Unveiling Novel Traits Associated with Ulcerative Colitis via Phenome-Wide Associations Enhanced by Polygenic Risk Statistics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collections
2.2. Quality Control
2.3. Genotype Imputation
2.4. Variant Annotation
2.5. Genome-Wide Association Study
2.6. Variant Collapsing Analysis
2.7. Pathway Analysis and Biological Distance Evaluation
2.8. Phenome-Wide Association Analysis
2.9. Polygenic Risk Score Based on Different SNP Sets
2.10. Genetic Correlation Between Traits
3. Results
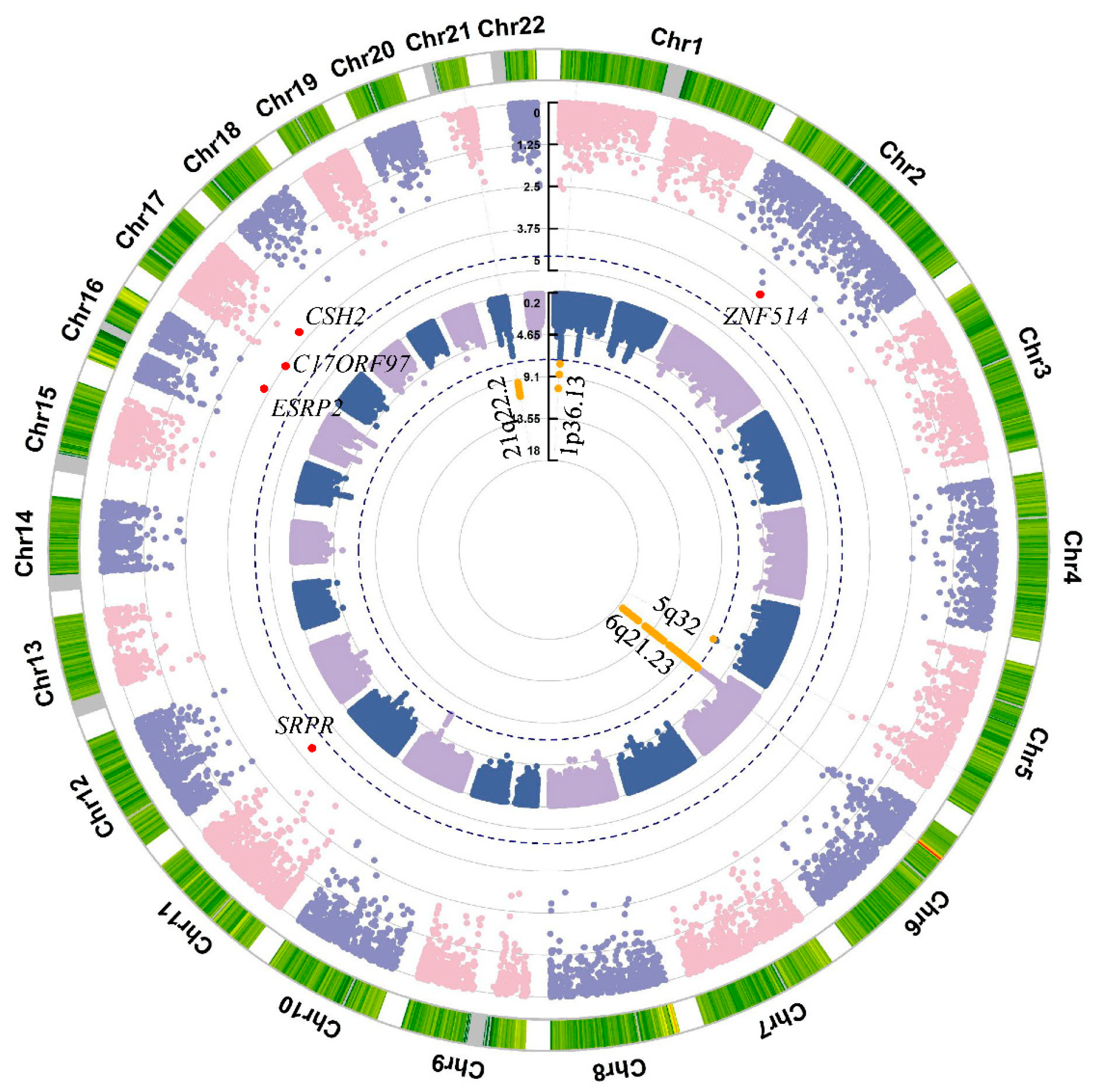
3.1. Variant-Level Association Analysis
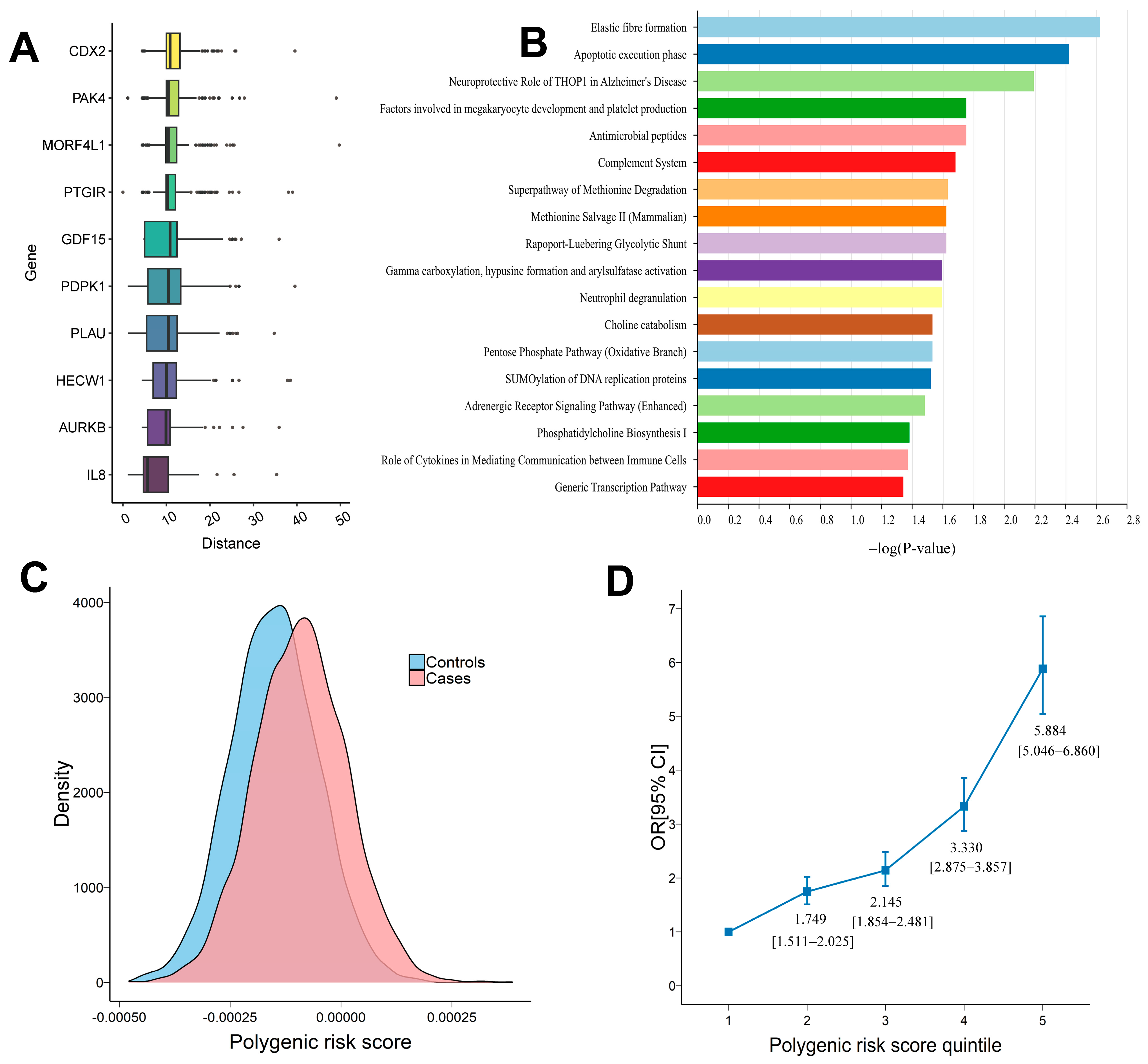
3.2. Gene-Level Association Analysis by Aggregating High Impact Rare Variants
3.3. Polygenic Risk Score Analysis
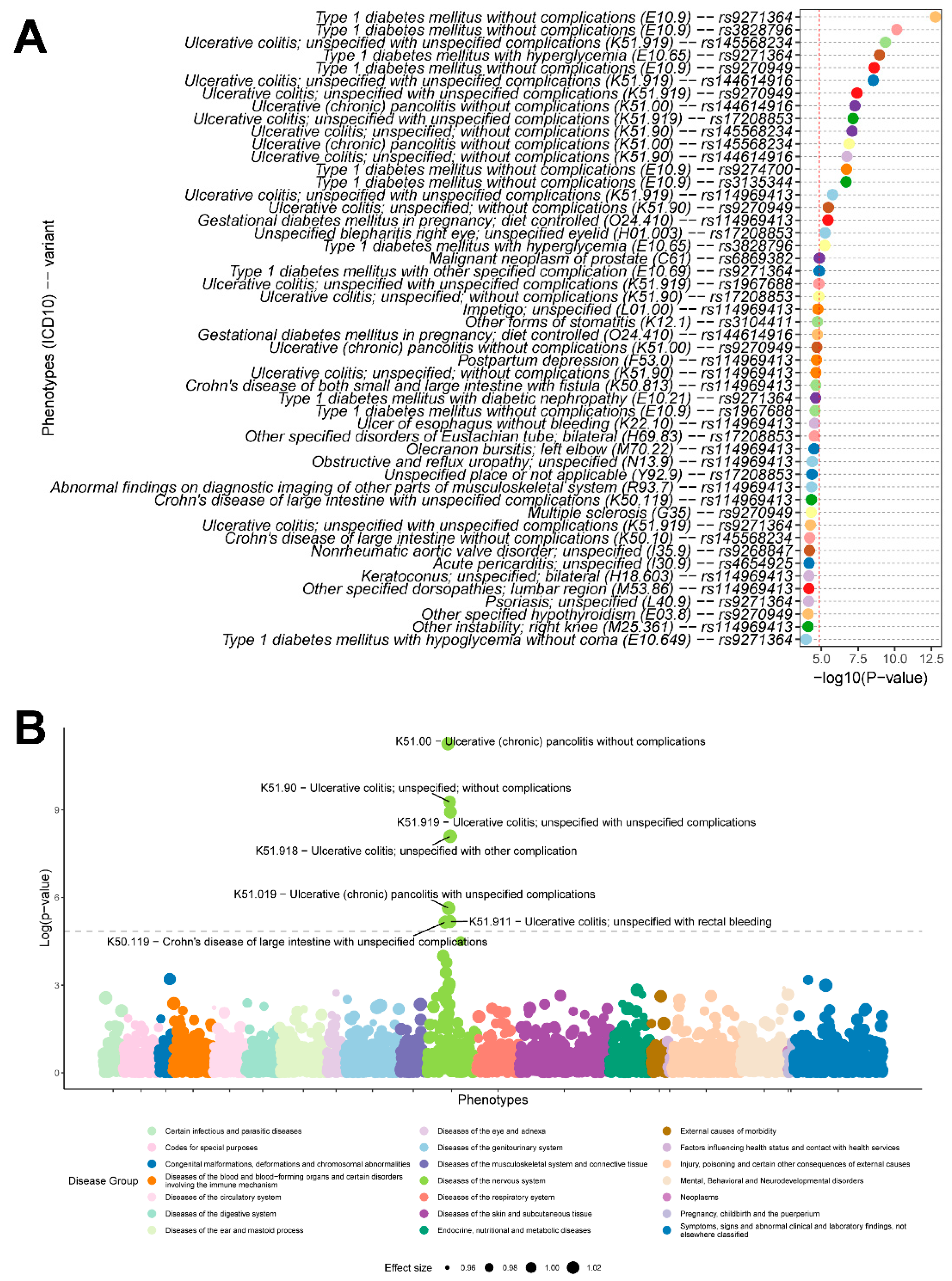
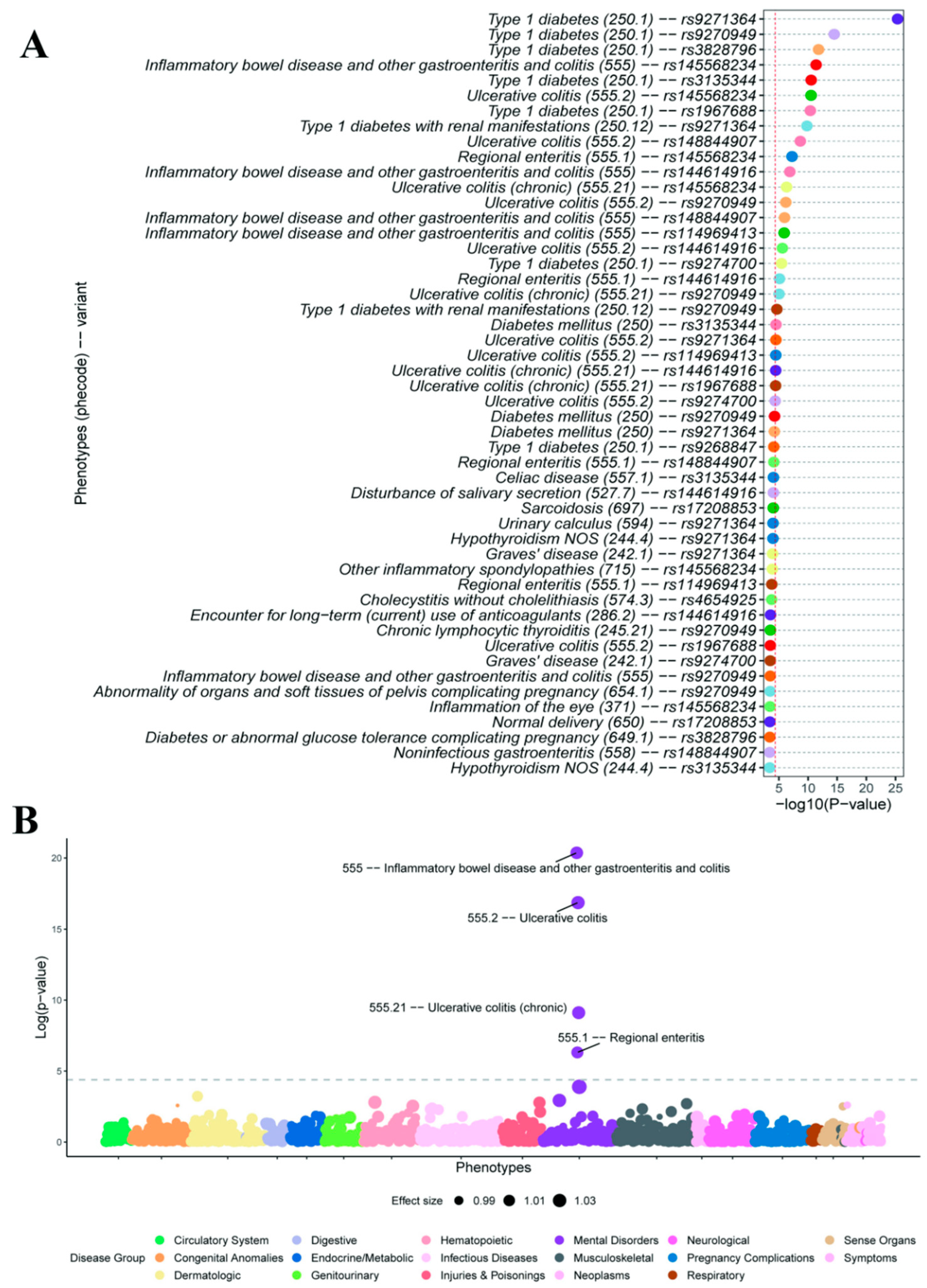
3.4. Variant-Level and PRS-Based Phenome-Wide Association Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UC | Ulcerative colitis |
| IBD | Inflammatory bowel disease |
| QC | Quality control |
| PCA | Principal component analysis |
| GWAS | Genome-wide association study |
| PheWAS | Phenome-wide association study |
| HGC | Human gene connectome |
| IPA | Ingenuity Pathways Analysis |
| PRS | Polygenic risk score |
| SNP | Single nucleotide polymorphism |
| eQTL | Expression quantitative trait locus |
| DEG | Differentially expressed gene |
| LD | Linkage disequilibrium |
| EHR | Electronic health records |
| HLA | Human leukocyte antigen |
| OR | Odds ratio |
| AUC | Area under the receiver operating characteristic curve |
References
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Primers. 2020, 6, 74. [Google Scholar] [CrossRef]
