soc. Trop. Med. Hyg. 1936, 30, 61–86. [Google Scholar] [CrossRef]
- Berry, G.P.; Kitchen, S.F. Yellow fever accidentally contracted in the laboratory. Am. J. Trop. Med. Hyg. 1931, s1-11, 365–434. [Google Scholar] [CrossRef]
- Monath, T.P. Yellow fever: A medically neglected disease. Report on a seminar. Rev. Infect. Dis. 1987, 9, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Barrett, A.D. Pathogenesis and pathophysiology of yellow fever. Adv. Virus Res. 2003, 60, 343–395. [Google Scholar] [PubMed]
- Klotz, O.; Belt, T.H. Regeneration of liver and kidney following yellow fever. Am. J. Pathol. 1930, 6, 689–697. [Google Scholar] [PubMed]
- Quaresma, J.A.; Pagliari, C.; Medeiros, D.B.; Duarte, M.I.; Vasconcelos, P.F. Immunity and immune response, pathology and pathologic changes: Progress and challenges in the immunopathology of yellow fever. Rev. Med. Virol. 2013, 23, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Klotz, O.; Belt, T.H. The pathology of the liver in yellow fiver. Am. J. Pathol. 1930, 6, 663–688. [Google Scholar] [PubMed]
- Klotz, O.; Belt, T.H. The pathology of the spleen in yellow fever. Am. J. Pathol. 1930, 6, 655–662. [Google Scholar] [PubMed]
- Engelmann, F.; Josset, L.; Girke, T.; Park, B.; Barron, A.; Dewane, J.; Hammarlund, E.; Lewis, A.; Axthelm, M.K.; Slifka, M.K.; et al. Pathophysiologic and transcriptomic analyses of viscerotropic yellow fever in a rhesus macaque model. PLoS Negl. Trop. Dis. 2014, 8, e3295. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; McArthur, M.A.; Cohen, M.; Jahrling, P.B.; Janosko, K.B.; Josleyn, N.; Kang, K.; Zhang, T.; Holbrook, M.R. Characterization of yellow fever virus infection of human and non-human primate antigen presenting cells and their interaction with CD4+ T cells. PLoS Negl. Trop. Dis. 2016, 10, e0004709. [Google Scholar] [CrossRef] [PubMed]
- Khaiboullina, S.F.; Rizvanov, A.A.; Holbrook, M.R.; St Jeor, S. Yellow fever virus strains asibi and 17d-204 infect human umbilical cord endothelial cells and induce novel changes in gene expression. Virology 2005, 342, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Lefeuvre, A.; Contamin, H.; Decelle, T.; Fournier, C.; Lang, J.; Deubel, V.; Marianneau, P. Host-cell interaction of attenuated and wild-type strains of yellow fever virus can be differentiated at early stages of hepatocyte infection. Microbes Infect. 2006, 8, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Woodson, S.E.; Freiberg, A.N.; Holbrook, M.R. Differential cytokine responses from primary human Kupffer cells following infection with wild-type or vaccine strain yellow fever virus. Virology 2011, 412, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Woodson, S.E.; Holbrook, M.R. Infection of hepatocytes with 17-D vaccine-strain yellow fever virus induces a strong pro-inflammatory host response. J. Gen. Virol. 2011, 92, 2262–2271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLinden, J.H.; Bhattarai, N.; Stapleton, J.T.; Chang, Q.; Kaufman, T.M.; Cassel, S.L.; Sutterwala, F.S.; Haim, H.; Houtman, J.C.; Xiang, J. Yellow fever virus, but not Zika virus or dengue virus, inhibits T-cell receptor-mediated T-cell function by an RNA-based mechanism. J. Infect. Dis. 2017, 216, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Ter Meulen, J.; Sakho, M.; Koulemou, K.; Magassouba, N.; Bah, A.; Preiser, W.; Daffis, S.; Klewitz, C.; Bae, H.G.; Niedrig, M.; et al. Activation of the cytokine network and unfavorable outcome in patients with yellow fever. J. Infect. Dis. 2004, 190, 1821–1827. [Google Scholar] [CrossRef] [PubMed]
- Quaresma, J.A.; Barros, V.L.; Pagliari, C.; Fernandes, E.R.; Guedes, F.; Takakura, C.F.; Andrade, H.F., Jr.; Vasconcelos, P.F.; Duarte, M.I. Revisiting the liver in human yellow fever: Virus-induced apoptosis in hepatocytes associated with TGF-β, TNF-α and NK cells activity. Virology 2006, 345, 22–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quaresma, J.A.; Duarte, M.I.; Vasconcelos, P.F. Midzonal lesions in yellow fever: A specific pattern of liver injury caused by direct virus action and in situ inflammatory response. Med. Hypotheses 2006, 67, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Quaresma, J.A.; Barros, V.L.; Fernandes, E.R.; Pagliari, C.; Guedes, F.; da Costa Vasconcelos, P.F.; de Andrade Junior, H.F.; Duarte, M.I. Immunohistochemical examination of the role of Fas ligand and lymphocytes in the pathogenesis of human liver yellow fever. Virus Res. 2006, 116, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Quaresma, J.A.; Barros, V.L.; Pagliari, C.; Fernandes, E.R.; Andrade, H.F., Jr.; Vasconcelos, P.F.; Duarte, M.I. Hepatocyte lesions and cellular immune response in yellow fever infection. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, M.D.; Meertens, L.; Chazal, M.; Hafirassou, M.L.; Dejarnac, O.; Zamborlini, A.; Despres, P.; Sauvonnet, N.; Arenzana-Seisdedos, F.; Jouvenet, N.; et al. Vaccine and wild-type strains of yellow fever virus engage distinct entry mechanisms and differentially stimulate antiviral immune responses. MBio 2016, 7, e01956-15. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Lobigs, M. E protein domain III determinants of yellow fever virus 17D vaccine strain enhance binding to glycosaminoglycans, impede virus spread, and attenuate virulence. J. Virol. 2008, 82, 6024–6033. [Google Scholar] [CrossRef] [PubMed]
- McElroy, K.L.; Girard, Y.A.; McGee, C.E.; Tsetsarkin, K.A.; Vanlandingham, D.L.; Higgs, S. Characterization of the antigen distribution and tissue tropisms of three phenotypically distinct yellow fever virus variants in orally infected Aedes aegypti mosquitoes. Vector Borne Zoonotic Dis. 2008, 8, 675–687. [Google Scholar] [CrossRef] [PubMed]
- McElroy, K.L.; Tsetsarkin, K.A.; Vanlandingham, D.L.; Higgs, S. Role of the yellow fever virus structural protein genes in viral dissemination from the Aedes aegypti mosquito midgut. J. Gen. Virol. 2006, 87, 2993–3001. [Google Scholar] [CrossRef] [PubMed]
- Sil, B.K.; Dunster, L.M.; Ledger, T.N.; Wills, M.R.; Minor, P.D.; Barrett, A.D. Identification of envelope protein epitopes that are important in the attenuation process of wild-type yellow fever virus. J. Virol. 1992, 66, 4265–4270. [Google Scholar] [PubMed]
- Beasley, D.W.; McAuley, A.J.; Bente, D.A. Yellow fever virus: Genetic and phenotypic diversity and implications for detection, prevention and therapy. Antivir. Res. 2015, 115, 48–70. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.D. Yellow fever in Angola and beyond—The problem of vaccine supply and demand. N. Engl. J. Med. 2016, 375, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Stock, N.K.; Laraway, H.; Faye, O.; Diallo, M.; Niedrig, M.; Sall, A.A. Biological and phylogenetic characteristics of yellow fever virus lineages from West Africa. J. Virol. 2013, 87, 2895–2907. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.E.; Vasconcelos, P.F.; Rijnbrand, R.C.; Mutebi, J.P.; Higgs, S.; Barrett, A.D. Size heterogeneity in the 3’ noncoding region of South American isolates of yellow fever virus. J. Virol. 2005, 79, 3807–3821. [Google Scholar] [CrossRef] [PubMed]
- Laemmert, H.W. Susceptibility of marmosets to different strains of yellow fever virus1. Am. J. Trop. Med. Hyg. 1944, s1-24, 71–81. [Google Scholar] [CrossRef]
- Liprandi, F.; Walder, R. Replication of virulent and attenuated strains of yellow fever virus in human monocytes and macrophage-like cells (U937). Arch. Virol. 1983, 76, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.D.; Gould, E.A. Comparison of neurovirulence of different strains of yellow fever virus in mice. J. Gen. Virol. 1986, 67, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Klitting, R.; Roth, L.; Rey, F.A.; de Lamballerie, X. Molecular determinants of yellow fever virus pathogenicity in Syrian golden hamsters: One mutation away from virulence. Emerg. Microbes Infect. 2018, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Tesh, R.B.; Wood, T.G.; Widen, S.G.; Ryman, K.D.; Barrett, A.D. Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing. J. Infect. Dis. 2014, 209, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Reed, W.; Carroll, J.; Agramonte, A.; Lazear, J.W. The etiology of yellow fever-a preliminary note. Public Health Pap. Rep. 1900, 26, 37–53. [Google Scholar] [PubMed]
- Theiler, M.; Smith, H.H. The use of yellow fever virus modified by in vitro cultivation for human immunization. J. Exp. Med. 1937, 65, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Garske, T.; Van Kerkhove, M.D.; Yactayo, S.; Ronveaux, O.; Lewis, R.F.; Staples, J.E.; Perea, W.; Ferguson, N.M.; Yellow Fever Expert Committee. Yellow fever in Africa: Estimating the burden of disease and impact of mass vaccination from outbreak and serological data. PLoS Med. 2014, 11, e1001638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Prince, J.A.; Orenstein, A.J. Mosquito Control in Panama: The Eradication of Malaria and Yellow Fever in Cuba and Panama; Putnam: New York, NY, USA, 1916. [Google Scholar]
- WHO. Winning the War against Yellow Fever. Available online: http://www.who.int/features/2016/winning-the-war-against-yellow-fever/en/ (accessed on 1 June 2018).