- Gros, B.; Kaplan, G.G. Ulcerative colitis in adults: A review. JAMA 2023, 330, 951–965. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef] [PubMed]
- McGovern, D.P.; Gardet, A.; Törkvist, L.; Goyette, P.; Essers, J.; Taylor, K.D.; Neale, B.M.; Ong, R.T.; Lagacé, C.; Li, C.; et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 2010, 42, 332–337. [Google Scholar] [CrossRef] [PubMed]
- de Lange, K.M.; Moutsianas, L.; Lee, J.C.; Lamb, C.A.; Luo, Y.; Kennedy, N.A.; Jostins, L.; Rice, D.L.; Gutierrez-Achury, J.; Ji, S.G.; et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 2017, 49, 256–261. [Google Scholar] [CrossRef]
- Yu, X.; Chen, Y.; Chen, J.; Fan, Y.; Lu, H.; Wu, D.; Xu, Y. Shared genetic architecture between autoimmune disorders and B-cell acute lymphoblastic leukemia: Insights from large-scale genome-wide cross-trait analysis. BMC Med. 2024, 22, 161. [Google Scholar] [CrossRef]
- Li, Y.R.; Li, J.; Zhao, S.D.; Bradfield, J.P.; Mentch, F.D.; Maggadottir, S.M.; Hou, C.; Abrams, D.J.; Chang, D.; Gao, F.; et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med. 2015, 21, 1018–1027. [Google Scholar] [CrossRef]
- Román, A.L.; Muñoz, F. Comorbidity in inflammatory bowel disease. World J. Gastroenterol. 2011, 17, 2723–2733. [Google Scholar] [CrossRef] [PubMed]
- Mosli, M.H.; Alsahafi, M.; Alsanea, M.N.; Alhasani, F.; Ahmed, M.; Saadah, O. Multimorbidity among inflammatory bowel disease patients in a tertiary care center: A retrospective study. BMC Gastroenterol. 2022, 22, 487. [Google Scholar] [CrossRef]
- Vadstrup, K.; Alulis, S.; Borsi, A.; Jørgensen, T.R.; Nielsen, A.; Munkholm, P.; Qvist, N. Extraintestinal Manifestations and Other Comorbidities in Ulcerative Colitis and Crohn Disease: A Danish Nationwide Registry Study 2003–2016. Crohn’s Colitis 360 2020, 2, otaa070. [Google Scholar] [CrossRef]
- Noor, N.M.; Sousa, P.; Paul, S.; Roblin, X. Early Diagnosis, Early Stratification, and Early Intervention to Deliver Precision Medicine in IBD. Inflamm. Bowel Dis. 2022, 28, 1254–1264. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Itan, Y.; Zhang, S.Y.; Vogt, G.; Abhyankar, A.; Herman, M.; Nitschke, P.; Fried, D.; Quintana-Murci, L.; Abel, L.; Casanova, J.L. The human gene connectome as a map of short cuts for morbid allele discovery. Proc. Natl. Acad. Sci. USA 2013, 110, 5558–5563. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Joo, Y.Y.; Actkins, K.; Pacheco, J.A.; Basile, A.O.; Carroll, R.; Crosslin, D.R.; Day, F.; Denny, J.C.; Velez Edwards, D.R.; Hakonarson, H.; et al. A polygenic and phenotypic risk prediction for polycystic ovary syndrome evaluated by phenome-wide association studies. J. Clin. Endocrinol. Metab. 2020, 105, 1918–1936. [Google Scholar] [CrossRef] [PubMed]
- Haberman, Y.; Tickle, T.L.; Dexheimer, P.J.; Kim, M.O.; Tang, D.; Karns, R.; Baldassano, R.N.; Noe, J.D.; Rosh, J.; Markowitz, J.; et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Investig. 2015, 125, 1363. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Uniken Venema, W.T.; Westra, H.J.; Vich Vila, A.; Barbieri, R.; Voskuil, M.D.; Blokzijl, T.; Jansen, B.H.; Li, Y.; Daly, M.J.; et al. Inflammation status modulates the effect of host genetic variation on intestinal gene expression in inflammatory bowel disease. Nat. Commun. 2021, 12, 1122. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.; O’Reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Franke, A.; Balschun, T.; Sina, C.; Ellinghaus, D.; Häsler, R.; Mayr, G.; Albrecht, M.; Wittig, M.; Buchert, E.; Nikolaus, S.; et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL). Nat. Genet. 