- MS Brasil. Monitoramento do Período Sazonal da Febre Amarela. Brasil—2017/2018: Informe nº 25; Ministério da Saúde: Brasília, Distrito Federal, Brasil, 2018. [Google Scholar]
- NCDC. Yellow Fever Outbreak in Nigeria; Nigerian Center for Disease Control: Abuja, Nigeria, 2018. [Google Scholar]
- Rice, C.M.; Lenches, E.M.; Eddy, S.R.; Shin, S.J.; Sheets, R.L.; Strauss, J.H. Nucleotide sequence of yellow fever virus: Implications for flavivirus gene expression and evolution. Science 1985, 229, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.R.; Kinney, R.M.; Trent, D.W.; Lenches, E.M.; Dalgarno, L.; Strauss, J.H. Amino-terminal amino acid sequences of structural proteins of three flaviviruses. Virology 1985, 143, 224–229. [Google Scholar] [CrossRef]
- Ray, D.; Shah, A.; Tilgner, M.; Guo, Y.; Zhao, Y.; Dong, H.; Deas, T.S.; Zhou, Y.; Li, H.; Shi, P.Y. West Nile virus 5’-cap structure is formed by sequential guanine N-7 and ribose 2’-O methylations by nonstructural protein 5. J. Virol. 2006, 80, 8362–8370. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ray, D.; Zhao, Y.; Dong, H.; Ren, S.; Li, Z.; Guo, Y.; Bernard, K.A.; Shi, P.Y.; Li, H. Structure and function of flavivirus ns5 methyltransferase. J. Virol. 2007, 81, 3891–3903. [Google Scholar] [CrossRef] [PubMed]
- Cleaves, G.R.; Dubin, D.T. Methylation status of intracellular dengue type 2 40 S RNA. Virology 1979, 96, 159–165. [Google Scholar] [CrossRef]
- Wengler, G.; Wengler, G.; Gross, H.J. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology 1978, 89, 423–437. [Google Scholar] [CrossRef]
- Corver, J.; Lenches, E.; Smith, K.; Robison, R.A.; Sando, T.; Strauss, E.G.; Strauss, J.H. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 2003, 77, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5’ and 3’ untranslated regions of the flaviviral genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; De Lella Ezcurra, A.L.; Fucito, S.; Gamarnik, A.V. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 2005, 339, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Holden, K.L.; Harris, E. Enhancement of dengue virus translation: Role of the 3’ untranslated region and the terminal 3’ stem-loop domain. Virology 2004, 329, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Holden, K.L.; Stein, D.A.; Pierson, T.C.; Ahmed, A.A.; Clyde, K.; Iversen, P.L.; Harris, E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3’ stem-loop structure. Virology 2006, 344, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Brinton, M.A. The 3’ stem loop of the West Nile virus genomic RNA can suppress translation of chimeric mRNAs. Virology 2001, 287, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Qin, C.; Jiang, T.; Li, X.; Zhao, H.; Liu, Z.; Deng, Y.; Liu, R.; Chen, S.; Yu, M.; et al. Translational regulation by the 3’ untranslated region of the dengue type 2 virus genome. Am. J. Trop Med. Hyg. 2009, 81, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Sanles, A.; Rios-Marco, P.; Romero-Lopez, C.; Berzal-Herranz, A. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front. Microbiol. 2017, 8, 546. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Lodeiro, M.F.; Alvarez, D.E.; Samsa, M.M.; Pietrasanta, L.; Gamarnik, A.V. A 5’ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006, 20, 2238–2249. [Google Scholar] [CrossRef] [PubMed]
- Lodeiro, M.F.; Filomatori, C.V.; Gamarnik, A.V. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 2009, 83, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Meka, H.; Guyatt, K.J.; Westaway, E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001, 75, 6719–6728. [Google Scholar] [CrossRef] [PubMed]
- Song, B.H.; Yun, S.I.; Choi, Y.J.; Kim, J.M.; Lee, C.H.; Lee, Y.M. A complex RNA motif defined by three discontinuous 5-nucleotide-long strands is essential for flavivirus RNA replication. RNA 2008, 14, 1791–1813. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Gamarnik, A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009, 139, 230–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bredenbeek, P.J.; Kooi, E.A.; Lindenbach, B.; Huijkman, N.; Rice, C.M.; Spaan, W.J. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 2003, 84, 1261–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polacek, C.; Foley, J.E.; Harris, E. Conformational changes in the solution structure of the dengue virus 5’ end in the presence and absence of the 3’ untranslated region. J. Virol. 2009, 83, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Davis, W.G.; Basu, M.; Elrod, E.J.; Germann, M.W.; Brinton, M.A. Identification of cis-acting nucleotides and a structural feature in West Nile virus 3’-terminus RNA that facilitate viral minus strand RNA synthesis. J. Virol. 2013, 87, 7622–7636. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhang, B.; Shi, P.Y. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 2008, 381, 123–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filomatori, C.V.; Iglesias, N.G.; Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Boil. Chem. 2011, 286, 6929–6939. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Harris, E. Interplay of RNA elements in the dengue virus 5’ and 3’ ends required for viral RNA replication. J. Virol. 2010, 84, 6103–6118. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Shi, P.Y.; Harris, E. The 5’ and 3’ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J. Virol. 2011, 85, 1900–1905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Dong, H.; Zhou, Y.; Shi, P.Y. Genetic interactions among the west nile virus methyltransferase, the RNA-dependent RNA polymerase, and the 5’ stem-loop of genomic RNA. J. Virol. 2008, 82, 7047–7058. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, T.S.; Gould, E.A. Origin and evolution of flavivirus 5’UTRs and panhandles: Trans-terminal duplications? Virology 2007, 366, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, T.S.; Gould, E.A. Direct repeats in the flavivirus 3’ untranslated region; a strategy for survival in the environment? Virology 2007, 358, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Thurner, C.; Witwer, C.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004, 85, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, T.S.; Gould, E.A. Direct repeats in the 3’ untranslated regions of mosquito-borne flaviviruses: Possible implications for virus transmission. J. Gen. Virol. 2006, 87, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Li, X.F.; Jiang, T.; Yu, X.D.; Deng, Y.Q.; Zhao, H.; Zhu, Q.Y.; Qin, E.D.; Qin, C.F. RNA elements within the 5’ untranslated region of the West Nile virus genome are critical for RNA synthesis and virus replication. J. Gen. Virol. 2010, 91, 1218–1223. [Google Scholar] [CrossRef] [PubMed]
- Mutebi, J.P.; Rijnbrand, R.C.; Wang, H.; Ryman, K.D.; Wang, E.; Fulop, L.D.; Titball, R.; Barrett, A.D. Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the flavivirus genus based on the 3’ noncoding region. J. Virol. 2004, 78, 9652–9665. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Weaver, S.C.; Shope, R.E.; Tesh, R.B.; Watts, D.M.; Barrett, A.D. Genetic variation in yellow fever virus: Duplication in the 3’ noncoding region of strains from africa. Virology 1996, 225, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Olsthoorn, R.C.; Bol, J.F. Sequence comparison and secondary structure analysis of the 3’ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 2001, 7, 1370–1377. [Google Scholar] [PubMed]
- Proutski, V.; Gould, E.A.; Holmes, E.C. Secondary structure of the 3’ untranslated region of flaviviruses: Similarities and differences. Nucleic Acids Res. 1997, 25, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Manzano, M.; Reichert, E.D.; Polo, S.; Falgout, B.; Kasprzak, W.; Shapiro, B.A.; Padmanabhan, R. Identification of cis-acting elements in the 3’-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J. Boil. Chem. 2011, 286, 22521–22534. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Balmori Melian, E.; Edmonds, J.; Dong, H.; Shi, P.Y.; Khromykh, A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solinska, J.; Teramoto, T.; Rausch, J.W.; Shapiro, B.A.; Padmanabhan, R.; Le Grice, S.F. Structural complexity of dengue virus untranslated regions: Cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 2013, 41, 5075–5089. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Costantino, D.A.; Rabe, J.L.; Moon, S.L.; Wilusz, J.; Nix, J.C.; Kieft, J.S. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014, 344, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Moon, S.L.; Wilusz, J.; Kieft, J.S. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. Elife 2014, 3, e01892. [Google Scholar] [CrossRef] [PubMed]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Charley, P.A.; Wilusz, J. Standing your ground to exoribonucleases: Function of flavivirus long non-coding rnas. Virus Res. 2016, 212, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.R.; Palacios, G.; Cardoso, J.F.; Martins, L.C.; Sousa, E.C., Jr.; de Lima, C.P.; Medeiros, D.B.; Savji, N.; Desai, A.; Rodrigues, S.G.; et al. Genomic and phylogenetic characterization of Brazilian yellow fever virus strains. J. Virol. 2012, 86, 13263–13271. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.L.; Brinton, M.A. BHK cell proteins that bind to the 3’ stem-loop structure of the West Nile virus genome RNA. J. Virol. 1995, 69, 5650–5658. [Google Scholar] [PubMed]
- Brinton, M.A.; Fernandez, A.V.; Dispoto, J.H. The 3’-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 1986, 153, 113–121. [Google Scholar] [CrossRef]
- Hahn, C.S.; Hahn, Y.S.; Rice, C.M.; Lee, E.; Dalgarno, L.; Strauss, E.G.; Strauss, J.H. Conserved elements in the 3’ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol 1987, 198, 33–41. [Google Scholar] [CrossRef]
- Khromykh, A.A.; Westaway, E.G. Subgenomic replicons of the flavivirus Kunjin: Construction and applications. J. Virol. 1997, 71, 1497–1505. [Google Scholar] [PubMed]
- Men, R.; Bray, M.; Clark, D.; Chanock, R.M.; Lai, C.J. Dengue type 4 virus mutants containing deletions in the 3’ noncoding region of the RNA genome: Analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996, 70, 3930–3937. [Google Scholar] [PubMed]
- Tilgner, M.; Deas, T.S.; Shi, P.Y. The flavivirus-conserved penta-nucleotide in the 3’ stem-loop of the west nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 2005, 331, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 2010, 16, 2325–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villordo, S.M.; Gamarnik, A.V. Differential RNA sequence requirement for dengue virus replication in mosquito and mammalian cells. J. Virol. 2013, 87, 9365–9372. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Markoff, L. The topology of bulges in the long stem of the flavivirus 3’ stem-loop is a major determinant of RNA replication competence. J. Virol. 2005, 79, 2309–2324. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nomaguchi, M.; Padmanabhan, R.; Markoff, L. Specific requirements for elements of the 5’ and 3’ terminal regions in flavivirus RNA synthesis and viral replication. Virology 2008, 374, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Falgout, B.; Markoff, L. Identification of specific nucleotide sequences within the conserved 3’-SL in the dengue type 2 virus genome required for replication. J. Virol. 1998, 72, 7510–7522. [Google Scholar] [PubMed]
- Khromykh, A.A.; Kondratieva, N.; Sgro, J.Y.; Palmenberg, A.; Westaway, E.G. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 2003, 77, 10623–10629. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.A.; Molenkamp, R.; Dalebout, T.J.; Charlier, N.; Neyts, J.H.; Spaan, W.J.; Bredenbeek, P.J. Conservation of the pentanucleotide motif at the top of the yellow fever virus 17D 3’ stem-loop structure is not required for replication. J. Gen. Virol. 2007, 88, 1738–1747. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Carballeda, J.M.; Filomatori, C.V.; Gamarnik, A.V. RNA structure duplications and flavivirus host adaptation. Trends Microbiol. 2016, 24, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Blaney, J.E., Jr.; Sathe, N.S.; Goddard, L.; Hanson, C.T.; Romero, T.A.; Hanley, K.A.; Murphy, B.R.; Whitehead, S.S. Dengue virus type 3 vaccine candidates generated by introduction of deletions in the 3’ untranslated region (3’-UTR) or by exchange of the DENV-3 3’-UTR with that of DENV-4. Vaccine 2008, 26, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W.; Charlesworth, B.; Charlesworth, D.; Crow, J.F. Rates of spontaneous mutation. Genetics 1998, 148, 1667–1686. [Google Scholar] [PubMed]
- Jenkins, G.M.; Rambaut, A.; Pybus, O.G.; Holmes, E.C. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J. Mol. Evol. 2002, 54, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Worobey, M.; Han, G.Z.; Rambaut, A. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature 2014, 508, 254–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Holmes, E.C. Avian influenza virus exhibits rapid evolutionary dynamics. Mol. Boil. Evol. 2006, 23, 2336–2341. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Rambaut, A.; Pybus, O.G. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006, 8, 125–140. [Google Scholar] [PubMed]
- Sall, A.A.; Faye, O.; Diallo, M.; Firth, C.; Kitchen, A.; Holmes, E.C. Yellow fever virus exhibits slower evolutionary dynamics than dengue virus. J. Virol. 2010, 84, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.E.; Holmes, E.C.; Barrett, A.D. Out of Africa: A molecular perspective on the introduction of yellow fever virus into the americas. PLoS Pathog. 2007, 3, e75. [Google Scholar] [CrossRef] [PubMed]
- Auguste, A.J.; Lemey, P.; Pybus, O.G.; Suchard, M.A.; Salas, R.A.; Adesiyun, A.A.; Barrett, A.D.; Tesh, R.B.; Weaver, S.C.; Carrington, C.V. Yellow fever virus maintenance in Trinidad and its dispersal throughout the Americas. J. Virol. 2010, 84, 9967–9977. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.L.; Voloch, C.M.; Schrago, C.G. Comparative evolutionary epidemiology of dengue virus serotypes. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2012, 12, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Klitting, R.; Gould, E.A.; Paupy, C.; de Lamballerie, X. What does the future hold for yellow fever virus? (I). Genes 2018, 9, 291. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.R.; Barrett, A.D. The enigma of yellow fever in east africa. Rev. Med. Virol. 2008, 18, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Descloux, E.; Cao-Lormeau, V.M.; Roche, C.; De Lamballerie, X. Dengue 1 diversity and microevolution, french polynesia 2001–2006: Connection with epidemiology and clinics. PLoS Negl. Trop. Dis. 2009, 3, e493. [Google Scholar] [CrossRef] [PubMed]
- Craig, S.; Thu, H.M.; Lowry, K.; Wang, X.F.; Holmes, E.C.; Aaskov, J. Diverse dengue type 2 virus populations contain recombinant and both parental viruses in a single mosquito host. J. Virol. 2003, 77, 4463–4467. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, Y.; Topel, M.; Elvang, A.; Melik, W.; Johansson, M. First dating of a recombination event in mammalian tick-borne flaviviruses. PLoS ONE 2012, 7, e31981. [Google Scholar] [CrossRef] [PubMed]
- Aaskov, J.; Buzacott, K.; Field, E.; Lowry, K.; Berlioz-Arthaud, A.; Holmes, E.C. Multiple recombinant dengue type 1 viruses in an isolate from a dengue patient. J. Gen. Virol. 2007, 88, 3334–3340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worobey, M.; Rambaut, A.; Holmes, E.C. Widespread intra-serotype recombination in natural populations of dengue virus. Proc. Natl. Acad. Sci. USA 1999, 96, 7352–7357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzcategui, N.Y.; Camacho, D.; Comach, G.; Cuello de Uzcategui, R.; Holmes, E.C.; Gould, E.A. Molecular epidemiology of dengue type 2 virus in Venezuela: Evidence for in situ virus evolution and recombination. J. Gen. Virol. 2001, 82, 2945–2953. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.K.; Chen, W.J. Experimental evidence that RNA recombination occurs in the Japanese encephalitis virus. Virology 2009, 394, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Carney, J.; Daly, J.M.; Nisalak, A.; Solomon, T. Recombination and positive selection identified in complete genome sequences of Japanese encephalitis virus. Arch. Virol. 2012, 157, 75–83. [Google Scholar] [CrossRef] [PubMed]
- McGee, C.E.; Tsetsarkin, K.A.; Guy, B.; Lang, J.; Plante, K.; Vanlandingham, D.L.; Higgs, S. Stability of yellow fever virus under recombinatory pressure as compared with Chikungunya virus. PLoS ONE 2011, 6, e23247. [Google Scholar] [CrossRef] [PubMed]
- De Wachter, R.; Fiers, W. Preparative two-dimensional polyacrylamide gel electrophoresis of 32 p-labeled RNA. Anal. Biochem. 1972, 49, 184–197. [Google Scholar] [CrossRef]
- Deubel, V.; Digoutte, J.P.; Monath, T.P.; Girard, M. Genetic heterogeneity of yellow fever virus strains from Africa and the Americas. J. Gen. Virol. 1986, 67, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Mutebi, J.P.; Wang, H.; Li, L.; Bryant, J.E.; Barrett, A.D. Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa. J. Virol. 2001, 75, 6999–7008. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Dalgarno, L.; Huong, V.T.; Monath, T.P.; Digoutte, J.P.; Deubel, V. Geographic distribution and evolution of yellow fever viruses based on direct sequencing of genomic cDNA fragments. J. Gen. Virol. 1994, 75, 417–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, G.J.; Cropp, B.C.; Kinney, R.M.; Trent, D.W.; Gubler, D.J. Nucleotide sequence variation of the envelope protein gene identifies two distinct genotypes of yellow fever virus. J. Virol. 1995, 69, 5773–5780. [Google Scholar] [PubMed]
- Wang, H.; Jennings, A.D.; Ryman, K.D.; Late, C.M.; Wang, E.; Ni, H.; Minor, P.D.; Barrett, A.D. Genetic variation among strains of wild-type yellow fever virus from Senegal. J. Gen. Virol. 1997, 78, 1349–1352. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.E.; Barrett, A.D. Comparative phylogenies of yellow fever isolates from peru and Brazil. FEMS Immunol. Med. Microbiol. 2003, 39, 103–118. [Google Scholar] [CrossRef]
- Vasconcelos, P.F.; Bryant, J.E.; da Rosa, T.P.; Tesh, R.B.; Rodrigues, S.G.; Barrett, A.D. Genetic divergence and dispersal of yellow fever virus, Brazil. Emerg. Infect. Dis. 2004, 10, 1578–1584. [Google Scholar] [CrossRef] [PubMed]
- Auguste, A.J.; Lemey, P.; Bergren, N.A.; Giambalvo, D.; Moncada, M.; Moron, D.; Hernandez, R.; Navarro, J.C.; Weaver, S.C. Enzootic transmission of yellow fever virus, venezuela. Emerg. Infect. Dis. 2015, 21, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Mir, D.; Delatorre, E.; Bonaldo, M.; Lourenco-de-Oliveira, R.; Vicente, A.C.; Bello, G. Phylodynamics of yellow fever virus in the americas: New insights into the origin of the 2017 Brazilian outbreak. Sci. Rep. 2017, 7, 7385. [Google Scholar] [CrossRef] [PubMed]
- Moreira-Soto, A.; Torres, M.C.; Lima de Mendonça, M.C.; Mares-Guia, M.A.; Dos Santos Rodrigues, C.D.; Fabri, A.A.; Dos Santos, C.C.; Machado Araújo, E.S.; Fischer, C.; Ribeiro Nogueira, R.M.; et al. Evidence for multiple sylvatic transmission cycles during the 2016–2017 yellow fever virus outbreak, Brazil. Clin. Microbiol. Infect. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hughes, H.R.; Kayiwa, J.; Mossel, E.C.; Lutwama, J.; Staples, J.E.; Lambert, A.J. Phylogeny of yellow fever virus, Uganda, 2016. Emerg. Infect. Dis. 2018, 24, 1598. [Google Scholar] [CrossRef] [PubMed]
- Stock, N.K.; Boschetti, N.; Herzog, C.; Appelhans, M.S.; Niedrig, M. The phylogeny of yellow fever virus 17D vaccines. Vaccine 2012, 30, 989–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goenaga, S.; Fabbri, C.; Duenas, J.C.; Gardenal, C.N.; Rossi, G.C.; Calderon, G.; Morales, M.A.; Garcia, J.B.; Enria, D.A.; Levis, S. Isolation of yellow fever virus from mosquitoes in Misiones province, Argentina. Vector Borne Zoonotic Dis. 2012, 12, 986–993. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.P.; Foster, P.G.; Sallum, M.A.; Coimbra, T.L.; Maeda, A.Y.; Silveira, V.R.; Moreno, E.S.; da Silva, F.G.; Rocco, I.M.; Ferreira, I.B.; et al. Detection of a new yellow fever virus lineage within the south american genotype I in Brazil. J. Med. Virol. 2010, 82, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Pan, Y.; Lyu, Y.; Liang, Z.; Li, J.; Sun, Y.; Dou, X.; Tian, L.; Huo, D.; Chen, L.; et al. Detection of yellow fever virus genomes from four imported cases in China. Int. J. Infect. Dis. 2017, 60, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, M.C.; Gomez, M.M.; Dos Santos, A.A.; Abreu, F.V.S.; Ferreira-de-Brito, A.; Miranda, R.M.; Castro, M.G.; Lourenco-de-Oliveira, R. Genome analysis of yellow fever virus of the ongoing outbreak in Brazil reveals polymorphisms. Mem. Do Inst. Oswaldo Cruz 2017, 112, 447–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baronti, C.; Goitia, N.J.; Cook, S.; Roca, Y.; Revollo, J.; Flores, J.V.; de Lamballerie, X. Molecular epidemiology of yellow fever in Bolivia from 1999 to 2008. Vector Borne Zoonotic Dis. 2011, 11, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, Z. Adaptive diversification between yellow fever virus west African and south American lineages: A genome-wide study. Am. J. Trop. Med. Hyg. 2017, 96, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Carrington, C.V.; Auguste, A.J. Evolutionary and ecological factors underlying the tempo and distribution of yellow fever virus activity. Infect. Genet. Evol. 2013, 13, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003, 59, 23–61. [Google Scholar] [PubMed]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Douam, F.; Ploss, A. Yellow fever virus: Knowledge gaps impeding the fight against an old foe. Trends Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Klema, V.J.; Padmanabhan, R.; Choi, K.H. Flaviviral replication complex: Coordination between RNA synthesis and 5’-RNA capping. Viruses 2015, 7, 4640–4656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Corver, J.; Chipman, P.R.; Zhang, W.; Pletnev, S.V.; Sedlak, D.; Baker, T.S.; Strauss, J.H.; Kuhn, R.J.; Rossmann, M.G. Structures of immature flavivirus particles. EMBO J. 2003, 22, 2604–2613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, R.J.; Zhang, W.; Rossmann, M.G.; Pletnev, S.V.; Corver, J.; Lenches, E.; Jones, C.T.; Mukhopadhyay, S.; Chipman, P.R.; Strauss, E.G.; et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 2002, 108, 717–725. [Google Scholar] [CrossRef]
- Samuel, G.H.; Wiley, M.R.; Badawi, A.; Adelman, Z.N.; Myles, K.M. Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc. Natl. Acad. Sci. USA 2016, 113, 13863–13868. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, I.C.; Allison, S.L.; Heinz, F.X.; Helenius, A. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 2002, 76, 5480–5491. [Google Scholar] [CrossRef] [PubMed]
- Konishi, E.; Mason, P.W. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J. Virol. 1993, 67, 1672–1675. [Google Scholar] [PubMed]
- Li, L.; Lok, S.M.; Yu, I.M.; Zhang, Y.; Kuhn, R.J.; Chen, J.; Rossmann, M.G. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science 2008, 319, 1830–1834. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.A.; Stiasny, K.; Heinz, F.X. Flavivirus structural heterogeneity: Implications for cell entry. Curr. Opin. Virol. 2017, 24, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Plevka, P.; Battisti, A.J.; Junjhon, J.; Winkler, D.C.; Holdaway, H.A.; Keelapang, P.; Sittisombut, N.; Kuhn, R.J.; Steven, A.C.; Rossmann, M.G. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 2011, 12, 602–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Lin, T.Y.; Dowd, K.A.; Manhart, C.J.; Pierson, T.C. The infectivity of prM-containing partially mature west nile virus does not require the activity of cellular furin-like proteases. J. Virol. 2011, 85, 12067–12072. [Google Scholar] [CrossRef] [PubMed]
- Junjhon, J.; Lausumpao, M.; Supasa, S.; Noisakran, S.; Songjaeng, A.; Saraithong, P.; Chaichoun, K.; Utaipat, U.; Keelapang, P.; Kanjanahaluethai, A.; et al. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J. Virol. 2008, 82, 10776–10791. [Google Scholar] [CrossRef] [PubMed]