2010, 42, 292–294. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, R.; Gao, H.; Jung, S.; Gao, X.; Sun, R.; Liu, X.; Kim, Y.; Lee, H.S.; Kawai, Y.; et al. Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat. Genet. 2023, 55, 796–806. [Google Scholar] [CrossRef]
- Thoo, L.; Noti, M.; Krebs, P. Keep calm: The intestinal barrier at the interface of peace and war. Cell Death Dis. 2019, 10, 849. [Google Scholar] [CrossRef]
- Meyers, T.J.; Weiner, A.B.; Graff, R.E.; Desai, A.S.; Cooley, L.F.; Catalona, W.J.; Hanauer, S.B.; Wu, J.D.; Schaeffer, E.M.; Abdulkadir, S.A.; et al. Association between inflammatory bowel disease and prostate cancer: A large-scale, prospective, population-based study. Int. J. Cancer 2020, 147, 2735–2742. [Google Scholar] [CrossRef]
- Troncoso, L.L.; Biancardi, A.L.; de Moraes, H.V., Jr.; Zaltman, C. Ophthalmic manifestations in patients with inflammatory bowel disease: A Review. World J. Gastroenterol. 2017, 23, 5836–5848. [Google Scholar] [CrossRef]
- Tandon, P.; Govardhanam, V.; Leung, K.; Maxwell, C.; Huang, V. Systematic review with meta-analysis: Risk of adverse pregnancy-related outcomes in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2020, 51, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Nikolaus, S.; Schreiber, S. Diagnostics of inflammatory bowel disease. Gastroenterology 2007, 133, 1670–1689. [Google Scholar] [CrossRef]
- Wendorff, M.; ElAbd, H.; Degenhardt, F.; Höppner, M.; Uellendahl-Werth, F.; Wacker, E.M.; Wienbrandt, L.; Juzenas, S.; Center, R.G.; Koudelka, T.; et al. Genome-wide analysis of individual coding variants and HLA-II-associated self-immunopeptidomes in ulcerative colitis. MedRxiv 2023. [Google Scholar] [CrossRef]