- Fibriansah, G.; Ng, T.S.; Kostyuchenko, V.A.; Lee, J.; Lee, S.; Wang, J.; Lok, S.M. Structural changes in dengue virus when exposed to a temperature of 37 °C. J. Virol. 2013, 87, 7585–7592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sheng, J.; Plevka, P.; Kuhn, R.J.; Diamond, M.S.; Rossmann, M.G. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. USA 2013, 110, 6795–6799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germi, R.; Crance, J.M.; Garin, D.; Guimet, J.; Lortat-Jacob, H.; Ruigrok, R.W.; Zarski, J.P.; Drouet, E. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 2002, 292, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Van der Most, R.G.; Corver, J.; Strauss, J.H. Mutagenesis of the RGD motif in the yellow fever virus 17D envelope protein. Virology 1999, 265, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.L.; Schalich, J.; Stiasny, K.; Mandl, C.W.; Kunz, C.; Heinz, F.X. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995, 69, 695–700. [Google Scholar] [PubMed]
- Stiasny, K.; Allison, S.L.; Marchler-Bauer, A.; Kunz, C.; Heinz, F.X. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 1996, 70, 8142–8147. [Google Scholar] [PubMed]
- Stiasny, K.; Allison, S.L.; Schalich, J.; Heinz, F.X. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 2002, 76, 3784–3790. [Google Scholar] [CrossRef] [PubMed]
- Bressanelli, S.; Stiasny, K.; Allison, S.L.; Stura, E.A.; Duquerroy, S.; Lescar, J.; Heinz, F.X.; Rey, F.A. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004, 23, 728–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature 2004, 427, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Gollins, S.W.; Porterfield, J.S. Flavivirus infection enhancement in macrophages: An electron microscopic study of viral cellular entry. J. Gen. Virol. 1985, 66 Pt 9, 1969–1982. [Google Scholar] [CrossRef]
- Gollins, S.W.; Porterfield, J.S. The uncoating and infectivity of the flavivirus West Nile on interaction with cells: Effects of pH and ammonium chloride. J. Gen. Virol. 1986, 67 Pt 9, 1941–1950. [Google Scholar] [CrossRef]
- Jennings, A.D.; Whitby, J.E.; Minor, P.D.; Barrett, A.D. Comparison of the nucleotide and deduced amino acid sequences of the structural protein genes of the yellow fever 17DD vaccine strain from senegal with those of other yellow fever vaccine viruses. Vaccine 1993, 11, 679–681. [Google Scholar] [CrossRef]
- Ryman, K.D.; Ledger, T.N.; Campbell, G.A.; Watowich, S.J.; Barrett, A.D. Mutation in a 17D-204 vaccine substrain-specific envelope protein epitope alters the pathogenesis of yellow fever virus in mice. Virology 1998, 244, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Arroyo, J.; Levenbook, I.; Zhang, Z.X.; Catalan, J.; Draper, K.; Guirakhoo, F. Single mutation in the flavivirus envelope protein hinge region increases neurovirulence for mice and monkeys but decreases viscerotropism for monkeys: Relevance to development and safety testing of live, attenuated vaccines. J. Virol. 2002, 76, 1932–1943. [Google Scholar] [CrossRef] [PubMed]
- McArthur, M.A.; Suderman, M.T.; Mutebi, J.P.; Xiao, S.Y.; Barrett, A.D. Molecular characterization of a hamster viscerotropic strain of yellow fever virus. J. Virol. 2003, 77, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Blanco, M.A.; Vasudevan, S.G.; Bradrick, S.S.; Nicchitta, C. Flavivirus RNA transactions from viral entry to genome replication. Antivir. Res. 2016, 134, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Grakoui, A.; Rice, C.M. Processing of the yellow fever virus nonstructural polyprotein: A catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 1991, 65, 6042–6050. [Google Scholar] [PubMed]
- Chambers, T.J.; Droll, D.A.; Tang, Y.; Liang, Y.; Ganesh, V.K.; Murthy, K.H.; Nickells, M. Yellow fever virus NS2B-NS3 protease: Characterization of charged-to-alanine mutant and revertant viruses and analysis of polyprotein-cleavage activities. J. Gen. Virol. 2005, 86, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Nestorowicz, A.; Rice, C.M. Mutagenesis of the yellow fever virus NS2B/3 cleavage site: Determinants of cleavage site specificity and effects on polyprotein processing and viral replication. J. Virol. 1995, 69, 1600–1605. [Google Scholar] [PubMed]
- Lin, C.; Amberg, S.M.; Chambers, T.J.; Rice, C.M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4a/4b signalase site. J. Virol. 1993, 67, 2327–2335. [Google Scholar] [PubMed]
- Saeedi, B.J.; Geiss, B.J. Regulation of flavivirus RNA synthesis and capping. Wiley Interdiscip. Rev. RNA 2013, 4, 723–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roosendaal, J.; Westaway, E.G.; Khromykh, A.; Mackenzie, J.M. Regulated cleavages at the west nile virus NS4A-2k-NS4B junctions play a major role in rearranging cytoplasmic membranes and golgi trafficking of the NS4A protein. J. Virol. 2006, 80, 4623–4632. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Buhler, S.; Bartenschlager, R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2k-regulated manner. J. Boil. Chem. 2007, 282, 8873–8882. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Trans-complementation of yellow fever virus ns1 reveals a role in early RNA replication. J. Virol. 1997, 71, 9608–9617. [Google Scholar] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 1999, 73, 4611–4621. [Google Scholar] [PubMed]
- Le Sommer, C.; Barrows, N.J.; Bradrick, S.S.; Pearson, J.L.; Garcia-Blanco, M.A. G protein-coupled receptor kinase 2 promotes Flaviviridae entry and replication. PLoS Negl. Trop. Dis. 2012, 6, e1820. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus NS1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Umareddy, I.; Chao, A.; Sampath, A.; Gu, F.; Vasudevan, S.G. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 2006, 87, 2605–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiryaev, S.A.; Chernov, A.V.; Aleshin, A.E.; Shiryaeva, T.N.; Strongin, A.Y. NS4A regulates the atpase activity of the NS3 helicase: A novel cofactor role of the non-structural protein NS4A from West Nile virus. J. Gen. Virol. 2009, 90, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Jordan, J.L.; Laurent-Rolle, M.; Ashour, J.; Martinez-Sobrido, L.; Ashok, M.; Lipkin, W.I.; Garcia-Sastre, A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005, 79, 8004–8013. [Google Scholar] [CrossRef] [PubMed]
- Apte-Sengupta, S.; Sirohi, D.; Kuhn, R.J. Coupling of replication and assembly in flaviviruses. Curr. Opin. Virol. 2014, 9, 134–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pong, W.L.; Huang, Z.S.; Teoh, P.G.; Wang, C.C.; Wu, H.N. RNA binding property and RNA chaperone activity of dengue virus core protein and other viral RNA-interacting proteins. FEBS Lett. 2011, 585, 2575–2581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teoh, P.G.; Huang, Z.S.; Pong, W.L.; Chen, P.C.; Wu, H.N. Maintenance of dimer conformation by the dengue virus core protein α4-α4’ helix pair is critical for nucleocapsid formation and virus production. J. Virol. 2014, 88, 7998–8015. [Google Scholar] [CrossRef] [PubMed]
- Deubel, V.; Digoutte, J.P.; Mattei, X.; Pandare, D. Morphogenesis of yellow fever virus in Aedes aegypti cultured cells. II. An ultrastructural study. Am. J. Trop. Med. Hyg. 1981, 30, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Elshuber, S.; Allison, S.L.; Heinz, F.X.; Mandl, C.W. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 2003, 84, 183–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadler, K.; Allison, S.L.; Schalich, J.; Heinz, F.X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 1997, 71, 8475–8481. [Google Scholar] [PubMed]
- Hoffmann, H.H.; Schneider, W.M.; Blomen, V.A.; Scull, M.A.; Hovnanian, A.; Brummelkamp, T.R.; Rice, C.M. Diverse viruses require the calcium transporter SPCA1 for maturation and spread. Cell Host Microbe 2017, 22, 460–470.e5. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.M.; Zhang, W.; Holdaway, H.A.; Li, L.; Kostyuchenko, V.A.; Chipman, P.R.; Kuhn, R.J.; Rossmann, M.G.; Chen, J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 2008, 319, 1834–1837. [Google Scholar] [CrossRef] [PubMed]
- Stiasny, K.; Heinz, F.X. Flavivirus membrane fusion. J. Gen. Virol. 2006, 87, 2755–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Denman, A.J.; Mackenzie, J.M. The importance of the nucleus during flavivirus replication. Viruses 2017, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Kraemer, M.U.G.; Hill, S.; de Jesus, J.G.; Aguiar, R.S.D.; Iani, F.C.M.; Xavier, J.; Quick, J.; Plessis, L.D.; Dellicour, S.; et al. Genomic and epidemiological monitoring of yellow fever virus transmission potential. bioRxiv 2018. [Google Scholar] [CrossRef]
- Chaves, T.D.S.S.; Orduna, T.; Lepetic, A.; Macchi, A.; Verbanaz, S.; Risquez, A.; Perret, C.; Echazarreta, S.; Rodríguez-Morales, A.J.; Lloveras, S.C. Yellow fever in Brazil: Epidemiological aspects and implications for travelers. Travel Med. Infect. Dis. 2018, 23, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, M.U.; Faria, N.R.; Reiner, R.C., Jr.; Golding, N.; Nikolay, B.; Stasse, S.; Johansson, M.A.; Salje, H.; Faye, O.; Wint, G.R.; et al. Spread of yellow fever virus outbreak in angola and the democratic republic of the congo 2015–16: A modelling study. Lancet Infect. Dis. 2017, 17, 330–338. [Google Scholar] [CrossRef]
- Johansson, M.A.; Vasconcelos, P.F.; Staples, J.E. The whole iceberg: Estimating the incidence of yellow fever virus infection from the number of severe cases. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 482–487. [Google Scholar] [CrossRef] [PubMed]
- WHO. Yellow Fever. Available online: http://www.who.int/mediacentre/factsheets/fs100/en/ (accessed on 1 July 2018).
- Cleton, N.; Koopmans, M.; Reimerink, J.; Godeke, G.J.; Reusken, C. Come fly with me: Review of clinically important arboviruses for global travelers. J. Clin. Virol. 2012, 55, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.M.; Chang, G.J.; Cropp, C.B.; Robbins, K.E.; Tsai, T.F. Detection of yellow fever virus by polymerase chain reaction. Clin. Diagn. Virol. 1994, 2, 41–51. [Google Scholar] [CrossRef]
- Pierre, V.; Drouet, M.T.; Deubel, V. Identification of mosquito-borne flavivirus sequences using universal primers and reverse transcription/polymerase chain reaction. Res. Virol. 1994, 145, 93–104. [Google Scholar] [CrossRef]
- Sanchez-Seco, M.P.; Rosario, D.; Hernandez, L.; Domingo, C.; Valdes, K.; Guzman, M.G.; Tenorio, A. Detection and subtyping of dengue 1–4 and yellow fever viruses by means of a multiplex RT-nested-PCR using degenerated primers. Trop. Med. Int. Health 2006, 11, 1432–1441. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.R.; Vianez, J.L., Jr.; Nunes, K.N.; da Silva, S.P.; Lima, C.P.; Guzman, H.; Martins, L.C.; Carvalho, V.L.; Tesh, R.B.; Vasconcelos, P.F. Analysis of a reverse transcription loop-mediated isothermal amplification (RT-LAMP) for yellow fever diagnostic. J. Virol. Methods 2015, 226, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Kwallah, A.; Inoue, S.; Muigai, A.W.; Kubo, T.; Sang, R.; Morita, K.; Mwau, M. A real-time reverse transcription loop-mediated isothermal amplification assay for the rapid detection of yellow fever virus. J. Virol. Methods 2013, 193, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Escadafal, C.; Faye, O.; Sall, A.A.; Faye, O.; Weidmann, M.; Strohmeier, O.; von Stetten, F.; Drexler, J.; Eberhard, M.; Niedrig, M.; et al. Rapid molecular assays for the detection of yellow fever virus in low-resource settings. PLoS Negl. Trop. Dis. 2014, 8, e2730. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.R.; Palacios, G.; Nunes, K.N.; Casseb, S.M.; Martins, L.C.; Quaresma, J.A.; Savji, N.; Lipkin, W.I.; Vasconcelos, P.F. Evaluation of two molecular methods for the detection of yellow fever virus genome. J. Virol. Methods 2011, 174, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Boutonnier, A.; Prina, E.; Sharma, S.; Reiter, P. Development of a SYBR green I based RT-PCR assay for yellow fever virus: Application in assessment of YFV infection in Aedes aegypti. Virol. J. 2012, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Domingo, C.; Patel, P.; Yillah, J.; Weidmann, M.; Mendez, J.A.; Nakoune, E.R.; Niedrig, M. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. J. Clin. Microbiol. 2012, 50, 4054–4060. [Google Scholar] [CrossRef] [PubMed]