- Li, B.; Sakaguchi, T.; Tani, H.; Ito, T.; Murakami, M.; Okumura, R.; Kobayashi, M.; Okuzaki, D.; Motooka, D.; Ikeuchi, H.; et al. OTUD3 prevents ulcerative colitis by inhibiting microbiota-mediated STING activation. Sci. Immunol. 2025, 10, eadm6843. [Google Scholar] [CrossRef]
- Kugathasan, S.; Baldassano, R.N.; Bradfield, J.P.; Sleiman, P.M.; Imielinski, M.; Guthery, S.L.; Cucchiara, S.; Kim, C.E.; Frackelton, E.C.; Annaiah, K.; et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 2008, 40, 1211–1215. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, C.J.; Wei, Z.; Panossian, S.; Wang, F.; Kim, C.E.; Mentch, F.D.; Chiavacci, R.M.; Kachelries, K.E.; Pandey, R.; Grant, S.F.; et al. Targeted resequencing identifies defective variants of decoy receptor 3 in pediatric-onset inflammatory bowel disease. Genes. Immun. 2013, 14, 447–452. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, R.; Consortium, I.I.B.D.G. OP20 Expanding the genetic spectrum of inflammatory bowel disease through large-scale exome sequencing. J. Crohn’s Colitis 2025, 19, i41–i42. [Google Scholar] [CrossRef]
- Scherr, R.; Essers, J.; Hakonarson, H.; Kugathasan, S. Genetic determinants of pediatric inflammatory bowel disease: Is age of onset genetically determined? Dig. Dis. 2009, 27, 236–239. [Google Scholar] [CrossRef]
- Ippolito, C.; Colucci, R.; Segnani, C.; Errede, M.; Girolamo, F.; Virgintino, D.; Dolfi, A.; Tirotta, E.; Buccianti, P.; Di Candio, G.; et al. Fibrotic and vascular remodelling of colonic wall in patients with active ulcerative colitis. J. Crohns Colitis 2016, 10, 1194–1204. [Google Scholar] [CrossRef]
- Gu, S.-C.; Zeng, S.-l.; Zhang, W.; Shen, C.-Y.; Yuan, C.-X.; Song, Y.; Ye, Q. Integrative multi-omics analysis identifies genetically supported druggable targets for inflammatory bowel disease. J. Genet. Eng. Biotechnol. 2025, 23, 100608. [Google Scholar] [CrossRef]
- Fan, J.-C.; Lu, Y.; Gan, J.-H.; Lu, H. Identification of potential novel targets for treating inflammatory bowel disease using Mendelian randomization analysis. Int. J. Colorectal Dis. 2024, 39, 165. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Kanda, T.; Ueda, N.; Ikebuchi, Y.; Hashiguchi, K.; Nakao, K.; Isomoto, H. IL-8 and LYPD8 expression levels are associated with the inflammatory response in the colon of patients with ulcerative colitis. Biomed. Rep. 2020, 12, 193–198. [Google Scholar] [CrossRef]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Polygenic Risk Score Task Force of the International Common Disease Alliance. Responsible use of polygenic risk scores in the clinic: Potential benefits, risks and gaps. Nat. Med. 2021, 27, 1876–1884. [Google Scholar] [CrossRef]
- Torkamani, A.; Wineinger, N.E.; Topol, E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018, 19, 581–590. [Google Scholar] [CrossRef]
- Chatterjee, N.; Shi, J.; García-Closas, M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016, 17, 392–406. [Google Scholar] [CrossRef]
- Middha, P.; Thummalapalli, R.; Betti, M.J.; Yao, L.; Quandt, Z.; Balaratnam, K.; Bejan, C.A.; Cardenas, E.; Falcon, C.J.; Faleck, D.M.; et al. Polygenic risk score for ulcerative colitis predicts immune checkpoint inhibitor-mediated colitis. Nat. Commun. 2024, 15, 2568. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Janssens, A.; Joyner, M.J. Polygenic risk scores that predict common diseases using millions of single nucleotide polymorphisms: Is more, better? Clin. Chem. 2019, 65, 609–611. [Google Scholar] [CrossRef]