- Drosten, C.; Gottig, S.; Schilling, S.; Asper, M.; Panning, M.; Schmitz, H.; Gunther, S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002, 40, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, M.; Faye, O.; Faye, O.; Kranaster, R.; Marx, A.; Nunes, M.R.; Vasconcelos, P.F.; Hufert, F.T.; Sall, A.A. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J. Clin. Virol. 2010, 48, 187–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantel, N.; Aguirre, M.; Gulia, S.; Girerd-Chambaz, Y.; Colombani, S.; Moste, C.; Barban, V. Standardized quantitative RT-PCR assays for quantitation of yellow fever and chimeric yellow fever-dengue vaccines. J. Virol. Methods 2008, 151, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Monteiro, A.G.; Trindade, G.F.; Yamamura, A.M.; Moreira, O.C.; de Paula, V.S.; Duarte, A.C.; Britto, C.; Lima, S.M. New approaches for the standardization and validation of a real-time qPCR assay using taqman probes for quantification of yellow fever virus on clinical samples with high quality parameters. Hum. Vaccines Immunother. 2015, 11, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.G.; Nitsche, A.; Teichmann, A.; Biel, S.S.; Niedrig, M. Detection of yellow fever virus: A comparison of quantitative real-time PCR and plaque assay. J. Virol. Methods 2003, 110, 185–191. [Google Scholar] [CrossRef]
- Chao, D.Y.; Davis, B.S.; Chang, G.J. Development of multiplex real-time reverse transcriptase PCR assays for detecting eight medically important flaviviruses in mosquitoes. J. Clin. Microbiol. 2007, 45, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Torres, M.C.; Patel, P.; Moreira-Soto, A.; Gould, E.A.; Charrel, R.N.; de Lamballerie, X.; Nogueira, R.M.R.; Sequeira, P.C.; Rodrigues, C.D.S.; et al. Lineage-specific real-time RT-PCR for yellow fever virus outbreak surveillance, Brazil. Emerg. Infect. Dis. 2017, 23, 1867. [Google Scholar] [CrossRef] [PubMed]
- Hughes, H.R.; Russell, B.J.; Mossel, E.C.; Kayiwa, J.; Lutwama, J.; Lambert, A.J. Development of a real-time RT-PCR assay for the global differentiation of yellow fever virus vaccine adverse events from natural infections. J. Clin. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Diagne, C.T.; Stittleburg, V.D.; Mohamed-Hadley, A.; de Guillen, Y.A.; Balmaseda, A.; Faye, O.; Faye, O.; Sall, A.A.; Harris, E.; et al. Internally controlled, multiplex real-time reverse transcription PCR for dengue virus and yellow fever virus detection. Am. J. Trop. Med. Hyg. 2018, 98, 1833–1836. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, And Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Rasche, A.; Baronti, C.; Aldabbagh, S.; Cadar, D.; Reusken, C.B.; Pas, S.D.; Goorhuis, A.; Schinkel, J.; Molenkamp, R.; et al. Assay optimization for molecular detection of Zika virus. Bull. World Health Organ. 2016, 94, 880–892. [Google Scholar] [CrossRef] [PubMed]
- Cleton, N.B.; Reusken, C.B.; Wagenaar, J.F.; van der Vaart, E.E.; Reimerink, J.; van der Eijk, A.A.; Koopmans, M.P. Syndromic approach to arboviral diagnostics for global travelers as a basis for infectious disease surveillance. PLoS Negl. Trop. Dis. 2015, 9, e0004073. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.M.; Di Paola, N.; Cunha, M.P.; Rodrigues-Jesus, M.J.; Araujo, D.B.; Silveira, V.B.; Leal, F.B.; Mesquita, F.S.; Botosso, V.F.; Zanotto, P.M.A.; et al. Yellow fever virus RNA in urine and semen of convalescent patient, Brazil. Emerg Infect. Dis. 2018, 24, 176. [Google Scholar] [CrossRef] [PubMed]
- Domingo, C.; Yactayo, S.; Agbenu, E.; Demanou, M.; Schulz, A.R.; Daskalow, K.; Niedrig, M. Detection of yellow fever 17D genome in urine. J. Clin. Microbiol. 2011, 49, 760–762. [Google Scholar] [CrossRef] [PubMed]
- Hamer, D.H.; Angelo, K.; Caumes, E.; van Genderen, P.J.J.; Florescu, S.A.; Popescu, C.P.; Perret, C.; McBride, A.; Checkley, A.; Ryan, J.; et al. Fatal yellow fever in travelers to Brazil, 2018. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 340–341. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P. Yellow fever: An update. Lancet Infect. Dis. 2001, 1, 11–20. [Google Scholar] [CrossRef]
- Gardner, C.L.; Ryman, K.D. Yellow fever: A reemerging threat. Clin. Lab. Med. 2010, 30, 237–260. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, L.; Lv, Y.; Zhang, W.; Li, J.; Zhang, Y.; Di, T.; Zhang, S.; Liu, J.; Li, J.; et al. A fatal yellow fever virus infection in China: Description and lessons. Emerg. Microbes Infect. 2016, 5, e69. [Google Scholar] [CrossRef] [PubMed]
- Wouthuyzen-Bakker, M.; Knoester, M.; van den Berg, A.P.; GeurtsvanKessel, C.H.; Koopmans, M.P.; Van Leer-Buter, C.; Oude Velthuis, B.; Pas, S.D.; Ruijs, W.L.; Schmidt-Chanasit, J.; et al. Yellow fever in a traveller returning from suriname to The Netherlands, March 2017. Euro Surveill. 2017, 22, 30488. [Google Scholar] [CrossRef] [PubMed]
- Barnett, E.D. Yellow fever: Epidemiology and prevention. Clin. Infect. Dis. 2007, 44, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Colebunders, R.; Mariage, J.L.; Coche, J.C.; Pirenne, B.; Kempinaire, S.; Hantson, P.; Van Gompel, A.; Niedrig, M.; Van Esbroeck, M.; Bailey, R.; et al. A Belgian traveler who acquired yellow fever in the Gambia. Clin. Infect. Dis. 2002, 35, e113–e116. [Google Scholar] [CrossRef] [PubMed]
- Akondy, R.S.; Johnson, P.L.; Nakaya, H.I.; Edupuganti, S.; Mulligan, M.J.; Lawson, B.; Miller, J.D.; Pulendran, B.; Antia, R.; Ahmed, R. Initial viral load determines the magnitude of the human CD8 T cell response to yellow fever vaccination. Proc. Natl. Acad. Sci. USA 2015, 112, 3050–3055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanteri, M.C.; Lee, T.H.; Wen, L.; Kaidarova, Z.; Bravo, M.D.; Kiely, N.E.; Kamel, H.T.; Tobler, L.H.; Norris, P.J.; Busch, M.P. West Nile virus nucleic acid persistence in whole blood months after clearance in plasma: Implication for transfusion and transplantation safety. Transfusion 2014, 54, 3232–3241. [Google Scholar] [CrossRef] [PubMed]
- Mead, P.S.; Duggal, N.K.; Hook, S.A.; Delorey, M.; Fischer, M.; Olzenak McGuire, D.; Becksted, H.; Max, R.J.; Anishchenko, M.; Schwartz, A.M.; et al. Zika virus shedding in semen of symptomatic infected men. N. Engl. J. Med. 2018, 378, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- St George, K.; Sohi, I.S.; Dufort, E.M.; Dean, A.B.; White, J.L.; Limberger, R.; Sommer, J.N.; Ostrowski, S.; Wong, S.J.; Backenson, P.B.; et al. Zika virus testing considerations: Lessons learned from the first 80 real-time reverse transcription-PCR-positive cases diagnosed in new york state. J. Clin. Microbiol. 2017, 55, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Bingham, A.M.; Cone, M.; Mock, V.; Heberlein-Larson, L.; Stanek, D.; Blackmore, C.; Likos, A. Comparison of test results for Zika virus RNA in urine, serum, and saliva specimens from persons with travel-associated Zika virus disease—Florida, 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Pessoa, R.; Patriota, J.V.; de Souza Mde, L.; Abd El Wahed, A.; Sanabani, S.S. Detection of Zika virus in Brazilian patients during the first five days of infection—Urine versus plasma. Euro Surveill. 2016, 21, 30302. [Google Scholar] [CrossRef] [PubMed]
- Paz-Bailey, G.; Rosenberg, E.S.; Doyle, K.; Munoz-Jordan, J.; Santiago, G.A.; Klein, L.; Perez-Padilla, J.; Medina, F.A.; Waterman, S.H.; Gubern, C.G.; et al. Persistence of Zika virus in body fluids—Preliminary report. N. Engl. J. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Joguet, G.; Mansuy, J.M.; Matusali, G.; Hamdi, S.; Walschaerts, M.; Pavili, L.; Guyomard, S.; Prisant, N.; Lamarre, P.; Dejucq-Rainsford, N.; et al. Effect of acute Zika virus infection on sperm and virus clearance in body fluids: A prospective observational study. Lancet Infect. Dis. 2017, 17, 1200–1208. [Google Scholar] [CrossRef]
- Nicastri, E.; Castilletti, C.; Liuzzi, G.; Iannetta, M.; Capobianchi, M.R.; Ippolito, G. Persistent detection of Zika virus RNA in semen for six months after symptom onset in a traveller returning from Haiti to Italy, February 2016. Euro Surveill. 2016, 21, 30314. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, D.W.; Green, S.; Kalayanarooj, S.; Innis, B.L.; Nimmannitya, S.; Suntayakorn, S.; Endy, T.P.; Raengsakulrach, B.; Rothman, A.L.; Ennis, F.A.; et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 2000, 181, 2–9. [Google Scholar] [CrossRef] [PubMed]
- WHO. Yellow Fever Laboratory Diagnostic Testing in Africa; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- PAHO. Laboratory Diagnosis of Yellow Fever Virus Infection February 2018; Pan American Health Organization: Washington, DC, USA, 2018. [Google Scholar]
- Domingo, C.; Ellerbrok, H.; Koopmans, M.; Nitsche, A.; Leitmeyer, K.; Charrel, R.N.; Reusken, C. Need for additional capacity and improved capability for molecular detection of yellow fever virus in European expert laboratories: External quality assessment, March 2018. Euro Surveill. 2018, 23, 1800341. [Google Scholar] [CrossRef] [PubMed]
- Domingo, C.; Escadafal, C.; Rumer, L.; Mendez, J.A.; Garcia, P.; Sall, A.A.; Teichmann, A.; Donoso-Mantke, O.; Niedrig, M. First international external quality assessment study on molecular and serological methods for yellow fever diagnosis. PLoS ONE 2012, 7, e36291. [Google Scholar] [CrossRef] [PubMed]
- Prince, H.E.; Matud, J.L. Estimation of dengue virus IGM persistence using regression analysis. Clin. Vaccine Immunol. 2011, 18, 2183–2185. [Google Scholar] [CrossRef] [PubMed]
- Andries, A.C.; Duong, V.; Ly, S.; Cappelle, J.; Kim, K.S.; Lorn Try, P.; Ros, S.; Ong, S.; Huy, R.; Horwood, P.; et al. Value of routine dengue diagnostic tests in urine and saliva specimens. PLoS Negl. Trop. Dis. 2015, 9, e0004100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veje, M.; Studahl, M.; Johansson, M.; Johansson, P.; Nolskog, P.; Bergstrom, T. Diagnosing tick-borne encephalitis: A re-evaluation of notified cases. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Pires-Marczeski, F.C.; Martinez, V.P.; Nemirovsky, C.; Padula, P.J. Intrathecal antibody production in two cases of yellow fever vaccine associated neurotropic disease in argentina. J. Med. Virol. 2011, 83, 2208–2212. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.; Riccio, P.; Patrucco, L.; Rojas, J.I.; Cristiano, E. Longitudinal myelitis associated with yellow fever vaccination. J. Neurovirol. 2009, 15, 348–350. [Google Scholar] [CrossRef] [PubMed]
- Basile, A.J.; Goodman, C.; Horiuchi, K.; Laven, J.; Panella, A.J.; Kosoy, O.; Lanciotti, R.S.; Johnson, B.W. Development and validation of an ELISA kit (YF MAC-HD) to detect igm to yellow fever virus. J. Virol. Methods 2015, 225, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Adungo, F.; Yu, F.; Kamau, D.; Inoue, S.; Hayasaka, D.; Posadas-Herrera, G.; Sang, R.; Mwau, M.; Morita, K. Development and characterization of monoclonal antibodies to yellow fever virus and application in antigen detection and igm capture enzyme-linked immunosorbent assay. Clin. Vaccine Immunol. 2016, 23, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Niedrig, M.; Kursteiner, O.; Herzog, C.; Sonnenberg, K. Evaluation of an indirect immunofluorescence assay for detection of immunoglobulin M (IgM) and IgG antibodies against yellow fever virus. Clin. Vaccine Immunol. 2008, 15, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Cropp, C.B.; Muth, D.J.; Calisher, C.H. Indirect fluorescent antibody test for the diagnosis of yellow fever. Trans. R. Soc. Trop. Med. Hyg. 1981, 75, 282–286. [Google Scholar] [CrossRef]
- Cleton, N.B.; Godeke, G.J.; Reimerink, J.; Beersma, M.F.; Doorn, H.R.; Franco, L.; Goeijenbier, M.; Jimenez-Clavero, M.A.; Johnson, B.W.; Niedrig, M.; et al. Spot the difference-development of a syndrome based protein microarray for specific serological detection of multiple flavivirus infections in travelers. PLoS Negl. Trop. Dis. 2015, 9, e0003580. [Google Scholar] [CrossRef] [PubMed]
- Spector, S.; Tauraso, N.M. Yellow fever virus. I. Development and evaluation of a plaque neutralization test. Appl. Microbiol. 1968, 16, 1770–1775. [Google Scholar] [PubMed]
- Simoes, M.; Camacho, L.A.; Yamamura, A.M.; Miranda, E.H.; Cajaraville, A.C.; da Silva Freire, M. Evaluation of accuracy and reliability of the plaque reduction neutralization test (micro-PRNT) in detection of yellow fever virus antibodies. Biologicals 2012, 40, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.L.; Horton, D.L.; Johnson, N.; Li, L.; Barrett, A.D.; Smith, D.J.; Galbraith, S.E.; Solomon, T.; Fooks, A.R. Flavivirus-induced antibody cross-reactivity. J. Gen. Virol. 2011, 92, 2821–2829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morens, D.M.; Halstead, S.B.; Larsen, L.K. Comparison of dengue virus plaque reduction neutralization by macro and “semi-micro’ methods in LLC-MK2 cells. Microbiol. Immunol. 1985, 29, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Halstead, S.B.; Repik, P.M.; Putvatana, R.; Raybourne, N. Simplified plaque reduction neutralization assay for dengue viruses by semimicro methods in BHK-21 cells: Comparison of the BHK suspension test with standard plaque reduction neutralization. J. Clin. Microbiol. 1985, 22, 250–254. [Google Scholar] [PubMed]
- Buckley, A.; Dawson, A.; Moss, S.R.; Hinsley, S.A.; Bellamy, P.E.; Gould, E.A. Serological evidence of West Nile virus, Usutu virus and Sindbis virus infection of birds in the UK. J. Gen. Virol. 2003, 84, 2807–2817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardekian, S.K.; Roberts, A.L. Diagnostic options and challenges for dengue and chikungunya viruses. BioMed Res. Int. 2015, 2015, 834371. [Google Scholar] [CrossRef] [PubMed]
- Mercier-Delarue, S.; Durier, C.; Colin de Verdiere, N.; Poveda, J.D.; Meiffredy, V.; Fernandez Garcia, M.D.; Lastere, S.; Cesaire, R.; Manuggera, J.C.; Molina, J.M.; et al. Screening test for neutralizing antibodies against yellow fever virus, based on a flavivirus pseudotype. PLoS ONE 2017, 12, e0177882. [Google Scholar] [CrossRef] [PubMed]
- Houghton-Trivino, N.; Montana, D.; Castellanos, J. Dengue-yellow fever sera cross-reactivity; challenges for diagnosis. Rev. Salud Publica (Bogota) 2008, 10, 299–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamieson, D.J.; Theiler, R.N.; Rasmussen, S.A. Emerging infections and pregnancy. Emerg. Infect. Dis. 2006, 12, 1638–1643. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N. Diagnosis of arboviral infections—A quagmire of cross reactions and complexities. Travel Med. Infect. Dis. 2016, 14, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Gabor, J.J.; Schwarz, N.G.; Esen, M.; Kremsner, P.G.; Grobusch, M.P. Dengue and chikungunya seroprevalence in Gabonese infants prior to major outbreaks in 2007 and 2010: A sero-epidemiological study. Travel Med. Infect. Dis. 2016, 14, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Domingo, C.; Alves, M.J.; de Ory, F.; Teichmann, A.; Schmitz, H.; Muller, R.; Niedrig, M. International external quality control assessment for the serological diagnosis of dengue infections. BMC Infect. Dis. 2015, 15, 167. [Google Scholar] [CrossRef] [PubMed]
- Sanchini, A.; Donoso-Mantke, O.; Papa, A.; Sambri, V.; Teichmann, A.; Niedrig, M. Second international diagnostic accuracy study for the serological detection of West Nile virus infection. PLoS Negl. Trop. Dis. 2013, 7, e2184. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Vasconcelos, P.F. Yellow fever. J. Clin. Virol. 2015, 64, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.E. Yellow fever vaccine-associated viscerotropic disease: Current perspectives. Drug Des. Dev. Ther. 2016, 10, 3345–3353. [Google Scholar] [CrossRef] [PubMed]
- WHO. Dengue vaccine: WHO position paper—July 2016. Vaccine 2017, 35, 1200–1201. [Google Scholar] [CrossRef] [PubMed]
- Skipetrova, A.; Wartel, T.A.; Gailhardou, S. Dengue vaccination during pregnancy—An overview of clinical trials data. Vaccine 2018, 36, 3345–3350. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Halasa-Rappel, Y.A.; Baurin, N.; Coudeville, L.; Shepard, D.S. Cost-effectiveness of dengue vaccination in ten endemic countries. Vaccine 2018, 36, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Guy, B.; Briand, O.; Lang, J.; Saville, M.; Jackson, N. Development of the Sanofi Pasteur tetravalent dengue vaccine: One more step forward. Vaccine 2015, 33, 7100–7111. [Google Scholar] [CrossRef] [PubMed]
- Vannice, K.S.; Durbin, A.; Hombach, J. Status of vaccine research and development of vaccines for dengue. Vaccine 2016, 34, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; Rossini, G. An overview of Usutu virus. Microbes Infect. 2017, 19, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; de Souza Luna, L.K.; Pedroso, C.; Pedral-Sampaio, D.B.; Queiroz, A.T.; Brites, C.; Netto, E.M.; Drosten, C. Rates of and reasons for failure of commercial human immunodeficiency virus type 1 viral load assays in Brazil. J. Clin. Microbiol. 2007, 45, 2061–2063. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, J.J.; Brandriss, M.W.; Monath, T.P. Monoclonal antibodies distinguish between wild and vaccine strains of yellow fever virus by neutralization, hemagglutination inhibition, and immune precipitation of the virus envelope protein. Virology 1983, 125, 8–17. [Google Scholar] [CrossRef]
- Ryman, K.D.; Ledger, T.N.; Weir, R.C.; Schlesinger, J.J.; Barrett, A.D. Yellow fever virus envelope protein has two discrete type-specific neutralizing epitopes. J. Gen. Virol. 1997, 78 Pt 6, 1353–1356. [Google Scholar] [CrossRef]
- Ledger, T.N.; Sil, B.K.; Wills, M.R.; Lewis, G.; Kinney, R.M.; Jennings, A.D.; Stephenson, J.R.; Barrett, A.D. Variation in the biological function of envelope protein epitopes of yellow fever vaccine viruses detected with monoclonal antibodies. Boil. J. Int. Assoc. Boil. Stand. 1992, 20, 117–128. [Google Scholar] [CrossRef]
- Daffis, S.; Kontermann, R.E.; Korimbocus, J.; Zeller, H.; Klenk, H.D.; Ter Meulen, J. Antibody responses against wild-type yellow fever virus and the 17D vaccine strain: Characterization with human monoclonal antibody fragments and neutralization escape variants. Virology 2005, 337, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.H. Antigenic analysis of certain group B arthropodborne viruses by antibody absorption. J. Exp. Med. 1960, 111, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.D.; Pryde, A.; Medlen, A.R.; Ledger, T.N.; Whitby, J.E.; Gibson, C.A.; DeSilva, M.; Groves, D.J.; Langley, D.J.; Minor, P.D. Examination of the envelope glycoprotein of yellow fever vaccine viruses with monoclonal antibodies. Vaccine 1989, 7, 333–336. [Google Scholar] [CrossRef]
- Barrett, A.D.; Mathews, J.H.; Miller, B.R.; Medlen, A.R.; Ledger, T.N.; Roehrig, J.T. Identification of monoclonal antibodies that distinguish between 17D-204 and other strains of yellow fever virus. J. Gen. Virol. 1990, 71 Pt 1, 13–18. [Google Scholar] [CrossRef]
- Monath, T.P. Review of the risks and benefits of yellow fever vaccination including some new analyses. Expert Rev. Vaccines 2012, 11, 427–448. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P. 17D yellow fever virus vaccine. Am. J. Trop. Med. Hyg. 2013, 89, 1225. [Google Scholar] [CrossRef] [PubMed]
- Lilay, A.; Asamene, N.; Bekele, A.; Mengesha, M.; Wendabeku, M.; Tareke, I.; Girmay, A.; Wuletaw, Y.; Adossa, A.; Ba, Y.; et al. Reemergence of yellow fever in Ethiopia after 50 years, 2013: Epidemiological and entomological investigations. BMC Infect. Dis. 2017, 17, 343. [Google Scholar] [CrossRef] [PubMed]
- Wamala, J.F.; Malimbo, M.; Okot, C.L.; Atai-Omoruto, A.D.; Tenywa, E.; Miller, J.R.; Balinandi, S.; Shoemaker, T.; Oyoo, C.; Omony, E.O.; et al. Epidemiological and laboratory characterization of a yellow fever outbreak in Northern Uganda, October 2010-January 2011. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2012, 16, e536–e542. [Google Scholar] [CrossRef] [PubMed]
- Markoff, L. Yellow fever outbreak in Sudan. N. Engl. J. Med. 2013, 368, 689–691. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Q.A.; Memish, Z.A. Yellow fever from Angola and Congo: A storm gathers. Trop. Doct. 2017, 47, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Campi-Azevedo, A.C.; de Almeida Estevam, P.; Coelho-Dos-Reis, J.G.; Peruhype-Magalhaes, V.; Villela-Rezende, G.; Quaresma, P.F.; Maia Mde, L.; Farias, R.H.; Camacho, L.A.; Freire Mda, S.; et al. Subdoses of 17DD yellow fever vaccine elicit equivalent virological/immunological kinetics timeline. BMC Infect. Dis. 2014, 14, 391. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.M.; Maia Mde, L.; Farias, R.H.; Camacho, L.A.; Freire, M.S.; Galler, R.; Yamamura, A.M.; Almeida, L.F.; Lima, S.M.; Nogueira, R.M.; et al. 17DD yellow fever vaccine: A double blind, randomized clinical trial of immunogenicity and safety on a dose-response study. Hum. Vaccines Immunother. 2013, 9, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Ahuka-Mundeke, S.; Casey, R.M.; Harris, J.B.; Dixon, M.G.; Nsele, P.M.; Kizito, G.M.; Umutesi, G.; Laven, J.; Paluku, G.; Gueye, A.S.; et al. Immunogenicity of fractional-dose vaccine during a yellow fever outbreak—Preliminary report. N. Engl. J. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- De Menezes Martins, R.; Maia, M.L.S.; de Lima, S.M.B.; de Noronha, T.G.; Xavier, J.R.; Camacho, L.A.B.; de Albuquerque, E.M.; Farias, R.H.G.; da Matta de Castro, T.; Homma, A.; et al. Duration of post-vaccination immunity to yellow fever in volunteers eight years after a dose-response study. Vaccine 2018, 36, 4112–4117. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Hamer, D.H. Vaccination strategies during shortages of yellow fever vaccine—Reply. JAMA 2018, 319, 1280–1281. [Google Scholar] [CrossRef] [PubMed]
- Shearer, F.M.; Moyes, C.L.; Pigott, D.M.; Brady, O.J.; Marinho, F.; Deshpande, A.; Longbottom, J.; Browne, A.J.; Kraemer, M.U.G.; O’Reilly, K.M.; et al. Global yellow fever vaccination coverage from 1970 to 2016: An adjusted retrospective analysis. Lancet Infect. Dis. 2017, 17, 1209–1217. [Google Scholar] [CrossRef]
- Chen, L.H.; Hamer, D.H. Vaccination challenges in confronting the resurgent threat from yellow fever. JAMA 2017, 318, 1651–1652. [Google Scholar] [CrossRef] [PubMed]
- Norton, S.A.; Morens, D.M. Vaccination strategies during shortages of yellow fever vaccine. JAMA 2018, 319, 1280. [Google Scholar] [CrossRef] [PubMed]
- Roukens, A.H.; Gelinck, L.B.; Visser, L.G. Intradermal vaccination to protect against yellow fever and influenza. Curr. Top. Microbiol. Immunol. 2012, 351, 159–179. [Google Scholar] [PubMed]
- Roukens, A.H.; Vossen, A.C.; Bredenbeek, P.J.; van Dissel, J.T.; Visser, L.G. Intradermally administered yellow fever vaccine at reduced dose induces a protective immune response: A randomized controlled non-inferiority trial. PLoS ONE 2008, 3, e1993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, A.M.; Lam, L.K.; Klimstra, W.B.; Ryman, K.D. The 17d-204 vaccine strain-induced protection against virulent yellow fever virus is mediated by humoral immunity and CD4+ but not CD8+ T cells. PLoS Pathog. 2016, 12, e1005786. [Google Scholar] [CrossRef] [PubMed]
- Lok. Singapore’s Dengue Haemorraghic Fever Control Programme: A Case Study on the Succesful Control of Aedes Aegypti and Aedes Albopictus Using Mainly Environmental Measures as a Part of an Integrated Vector Control; Southeast Asian Medical Information Center: Tokyo, Japan, 1985. [Google Scholar]
- Armada Gessa, J.A.; Figueredo González, R. Application of environmental management principles in the program for eradication of Aedes (Stegomyia) aegypti (Linneus, 1762) in the Republic of Cuba, 1984. Bull. Pan Am. Health Organ. 1986, 20, 186–193. [Google Scholar] [PubMed]
- Morrison, A.C.; Zielinski-Gutierrez, E.; Scott, T.W.; Rosenberg, R. Defining challenges and proposing solutions for control of the virus vector Aedes aegypti. PLoS Med. 2008, 5, e68. [Google Scholar] [CrossRef] [PubMed]