- Nica, A.C.; Dermitzakis, E.T. Expression quantitative trait loci: Present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120362. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Baldassano, R.; Zhang, H.; Qu, H.Q.; Imielinski, M.; Kugathasan, S.; Annese, V.; Dubinsky, M.; Rotter, J.I.; Russell, R.K.; et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum. Mol. Genet. 2010, 19, 2059–2067. [Google Scholar] [CrossRef]
- Kang, E.A.; Han, K.; Chun, J.; Soh, H.; Park, S.; Im, J.P.; Kim, J.S. Increased risk of diabetes in inflammatory bowel disease patients: A nationwide population-based study in Korea. J. Clin. Med. 2019, 8, 343. [Google Scholar] [CrossRef]
- Sun, J.; Yao, J.; Olén, O.; Halfvarsson, J.; Bergman, D.; Ebrahimi, F.; Carlsson, S.; Ludvigsson, J.; Ludvigsson, J.F. Bidirectional association between inflammatory bowel disease and type 1 diabetes: A nationwide matched cohort and case-control study. Lancet Reg. Health-Eur. 2024, 46, 101056. [Google Scholar] [CrossRef] [PubMed]
- Tarar, Z.I.; Farooq, U.; Zafar, M.U.; Saleem, S.; Nawaz, A.; Kamal, F.; Ghous, G.; Inayat, F.; Ghouri, Y.A. A national study of pregnancy-related maternal and fetal outcomes in women with inflammatory bowel disease. Int. J. Colorectal Dis. 2022, 37, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Waterman, M.; Xu, W.; Stempak, J.M.; Milgrom, R.; Bernstein, C.N.; Griffiths, A.M.; Greenberg, G.R.; Steinhart, A.H.; Silverberg, M.S. Distinct and overlapping genetic loci in Crohn’s disease and ulcerative colitis: Correlations with pathogenesis. Inflamm. Bowel Dis. 2011, 17, 1936–1942. [Google Scholar] [CrossRef] [PubMed]