- Renganathan, E.; Parks, W.; LIoyd, L.; Nathan, M.B.; Hosein, E.; Odugleh, A.; Clark, G.G.; Gubler, D.J.; Prasittisuk, C.; Palmer, K.; et al. Towards sustaining behavioural impact in dengue prevention and control. Dengue Bull. 2003, 27, 6–12. [Google Scholar]
- Alvarado-Castro, V.; Paredes-Solís, S.; Nava-Aguilera, E.; Morales-Pérez, A.; Alarcón-Morales, L.; Balderas-Vargas, N.A.; Andersson, N. Assessing the effects of interventions for Aedes aegypti control: Systematic review and meta-analysis of cluster randomised controlled trials. BMC Public Health 2017, 17, 384. [Google Scholar] [CrossRef] [PubMed]
- Bowman, L.R.; Donegan, S.; McCall, P.J. Is dengue vector control deficient in effectiveness or evidence?: Systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2016, 10, e0004551. [Google Scholar] [CrossRef] [PubMed]
- Achee, N.L.; Gould, F.; Perkins, T.A.; Reiner, R.C., Jr.; Morrison, A.C.; Ritchie, S.A.; Gubler, D.J.; Teyssou, R.; Scott, T.W. A critical assessment of vector control for dengue prevention. PLoS Negl. Trop. Dis. 2015, 9, e0003655. [Google Scholar] [CrossRef] [PubMed]
- Ono, L.; Wollinger, W.; Rocco, I.M.; Coimbra, T.L.; Gorin, P.A.; Sierakowski, M.R. In vitro and in vivo antiviral properties of sulfated galactomannans against yellow fever virus (BeH111 strain) and dengue 1 virus (Hawaii strain). Antivir. Res. 2003, 60, 201–208. [Google Scholar] [CrossRef]
- Zhou, Z.; Khaliq, M.; Suk, J.E.; Patkar, C.; Li, L.; Kuhn, R.J.; Post, C.B. Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus e protein. ACS Chem. Biol. 2008, 3, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Kampmann, T.; Yennamalli, R.; Campbell, P.; Stoermer, M.J.; Fairlie, D.P.; Kobe, B.; Young, P.R. In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antivir. Res. 2009, 84, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Mayhoub, A.S.; Khaliq, M.; Kuhn, R.J.; Cushman, M. Design, synthesis, and biological evaluation of thiazoles targeting flavivirus envelope proteins. J. Med. Chem. 2011, 54, 1704–1714. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Q.; Dai, J.X.; Ji, G.H.; Jiang, T.; Wang, H.J.; Yang, H.O.; Tan, W.L.; Liu, R.; Yu, M.; Ge, B.X.; et al. A broadly flavivirus cross-neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of e protein. PLoS ONE 2011, 6, e16059. [Google Scholar] [CrossRef] [PubMed]
- Umamaheswari, A.; Kumar, M.M.; Pradhan, D.; Marisetty, H. Docking studies towards exploring antiviral compounds against envelope protein of yellow fever virus. Interdiscip. Sci. 2011, 3, 64–77. [Google Scholar] [CrossRef] [PubMed]
- De Burghgraeve, T.; Kaptein, S.J.; Ayala-Nunez, N.V.; Mondotte, J.A.; Pastorino, B.; Printsevskaya, S.S.; de Lamballerie, X.; Jacobs, M.; Preobrazhenskaya, M.; Gamarnik, A.V.; et al. An analogue of the antibiotic teicoplanin prevents flavivirus entry in vitro. PLoS ONE 2012, 7, e37244. [Google Scholar] [CrossRef] [PubMed]
- Assuncao-Miranda, I.; Cruz-Oliveira, C.; Neris, R.L.; Figueiredo, C.M.; Pereira, L.P.; Rodrigues, D.; Araujo, D.F.; Da Poian, A.T.; Bozza, M.T. Inactivation of Dengue and Yellow Fever viruses by heme, cobalt-protoporphyrin IX and tin-protoporphyrin IX. J. Appl. Microbiol. 2016, 120, 790–804. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Trent, D.W.; Monath, T.P. Immune correlates of protection against yellow fever determined by passive immunization and challenge in the hamster model. Vaccine 2011, 29, 6008–6016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacca, C.C.; Severino, A.A.; Mondini, A.; Rahal, P.; D’Avila, S.G.; Cordeiro, J.A.; Nogueira, M.C.; Bronzoni, R.V.; Nogueira, M.L. RNA interference inhibits yellow fever virus replication in vitro and in vivo. Virus Genes 2009, 38, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sakamuru, S.; Huang, R.; Brecher, M.; Koetzner, C.A.; Zhang, J.; Chen, H.; Qin, C.F.; Zhang, Q.Y.; Zhou, J.; et al. Erythrosin B is a potent and broad-spectrum orthosteric inhibitor of the flavivirus NS2B-NS3 protease. Antivir. Res. 2018, 150, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, E.; Pezzullo, M.; De Burghgraeve, T.; Kaptein, S.; Pastorino, B.; Dallmeier, K.; de Lamballerie, X.; Neyts, J.; Hanson, A.M.; Frick, D.N.; et al. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: New prospects for an old drug. J. Antimicrob. Chemother. 2012, 67, 1884–1894. [Google Scholar] [CrossRef] [PubMed]
- Patkar, C.G.; Larsen, M.; Owston, M.; Smith, J.L.; Kuhn, R.J. Identification of inhibitors of yellow fever virus replication using a replicon-based high-throughput assay. Antimicrob. Agents Chemother. 2009, 53, 4103–4114. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Wu, S.; Julander, J.; Ma, J.; Zhang, X.; Kulp, J.; Cuconati, A.; Block, T.M.; Du, Y.; Guo, J.T.; et al. A novel benzodiazepine compound inhibits yellow fever virus infection by specifically targeting NS4B protein. J. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pierra, C.; Amador, A.; Benzaria, S.; Cretton-Scott, E.; D’Amours, M.; Mao, J.; Mathieu, S.; Moussa, A.; Bridges, E.G.; Standring, D.N.; et al. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2’-C-methylcytidine. J. Med. Chem. 2006, 49, 6614–6620. [Google Scholar] [CrossRef] [PubMed]
- Stuyver, L.J.; McBrayer, T.R.; Tharnish, P.M.; Clark, J.; Hollecker, L.; Lostia, S.; Nachman, T.; Grier, J.; Bennett, M.A.; Xie, M.Y.; et al. Inhibition of hepatitis C replicon RNA synthesis by β-D-2’-deoxy-2’-fluoro-2’-C-methylcytidine: A specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 2006, 17, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Fogt, J.; Januszczyk, P.; Framski, G.; Onishi, T.; Izawa, K.; De Clercq, E.; Neyts, J.; Boryski, J. Synthesis and antiviral activity of novel derivatives of 2’-β-C-methylcytidine. Nucleic Acids Symp. Ser. (Oxf.) 2008, 52, 605–606. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Jha, A.K.; Choi, J.A.; Jung, K.H.; Smee, D.F.; Morrey, J.D.; Chu, C.K. Efficacy of 2’-C-methylcytidine against yellow fever virus in cell culture and in a hamster model. Antivir. Res. 2010, 86, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Furuta, Y.; Shafer, K.; Sidwell, R.W. Activity of T-1106 in a hamster model of yellow fever virus infection. Antimicrob. Agents Chemother. 2007, 51, 1962–1966. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Shafer, K.; Smee, D.F.; Morrey, J.D.; Furuta, Y. Activity of T-705 in a hamster model of yellow fever virus infection in comparison with that of a chemically related compound, T-1106. Antimicrob. Agents Chemother. 2009, 53, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhang, X.; Soloveva, V.; Warren, T.; Guo, F.; Wu, S.; Lu, H.; Guo, J.; Su, Q.; Shen, H.; et al. Enhancing the antiviral potency of ER α-glucosidase inhibitor IHVR-19029 against hemorrhagic fever viruses in vitro and in vivo. Antivir. Res. 2018, 150, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Bantia, S.; Taubenheim, B.R.; Minning, D.M.; Kotian, P.; Morrey, J.D.; Smee, D.F.; Sheridan, W.P.; Babu, Y.S. BCX4430, a novel nucleoside analog, effectively treats yellow fever in a hamster model. Antimicrob. Agents Chemother. 2014, 58, 6607–6614. [Google Scholar] [CrossRef] [PubMed]
- Malet, H.; Masse, N.; Selisko, B.; Romette, J.L.; Alvarez, K.; Guillemot, J.C.; Tolou, H.; Yap, T.L.; Vasudevan, S.; Lescar, J.; et al. The flavivirus polymerase as a target for drug discovery. Antivir. Res. 2008, 80, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.P.; Noble, C.G.; Shi, P.Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015, 119, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Bullard-Feibelman, K.M.; Govero, J.; Zhu, Z.; Salazar, V.; Veselinovic, M.; Diamond, M.S.; Geiss, B.J. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antivir. Res. 2017, 137, 134–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, A.C.; Zaverucha-do-Valle, C.; Reis, P.A.; Barbosa-Lima, G.; Vieira, Y.R.; Mattos, M.; Silva, P.P.; Sacramento, C.; de Castro Faria Neto, H.C.; Campanati, L.; et al. Sofosbuvir protects Zika virus-infected mice from mortality, preventing short- and long-term sequelae. Sci. Rep. 2017, 7, 9409. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.T.; Colby-Germinario, S.P.; Hassounah, S.A.; Fogarty, C.; Osman, N.; Palanisamy, N.; Han, Y.; Oliveira, M.; Quan, Y.; Wainberg, M.A. Evaluation of sofosbuvir (β-D-2’-deoxy-2’-α-fluoro-2’-β-C-methyluridine) as an inhibitor of Dengue virus replication. Sci. Rep. 2017, 7, 6345. [Google Scholar] [CrossRef] [PubMed]
- Leyssen, P.; Balzarini, J.; De Clercq, E.; Neyts, J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 2005, 79, 1943–1947. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Morrey, J.D.; Blatt, L.M.; Shafer, K.; Sidwell, R.W. Comparison of the inhibitory effects of interferon alfacon-1 and ribavirin on yellow fever virus infection in a hamster model. Antivir. Res. 2007, 73, 140–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sbrana, E.; Xiao, S.Y.; Guzman, H.; Ye, M.; Travassos da Rosa, A.P.; Tesh, R.B. Efficacy of post-exposure treatment of yellow fever with ribavirin in a hamster model of the disease. Am. J. Trop. Med. Hyg. 2004, 71, 306–312. [Google Scholar] [PubMed]
- Bhattacharya, D.; Ansari, I.H.; Striker, R. The flaviviral methyltransferase is a substrate of Casein Kinase 1. Virus Res. 2009, 141, 101–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwar, A.; Hosoya, T.; Leong, K.M.; Onogi, H.; Okuno, Y.; Hiramatsu, T.; Koyama, H.; Suzuki, M.; Hagiwara, M.; Garcia-Blanco, M.A. The kinase inhibitor SFV785 dislocates dengue virus envelope protein from the replication complex and blocks virus assembly. PLoS ONE 2011, 6, e23246. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Ennis, J.; Turner, J.; Morrey, J.D. Treatment of yellow fever virus with an adenovirus-vectored interferon, DEF201, in a hamster model. Antimicrob. Agents Chemother. 2011, 55, 2067–2073. [Google Scholar] [CrossRef] [PubMed]
- Vellozzi, C.; Mitchell, T.; Miller, E.; Casey, C.G.; Eidex, R.B.; Hayes, E.B. Yellow fever vaccine-associated viscerotropic disease (YEL-AVD) and corticosteroid therapy: Eleven united states cases, 1996–2004. Am. J. Trop. Med. Hyg. 2006, 75, 333–336. [Google Scholar] [PubMed]
- Shearer, F.M.; Longbottom, J.; Browne, A.J.; Pigott, D.M.; Brady, O.J.; Kraemer, M.U.G.; Marinho, F.; Yactayo, S.; de Araujo, V.E.M.; da Nobrega, A.A.; et al. Existing and potential infection risk zones of yellow fever worldwide: A modelling analysis. Lancet Glob. Health 2018, 6, e270–e278. [Google Scholar] [CrossRef]
- WHO. Eliminate Yellow Fever Epidemics (EYE): A Global Strategy, 2017–2026; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Gómez, M.M.; Abreu, F.V.S.; Santos, A.A.C.D.; Mello, I.S.; Santos, M.P.; Ribeiro, I.P.; Ferreira-de-Brito, A.; Miranda, R.M.; Castro, M.G.; Ribeiro, M.S.; et al. Genomic and structural features of the yellow fever virus from the 2016–2017 Brazilian outbreak. J. Gen. Virol. 2018, 99, 536–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Vasilakis, N. Dengue—Quo tu et quo vadis? Viruses 2011, 3, 1562–1608. [Google Scholar] [CrossRef] [PubMed]
- Hanley, K.A.; Monath, T.P.; Weaver, S.C.; Rossi, S.L.; Richman, R.L.; Vasilakis, N. Fever versus fever: The role of host and vector susceptibility and interspecific competition in shaping the current and future distributions of the sylvatic cycles of dengue virus and yellow fever virus. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013, 19, 292–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Pickering, P.; Duchêne, S.; Holmes, E.C.; Aaskov, J.G. Highly divergent dengue virus type 2 in traveler returning from Borneo to Australia. Emerg. Infect. Dis. 2016, 22, 2146–2148. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Cardosa, J.; Diallo, M.; Sall, A.A.; Holmes, E.C.; Hanley, K.A.; Weaver, S.C.; Mota, J.; Rico-Hesse, R. Sylvatic dengue viruses share the pathogenic potential of urban/endemic dengue viruses. J. Virol. 2010, 84, 3726–3727; author reply 3727–3728. [Google Scholar] [CrossRef] [PubMed]
- WHO. Eliminating Yellow Fever Epidemics (EYE) Strategy: Meeting Demand for Yellow Fever Vaccines. Available online: http://www.who.int/csr/disease/yellowfev/meeting-demand-for-vaccines/en/ (accessed on 1 July 2018).