| Models | AUC | Number of SNPs | |
|---|---|---|---|
| Covariates-only | 0.513 | ||
| Weighted PRS | Genome-wide significant SNPs | 0.590 | 47 |
| Genome-wide SNPs (r2 = 0.8, p = 5 × 10−2) | 0.665 | 164,107 | |
| Selected DEG-SNPs (r2 = 0.6, p = 5 × 10−4) | 0.532 | 73 | |
| Selected intestinal eQTLs (r2 = 0.6, p = 1) | 0.602 | 7822 | |
| Selected UC eQTLs (r2 = 0.1, p = 1) | 0.589 | 89 | |
| Unweighted PRS | Selected intestinal eQTLs (all sites) | 0.513 | 8327 |
| Selected UC eQTLs (r2 = 0.05) | 0.527 | 74 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Liu, L.; Kars, M.E.; Li, R.; Li, M.; Itan, Y. Unveiling Novel Traits Associated with Ulcerative Colitis via Phenome-Wide Associations Enhanced by Polygenic Risk Statistics. Genes 2025, 16, 1431. https://doi.org/10.3390/genes16121431
Wu Y, Liu L, Kars ME, Li R, Li M, Itan Y. Unveiling Novel Traits Associated with Ulcerative Colitis via Phenome-Wide Associations Enhanced by Polygenic Risk Statistics. Genes. 2025; 16(12):1431. https://doi.org/10.3390/genes16121431
Chicago/Turabian StyleWu, Yiming, Ling Liu, Meltem Ece Kars, Rui Li, Menglong Li, and Yuval Itan. 2025. "Unveiling Novel Traits Associated with Ulcerative Colitis via Phenome-Wide Associations Enhanced by Polygenic Risk Statistics" Genes 16, no. 12: 1431. https://doi.org/10.3390/genes16121431
APA StyleWu, Y., Liu, L., Kars, M. E., Li, R., Li, M., & Itan, Y. (2025). Unveiling Novel Traits Associated with Ulcerative Colitis via Phenome-Wide Associations Enhanced by Polygenic Risk Statistics. Genes, 16(12), 1431. https://doi.org/10.3390/genes16121431