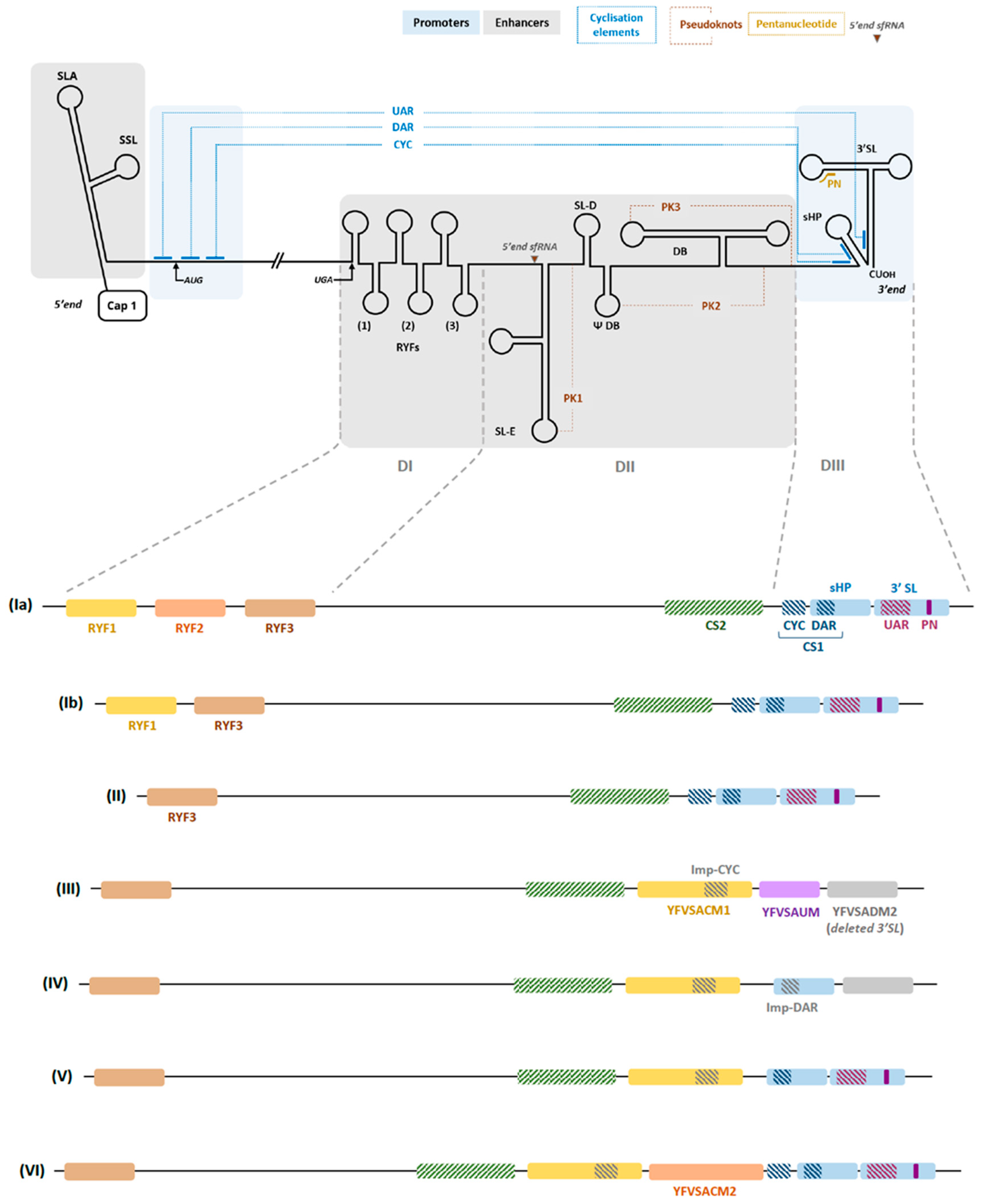
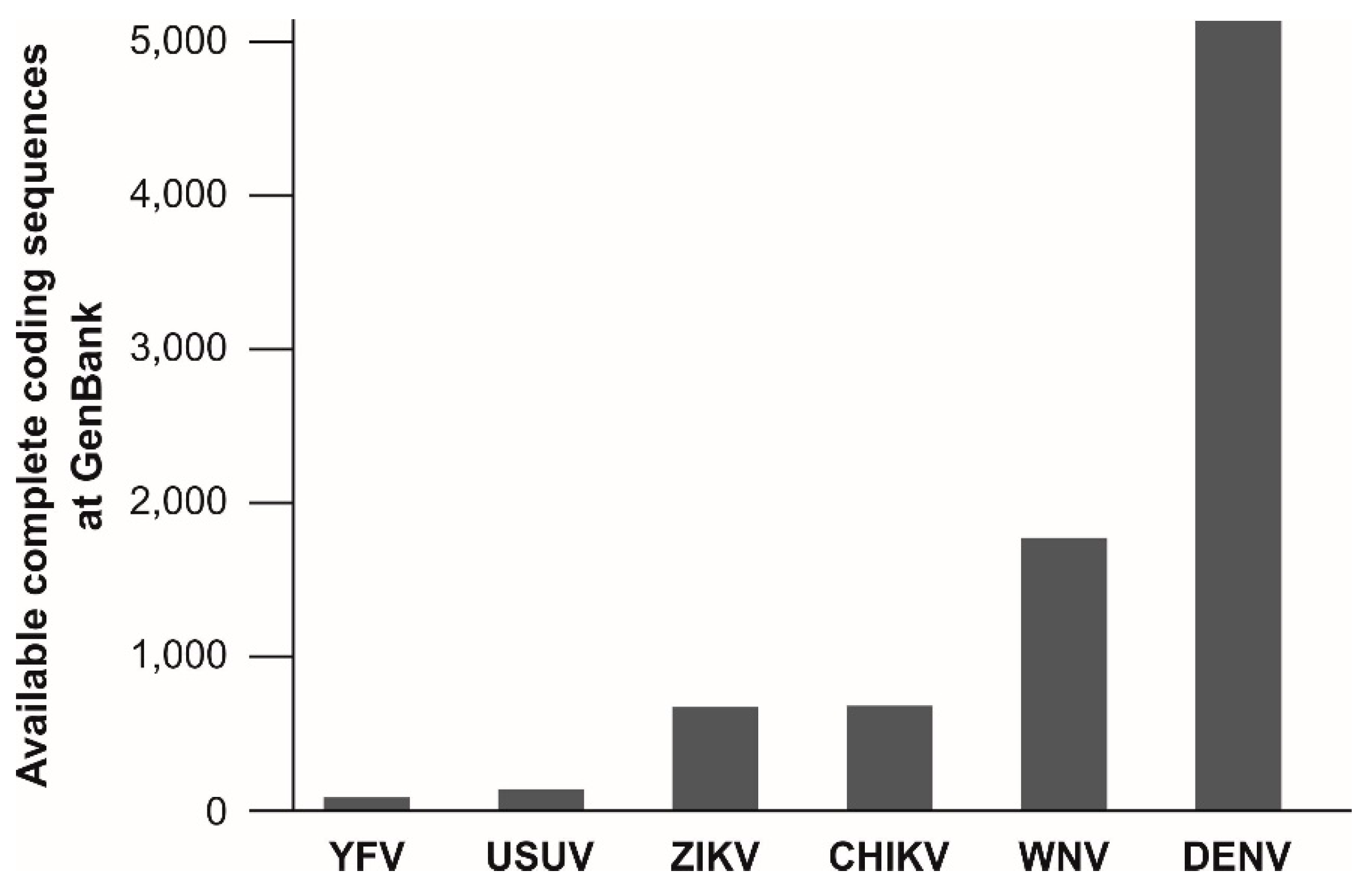

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klitting, R.; Fischer, C.; Drexler, J.F.; Gould, E.A.; Roiz, D.; Paupy, C.; De Lamballerie, X. What Does the Future Hold for Yellow Fever Virus? (II). Genes 2018, 9, 425. https://doi.org/10.3390/genes9090425
Klitting R, Fischer C, Drexler JF, Gould EA, Roiz D, Paupy C, De Lamballerie X. What Does the Future Hold for Yellow Fever Virus? (II). Genes. 2018; 9(9):425. https://doi.org/10.3390/genes9090425
Chicago/Turabian StyleKlitting, Raphaëlle, Carlo Fischer, Jan F. Drexler, Ernest A. Gould, David Roiz, Christophe Paupy, and Xavier De Lamballerie. 2018. "What Does the Future Hold for Yellow Fever Virus? (II)" Genes 9, no. 9: 425. https://doi.org/10.3390/genes9090425




